Novel Candidate Genes Involved in an Initial Stage of White Striping Development in Broiler Chickens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Histopathological Analysis
2.3. RNA Extraction, Preparation, and Sequencing of RNA-seq Libraries
2.4. RNA-seq Data Analysis
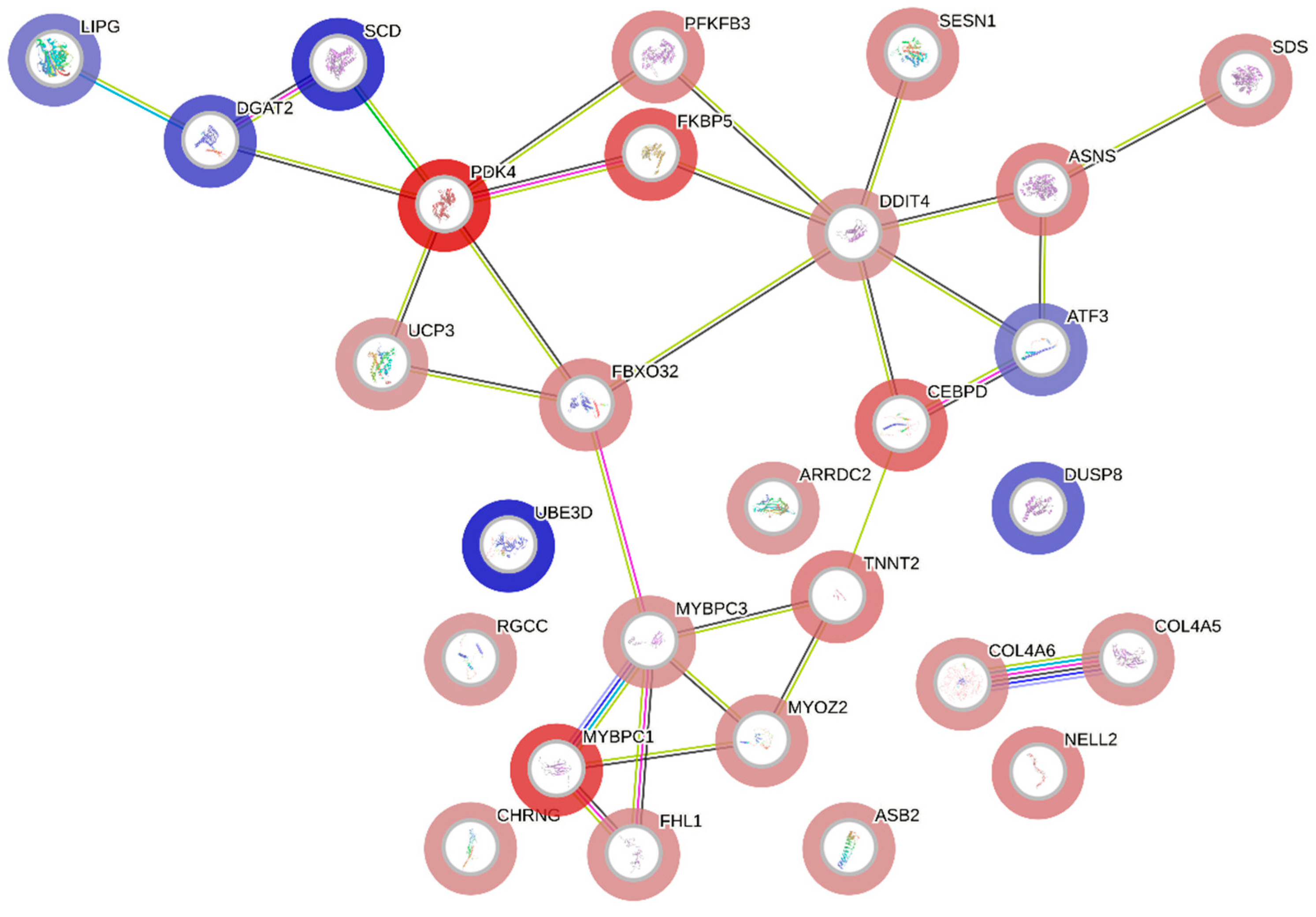
2.5. Functional Annotation and Gene Interaction Networks
2.6. qPCR Confirmation Analysis
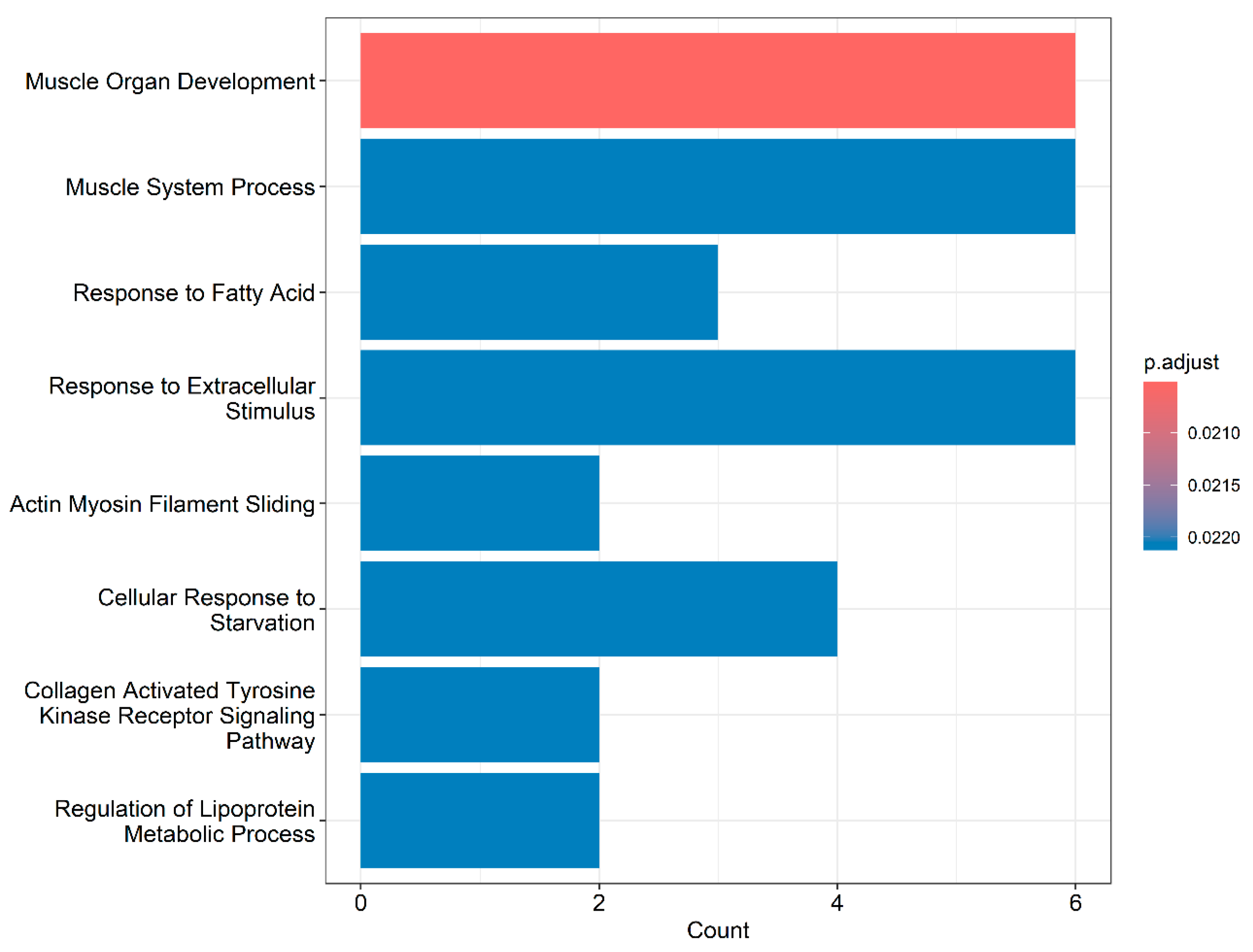
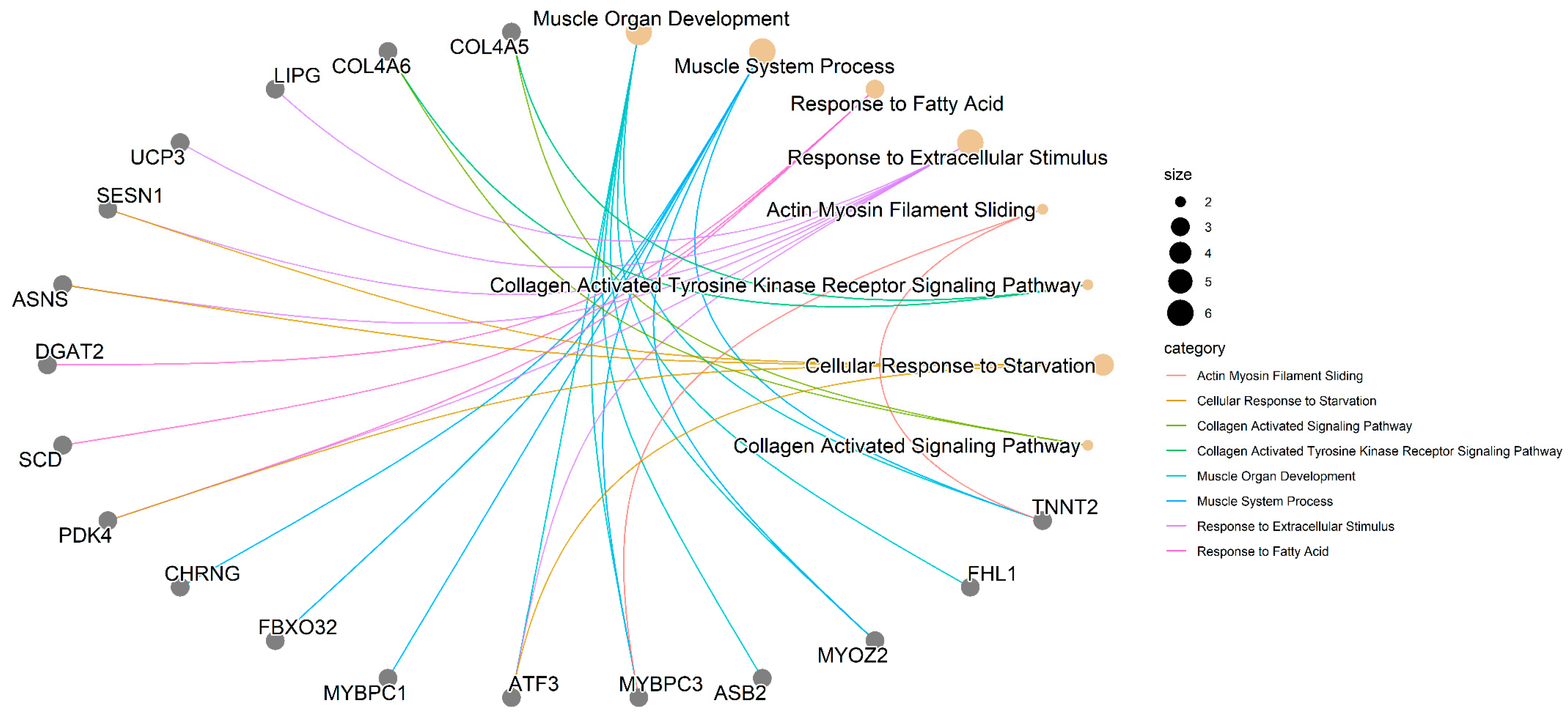
3. Results
4. Discussion
4.1. Muscle-Related Genes
4.2. Genes Related to Lipid Metabolism and Cellular Processes
4.3. Collagen-Related Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Praud, C.; Jimenez, J.; Pampouille, E.; Couroussé, N.; Godet, E.; Le Bihan-Duval, E.; Berri, C. Molecular Phenotyping of White Striping and Wooden Breast Myopathies in Chicken. Front. Physiol. 2020, 11, 549704. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G. Recent Developments in Breast Muscle Myopathies Associated with Growth in Poultry. Annu. Rev. Anim. Biosci. 2019, 7, 289–308. [Google Scholar] [CrossRef] [PubMed]
- Kuttappan, V.A.; Brewer, V.B.; Apple, J.K.; Waldroup, P.W.; Owens, C.M. Influence of Growth Rate on the Occurrence of White Striping in Broiler Breast Fillets. Poult. Sci. 2012, 91, 2677–2685. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, M.; Mudalal, S.; Cavani, C.; Petracci, M. Incidence of White Striping under Commercial Conditions in Medium and Heavy Broiler Chickens in Italy. J. Appl. Poult. Res. 2014, 23, 754–758. [Google Scholar] [CrossRef]
