A Comparative Analysis of the Gene Expression Profiles in the Mammary Glands of Lactating and Nonlactating Mares at the Second Month of Gestation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Equine Mammary Gland Samples
2.2. Histomorphology Examination
2.3. RNA Extraction, Library Preparation, and Sequencing
2.4. Quality Control and Alignment
2.5. Screening of Differentially Expressed Genes (DEGs)
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
2.7. Quantitative Real-Time PCR (qPCR)
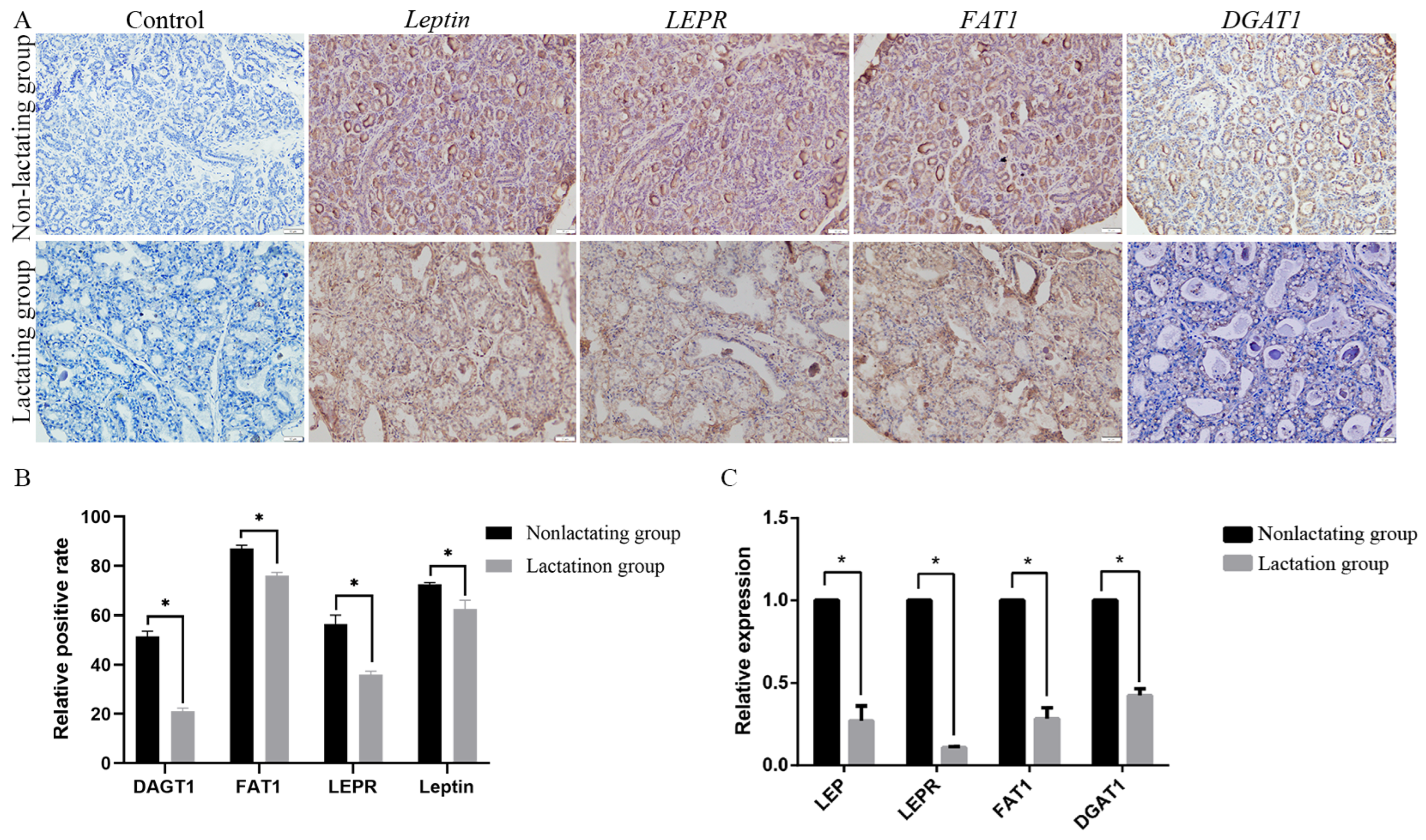
2.8. Immunohistochemistry
2.9. Statistical Analysis
3. Results
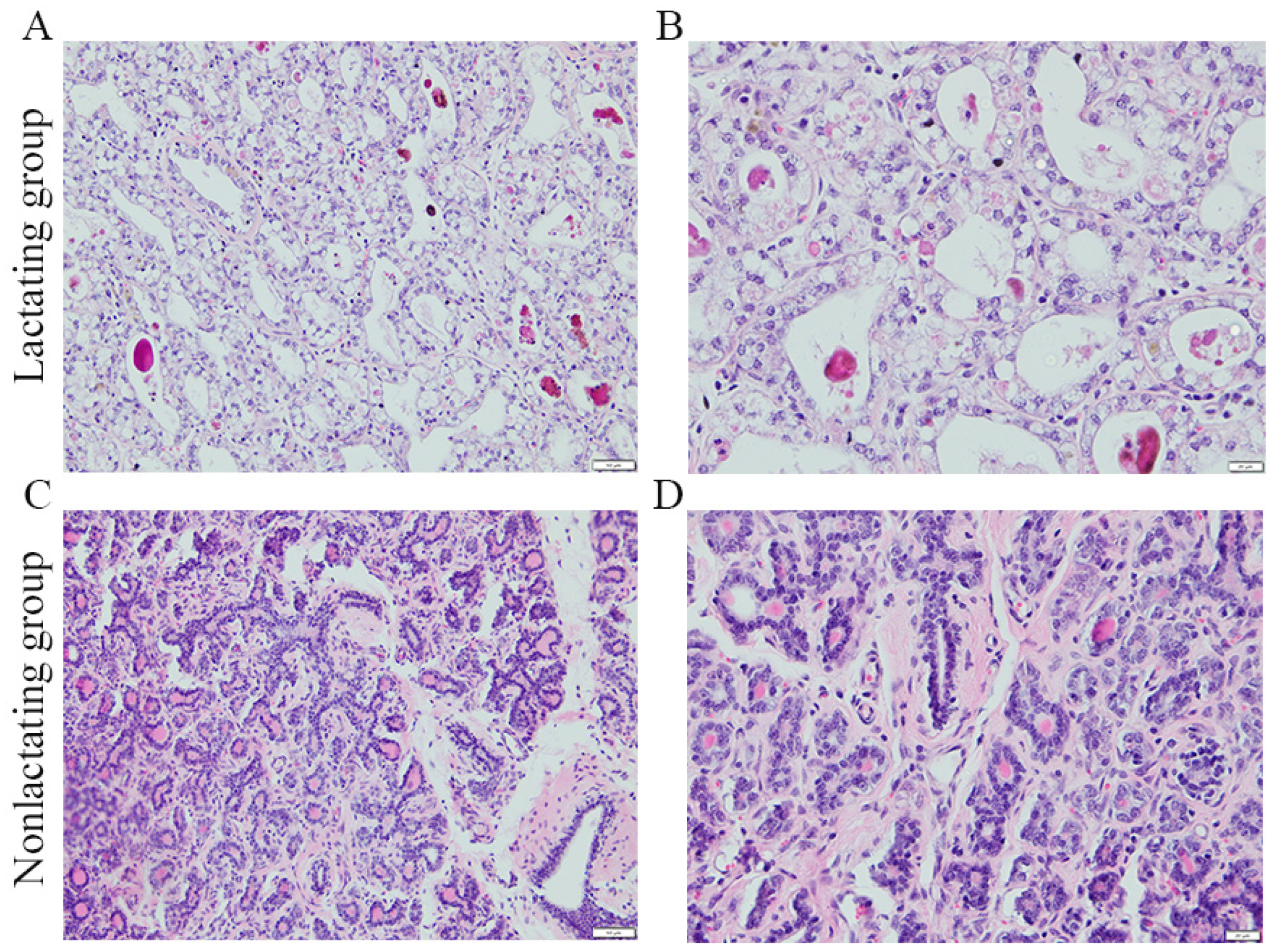
3.1. Histomorphometric Analysis of Horse Mammary Glands
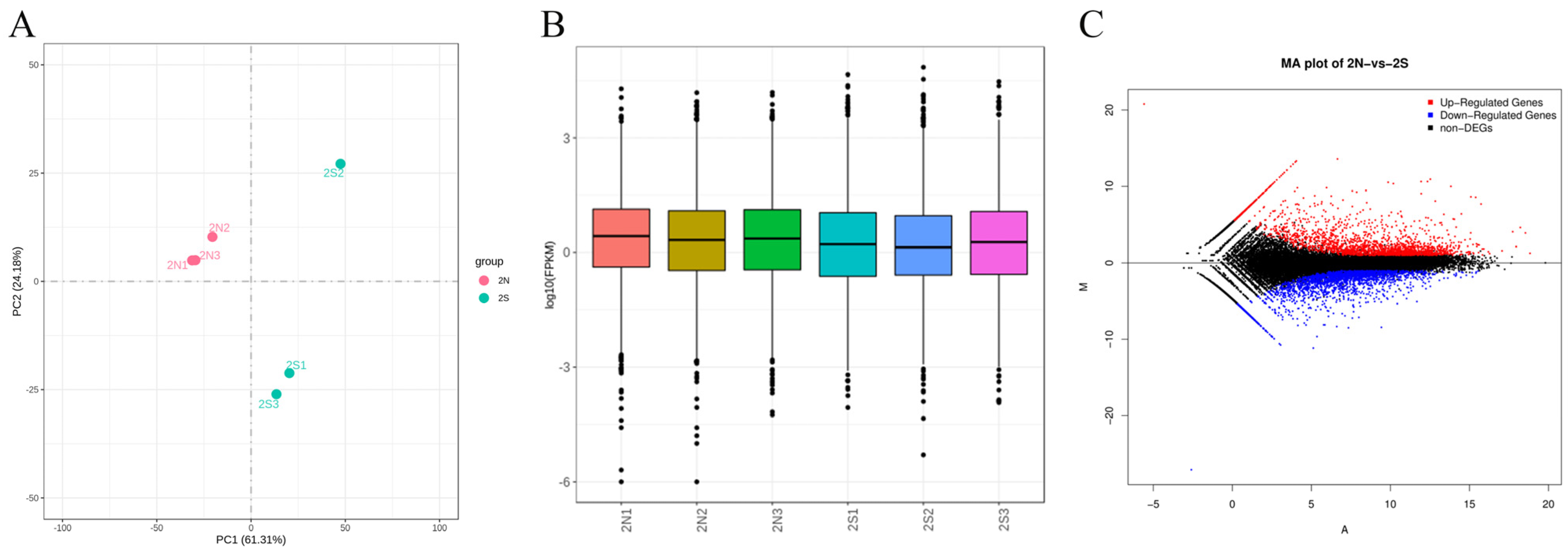
3.2. Basic Statistical Analysis Results of Sequencing Data
3.3. Differential Expression Analysis Results
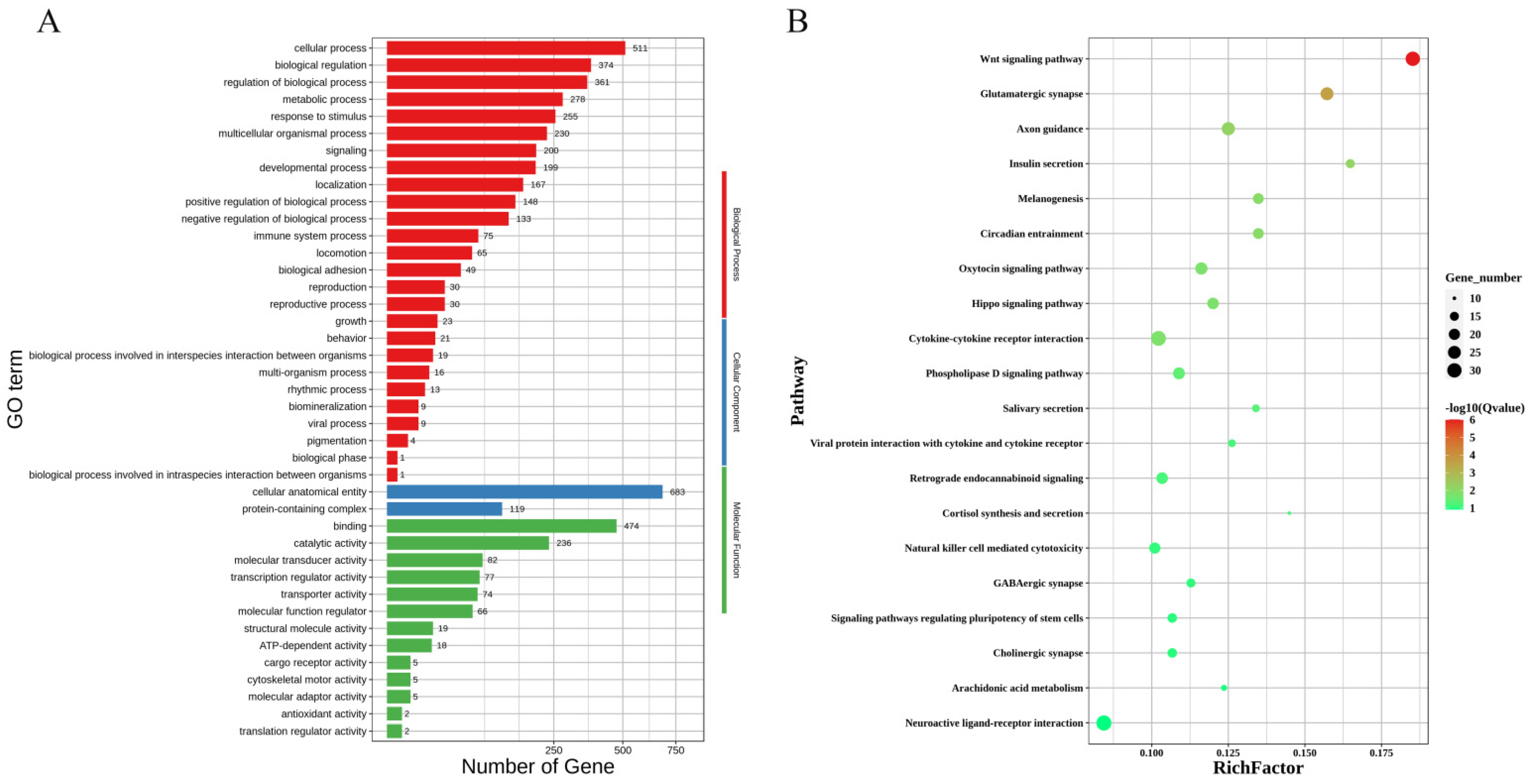
3.4. GO and KEGG Pathway Enrichment Analysis of DEGs
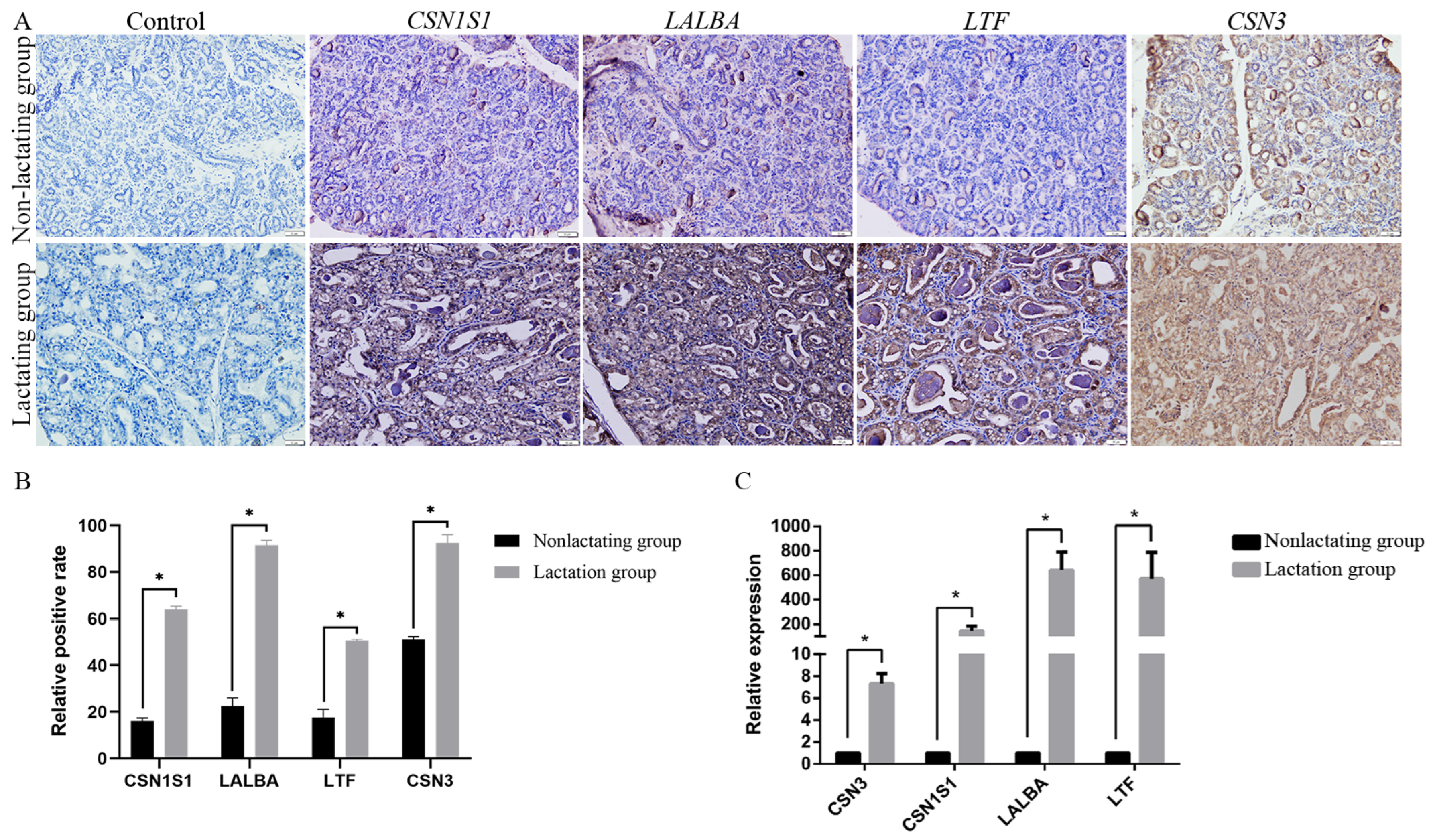
3.5. Gene Expression Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Guo, Z.; Zhang, Q.; Yan, L.; Chen, W.; Liu, X.M.; Zhang, H.P. Fermentation characteristics and transit tolerance of probiotic Lactobacillus casei Zhang in soymilk and bovine milk during storage. J. Dairy Sci. 2009, 92, 2468–2476. [Google Scholar] [CrossRef] [PubMed]
- Martuzzi, F.; Franceschi, P.; Formaggioni, P. Fermented Mare Milk and Its Microorganisms for Human Consumption and Health. Foods 2024, 13, 493. [Google Scholar] [CrossRef] [PubMed]
- Burenbayaer; Zhao, J.J.; Qiao, X.H. Nutrition and health care function of mare’s milk. Mod. Anim. Husb. 2023, 3, 43. (In Chinese) [Google Scholar] [CrossRef]
- Barreto, I.; Urbano, S.A.; Oliveira, C.A.A.; Macedo, C.S.; Borba, L.H.F.; Chags, B.M.E.; Rangel, A.H.N. Chemical composition and lipid profile of mare colostrum and milk of the quarter horse breed. PLoS ONE 2020, 15, e0238921. [Google Scholar] [CrossRef] [PubMed]
- Musaev, A.; Sadykova, S.; Anambayeva, A.; Saizhanova, M.; Balkanay, G.; Kolbaev, M. Mare’s Milk: Composition, Properties, and Application in Medicine. Arch. Razi Inst. 2021, 76, 1125–1135. [Google Scholar] [CrossRef]
- Xue, Q.G.; Schey, K.L.; Volety, A.K.; Chu, F.L.; La Peyre, J.F. Purification and characterization of lysozyme from plasma of the eastern oyster (Crassostrea virginica). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 139, 11–25. [Google Scholar] [CrossRef]
- Pecka, E.; Dobrzanski, Z.; Zachwieja, A.; Szulc, T.; Czyz, K. Studies of composition and major protein level in milk and colostrum of mares. Anim. Sci. J. 2012, 83, 162–168. [Google Scholar] [CrossRef]
- Reiter, A.S.; Reed, S.A. Lactation in horses. Anim. Front. Rev. Mag. Anim. Agric. 2023, 13, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.H.; Osbakk, S.A.; Vorland, L.H.; Traavik, T.; Gutteberg, T.J. Lactoferrin and cyclic lactoferricin inhibit the entry of human cytomegalovirus into human fibroblasts. Antivir. Res. 2001, 51, 141–149. [Google Scholar] [CrossRef]
- Superti, F. Lactoferrin from Bovine Milk: A Protective Companion for Life. Nutrients 2020, 12, 2562. [Google Scholar] [CrossRef]