- Ferreira, T.Z.; Casagrande, R.A.; Vieira, S.L.; Driemeier, D.; Kindlein, L. An Investigation of a Reported Case of White Striping in Broilers. J. Appl. Poult. Res. 2014, 23, 748–753. [Google Scholar] [CrossRef]
- Petracci, M.; Mudalal, S.; Bonfiglio, A.; Cavani, C. Occurrence of White Striping under Commercial Conditions and Its Impact on Breast Meat Quality in Broiler Chickens. Poult. Sci. 2013, 92, 1670–1675. [Google Scholar] [CrossRef] [PubMed]
- Alnahhas, N.; Berri, C.; Chabault, M.; Chartrin, P.; Boulay, M.; Bourin, M.C.; Bihan-Duval, E. Le Genetic Parameters of White Striping in Relation to Body Weight, Carcass Composition, and Meat Quality Traits in Two Broiler Lines Divergently Selected for the Ultimate PH of the Pectoralis Major Muscle. BMC Genet. 2016, 17, 61. [Google Scholar] [CrossRef]
- Bailey, R.A.; Souza, E.; Avendano, S. Characterising the Influence of Genetics on Breast Muscle Myopathies in Broiler Chickens. Front. Physiol. 2020, 11, 547817. [Google Scholar] [CrossRef]
- Bailey, R.A.; Watson, K.A.; Bilgili, S.F.; Avendano, S. The Genetic Basis of Pectoralis Major Myopathies in Modern Broiler Chicken Lines. Poult. Sci. 2015, 94, 2870–2879. [Google Scholar] [CrossRef]
- Lake, J.A.; Dekkers, J.C.M.; Abasht, B. Genetic Basis and Identification of Candidate Genes for Wooden Breast and White Striping in Commercial Broiler Chickens. Sci. Rep. 2021, 11, 6785. [Google Scholar] [CrossRef]
- Malila, Y.; Uengwetwanit, T.; Arayamethakorn, S.; Srimarut, Y.; Thanatsang, K.V.; Soglia, F.; Strasburg, G.M.; Rungrassamee, W.; Visessanguan, W. Transcriptional Profiles of Skeletal Muscle Associated with Increasing Severity of White Striping in Commercial Broilers. Front. Physiol. 2020, 11, 521734. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.A.P.; Ibelli, A.M.G.; Peixoto, J.O.; Cantão, M.E.; Pandolfi, J.R.C.; Marciano, C.M.M.; Zanella, R.; Settles, M.L.; Coutinho, L.L.; Ledur, M.C. Whole Transcriptome Analysis of the Pectoralis Major Muscle Reveals Molecular Mechanisms Involved with White Striping in Broiler Chickens. Poult. Sci. 2019, 98, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Vanderhout, R.J.; Abdalla, E.A.; Leishman, E.M.; Barbut, S.; Wood, B.J.; Baes, C.F. Genetic Architecture of White Striping in Turkeys (Meleagris gallopavo). Sci. Rep. 2024, 14, 9007. [Google Scholar] [CrossRef] [PubMed]
- Pampouille, E.; Berri, C.; Boitard, S.; Hennequet-Antier, C.; Beauclercq, S.A.; Godet, E.; Praud, C.; Jégo, Y.; Le Bihan-Duval, E. Mapping QTL for White Striping in Relation to Breast Muscle Yield and Meat Quality Traits in Broiler Chickens. BMC Genom. 2018, 19, 202. [Google Scholar] [CrossRef]
- Boerboom, G.; Van Kempen, T.; Navarro-Villa, A.; Pérez-Bonilla, A. Unraveling the Cause of White Striping in Broilers Using Metabolomics. Poult. Sci. 2018, 97, 3977–3986. [Google Scholar] [CrossRef] [PubMed]
- Bordini, M.; Soglia, F.; Davoli, R.; Zappaterra, M.; Petracci, M.; Meluzzi, A. Molecular Pathways and Key Genes Associated with Breast Width and Protein Content in White Striping and Wooden Breast Chicken Pectoral Muscle. Front. Physiol. 2022, 13, 936768. [Google Scholar] [CrossRef]
- Pizzol, M.S.D.; Ibelli, A.M.G.; Cantão, M.E.; Campos, F.G.; de Oliveira, H.C.; de Oliveira Peixoto, J.; Fernandes, L.T.; de Castro Tavernari, F.; Morés, M.A.Z.; Bastos, A.P.A.; et al. Differential Expression of MiRNAs Associated with Pectoral Myopathies in Young Broilers: Insights from a Comparative Transcriptome Analysis. BMC Genom. 2024, 25, 104. [Google Scholar] [CrossRef]