- Li, N.; Xie, Q.; Chen, Q.; Evivie, S.E.; Liu, D.; Dong, J.; Huo, G.; Li, B. Cow, Goat, and Mare Milk Diets Differentially Modulated the Immune System and Gut Microbiota of Mice Colonized by Healthy Infant Feces. J. Agric. Food Chem. 2020, 68, 15345–15357. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 11, Unit 11.7. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, X.; Fang, C.; Liu, L.; Zhao, X.; Liu, W.; Cao, H.; Lv, S. Transcriptome study underling difference of milk yield during peak lactation of Kazakh horse. J. Equine Vet. Sci. 2021, 102, 103424. [Google Scholar] [CrossRef] [PubMed]
- Chanat, E.; Martin, P.; Ollivier-Bousquet, M. Alpha(S1)-casein is required for the efficient transport of beta- and kappa-casein from the endoplasmic reticulum to the Golgi apparatus of mammary epithelial cells. J. Cell Sci. 1999, 112 Pt 19, 3399–3412. [Google Scholar] [CrossRef]
- Martin, P.; Szymanowska, M.; Zwierzchowski, L.; Leroux, C. The impact of genetic polymorphisms on the protein composition of ruminant milks. Reprod. Nutr. Dev. 2002, 42, 433–459. [Google Scholar] [CrossRef]
- Noce, A.; Pazzola, M.; Dettori, M.L.; Amills, M.; Castello, A.; Cecchinato, A.; Bittante, G.; Vacca, G.M. Variations at regulatory regions of the milk protein genes are associated with milk traits and coagulation properties in the Sarda sheep. Anim. Genet. 2016, 47, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Gai, N.; Uniacke-Lowe, T.; O’Regan, J.; Faulkner, H.; Kelly, A.L. Effect of Protein Genotypes on Physicochemical Properties and Protein Functionality of Bovine Milk: A Review. Foods 2021, 10, 2409. [Google Scholar] [CrossRef] [PubMed]
- Citek, J.; Samkova, E.; Brzakova, M.; Hanus, O.; Vecerek, L.; Hostickova, I.; Jozova, E.; Hasonova, L.; Halova, K. CSN1S1 and LALBA Polymorphisms and Other Factors Influencing Yield, Composition, Somatic Cell Score, and Technological Properties of Cow’s Milk. Animals 2023, 13, 2079. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Fu, Y.; Xu, L.; Li, H.; Li, Y.; Liu, L.; Ma, Z.; Sun, D.; Han, B. Associations between polymorphisms of SLC22A7, NGFR, ARNTL and PPP2R2B genes and Milk production traits in Chinese Holstein. BMC Genom. Data 2021, 22, 47. [Google Scholar] [CrossRef] [PubMed]
- Cieslak, J.; Wodas, L.; Borowska, A.; Pawlak, P.; Czyzak-Runowska, G.; Wojtowski, J.; Puppel, K.; Kuczynska, B.; Mackowski, M. 5’-flanking variants of equine casein genes (CSN1S1, CSN1S2, CSN2, CSN3) and their relationship with gene expression and milk composition. J. Appl. Genet. 2019, 60, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Moioli, B.; D’Andrea, M.; Pilla, F.J.S.R.R. Candidate genes affecting sheep and goat milk quality. Small Rumin. Res. 2007, 68, 179–192. [Google Scholar] [CrossRef]
- Bittante, G.; Penasa, M.; Cecchinato, A. Invited review: Genetics and modeling of milk coagulation properties. J. Dairy Sci. 2012, 95, 6843–6870. [Google Scholar] [CrossRef] [PubMed]
- Li, L. Expression Analysis of CSN1S2, CSN2 and CSN3 Genes in Mammary Glands of XINONG SAANEN Dairy Goat; Northwest Agriculture & Forestry University: Xianyang, China, 2009. [Google Scholar]
- Fan, B.L.; Li, N.; Hu, X.X.; Wu, C.X. Cloning, sequencing and polymorphism analyzing of the exon 4 of the kappa-casein gene in yak. Prog. Nat. Sci. 2000, 10, 769. (In Chinese) [Google Scholar]