- Marciano, C.M.M.; Ibelli, A.M.G.; Marchesi, J.A.P.; de Oliveira Peixoto, J.; Fernandes, L.T.; Savoldi, I.R.; do Carmo, K.B.; Ledur, M.C. Differential Expression of Myogenic and Calcium Signaling-Related Genes in Broilers Affected with White Striping. Front. Physiol. 2021, 12, 712464. [Google Scholar] [CrossRef]
- Fornari, M.B.; Zanella, R.; Ibelli, A.M.G.; Fernandes, L.T.; Cantão, M.E.; Thomaz-Soccol, V.; Ledur, M.C.; Peixoto, J.O. Unraveling the Associations of Osteoprotegerin Gene with Production Traits in a Paternal Broiler Line. Springerplus 2014, 3, 682. [Google Scholar] [CrossRef]
- Marchesi, J.A.P.; Buzanskas, M.E.; Cantão, M.E.; Ibelli, A.M.G.; Peixoto, J.O.; Joaquim, L.B.; Moreira, G.C.M.; Godoy, T.F.; Sbardella, A.P.; Figueiredo, E.A.P.; et al. Relationship of Runs of Homozygosity with Adaptive and Production Traits in a Paternal Broiler Line. Animal 2018, 12, 1126–1134. [Google Scholar] [CrossRef]
- Rostagno, H.S.; Albino, L.T.; Donzele, J.L.; Gomes, P.C.; Oliveira, R.D.; Lopes, D.C.; Ferreira, A.S.; Barreto, S.D.T.; Euclides, R.F. Tabelas Brasileiras Para Aves e Suínos: Composição de Alimentos e Exigências Nutricionais; Rostagno, H.S., Ed.; Universidade Federal de Viçosa: Viçosa, Brasil, 2017. [Google Scholar]