- Layman, D.K.; Lonnerdal, B.; Fernstrom, J.D. Applications for alpha-lactalbumin in human nutrition. Nutr. Rev. 2018, 76, 444–460. [Google Scholar] [CrossRef]
- Sharp, J.A.; Lefevre, C.; Nicholas, K.R. Lack of functional alpha-lactalbumin prevents involution in Cape fur seals and identifies the protein as an apoptotic milk factor in mammary gland involution. BMC Biol. 2008, 6, 48. [Google Scholar] [CrossRef]
- Bleck, G.T.; Bremel, R.D. Correlation of the alpha-lactalbumin (+15) polymorphism to milk production and milk composition of Holsteins. J. Dairy Sci. 1993, 76, 2292–2298. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gamez, E.; Gutierrez-Gil, B.; Sahana, G.; Sanchez, J.P.; Bayon, Y.; Arranz, J.J. GWA analysis for milk production traits in dairy sheep and genetic support for a QTN influencing milk protein percentage in the LALBA gene. PLoS ONE 2012, 7, e47782. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.F.; Milazzotto, M.P.; Feitosa, W.B.; Coutinho, A.R.; Simoes, R.; Marques, M.G.; Assumpcao, M.E.; Visintin, J.A. Sequence variation of the alpha-lactalbumin gene in Holstein and Nellore cows. Anim. Biotechnol. 2008, 19, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Ren, Y.; Lu, Q.; Wang, K.; Wu, Y.; Wang, Y.; Zhang, Y.; Cui, X.S.; Yang, Z.; Chen, Z. Lactoferrin: A glycoprotein that plays an active role in human health. Front Nutr 2022, 9, 1018336. [Google Scholar] [CrossRef] [PubMed]
- Cieslak, J.; Wodas, L.; Borowska, A.; Sadoch, J.; Pawlak, P.; Puppel, K.; Kuczynska, B.; Mackowski, M. Variability of lysozyme and lactoferrin bioactive protein concentrations in equine milk in relation to LYZ and LTF gene polymorphisms and expression. J. Sci. Food Agric. 2017, 97, 2174–2181. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Wang, X. Isolation and expression of porcine lactoferrin gene. Prog. Vet. Med. 1999, 20, 47. (In Chinese) [Google Scholar] [CrossRef]
- Yang, K.; Gao, Y.; Wu, M.L.; Lan, X.Y.; Lei, C.C.; Chen, H.; Liu, W.J.; Dang, R.H. Polymorphism of LTF gene and its Correlation with lactation performance in Yili horse. J. Domest. Anim. Ecol. 2018, 39, 24. (In Chinese) [Google Scholar]
- Li, Z.Q.; Ma, S.H.; Xu, J.; Wen, L.; Tian, L.; Liu, L.X.; Zhang, L. Genetic polymorphism of LF gene and its associations with milk traits and mastitis of Chinese Holstein cattle. Acta Agric. Zhejiangensis 2016, 28, 1142. (In Chinese) [Google Scholar] [CrossRef]
- Coleman, R.A.; Lee, D.P. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004, 43, 134–176. [Google Scholar] [CrossRef]
- Invernizzi, G.; Thering, B.J.; McGuire, M.A.; Savoini, G.; Loor, J.J. Sustained upregulation of stearoyl-CoA desaturase in bovine mammary tissue with contrasting changes in milk fat synthesis and lipogenic gene networks caused by lipid supplements. Funct. Integr. Genom. 2010, 10, 561–575. [Google Scholar] [CrossRef]
- Cases, S.; Zhou, P.; Shillingford, J.M.; Wiseman, B.S.; Fish, J.D.; Angle, C.S.; Hennighausen, L.; Werb, Z.; Farese, R.V., Jr. Development of the mammary gland requires DGAT1 expression in stromal and epithelial tissues. Development 2004, 131, 3047–3055. [Google Scholar] [CrossRef] [PubMed]
- Grisart, B.; Farnir, F.; Karim, L.; Cambisano, N.; Kim, J.J.; Kvasz, A.; Mni, M.; Simon, P.; Frere, J.M.; Coppieters, W.; et al. Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc. Natl. Acad. Sci. USA 2004, 101, 2398–2403. [Google Scholar] [CrossRef] [PubMed]
- Strucken, E.M.; Laurenson, Y.C.; Brockmann, G.A. Go with the flow-biology and genetics of the lactation cycle. Front. Genet. 2015, 6, 118. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, L.A. The leptin receptor. J. Biol. Chem. 1997, 272, 6093–6096. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Juneja, S.C.; Maihle, N.J.; Cleary, M.P. Leptin--a growth factor in normal and malignant breast cells and for normal mammary gland development. J. Natl. Cancer Inst. 2002, 94, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Baratta, M.; Grolli, S.; Tamanini, C. Effect of leptin in proliferating and differentiated HC11 mouse mammary cells. Regul. Pept. 2003, 113, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, Q. Expression and function of Leptin and its receptor in mammary gland of goat. Sci. Agric. Sin. 2008, 41, 4187–4193. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Nan, W. Study on the Regulation Effect of Leptin on Milk Fat Synthesis in Dairy Cows; Northeast Agricultural University: Harbin, China, 2021. [Google Scholar]
- Duquenne, M.; Folgueira, C.; Bourouh, C.; Millet, M.; Silva, A.; Clasadonte, J.; Imbernon, M.; Fernandois, D.; Martinez-Corral, I.; Kusumakshi, S.; et al. Leptin brain entry via a tanycytic LepR-EGFR shuttle controls lipid metabolism and pancreas function. Nat. Metab. 2021, 3, 1071–1090. [Google Scholar] [CrossRef]
- Kim, J.E.; Choi, Y.J.; Lee, S.J.; Gong, J.E.; Lim, Y.; Hong, J.T.; Hwang, D.Y. Molecular Characterization of Constipation Disease as Novel Phenotypes in CRISPR-Cas9-Generated Leptin Knockout Mice with Obesity. Int. J. Mol. Sci. 2020, 21, 9464. [Google Scholar] [CrossRef]
- Liu, L.; Fang, C.; Sun, Y.; Liu, W. Evaluation of key miRNAs during early pregnancy in Kazakh horse using RNA sequencing. PeerJ 2021, 9, e10796. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, Y.; Ma, H.; Cao, H.; Liu, W. Integrating genome-wide methylation and transcriptome-wide analyses to reveal the genetic mechanism of milk traits in Kazakh horses. Gene 2023, 856, 147143. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulaangerel, T.; Wang, M.; Zhao, B.; Yi, M.; Shen, Y.; Mengkh, Y.; Wen, X.; Dugarjav, M.; Bou, G. A Comparative Analysis of the Gene Expression Profiles in the Mammary Glands of Lactating and Nonlactating Mares at the Second Month of Gestation. Animals 2024, 14, 2319. https://doi.org/10.3390/ani14162319
Ulaangerel T, Wang M, Zhao B, Yi M, Shen Y, Mengkh Y, Wen X, Dugarjav M, Bou G. A Comparative Analysis of the Gene Expression Profiles in the Mammary Glands of Lactating and Nonlactating Mares at the Second Month of Gestation. Animals. 2024; 14(16):2319. https://doi.org/10.3390/ani14162319
Chicago/Turabian StyleUlaangerel, Tseweendolmaa, Min Wang, Bilig Zhao, Minna Yi, Yingchao Shen, Yibeeltu Mengkh, Xin Wen, Manglai Dugarjav, and Gerelchimeg Bou. 2024. "A Comparative Analysis of the Gene Expression Profiles in the Mammary Glands of Lactating and Nonlactating Mares at the Second Month of Gestation" Animals 14, no. 16: 2319. https://doi.org/10.3390/ani14162319