- Kuttappan, V.A.; Lee, Y.S.; Erf, G.F.; Meullenet, J.F.C.; Mckee, S.R.; Owens, C.M. Consumer Acceptance of Visual Appearance of Broiler Breast Meat with Varying Degrees of White Striping. Poult. Sci. 2012, 91, 1240–1247. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, L.; Lun, A.T.L.; Baldoni, P.L.; Smyth, G.K. EdgeR 4.0: Powerful Differential Analysis of Sequencing Data with Expanded Functionality and Improved Support for Small Counts and Larger Datasets. bioRxiv 2024. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org (accessed on 30 January 2024).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Carlson, M. Org.Gg.Eg.Db: Genome Wide Annotation for Chicken. R Package Version 3.8.2. 2019. Available online: https://doi.org/10.18129/B9.bioc.org.Gg.eg.db (accessed on 3 August 2024).
- Pagès, H.; Carlson, M.; Falcon, S.; Li, N. AnnotationDbi: Manipulation of SQLite-Based Annotations in Bioconductor. 2023. Available online: https://doi.org/10.18129/B9.bioc.AnnotationDbi (accessed on 3 August 2024).
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the Future: Meeting New Challenges and Providing Updated Services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef] [PubMed]
- Paludo, E.; Ibelli, A.M.G.; Peixoto, J.O.; Tavernari, F.C.; Lima-Rosa, C.A.V.; Pandolfi, J.R.C.; Ledur, M.C. The Involvement of RUNX2 and SPARC Genes in the Bacterial Chondronecrosis with Osteomyelitis in Broilers. Animal 2017, 11, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, C.S.; Barbosa, L.T.; Brito, C.; Fernandes, R.P.M.; Mann, R.S.; Pinto, A.P.G.; Oliveira, H.C.; Dodson, M.V.; Guimarães, S.E.F.; Duarte, M.S. Identification of Suitable Reference Genes for Real Time Quantitative Polymerase Chain Reaction Assays on Pectoralis Major Muscle in Chicken (Gallus gallus). PLoS ONE 2015, 10, e0127935. [Google Scholar] [CrossRef]
- Petry, B.; Savoldi, I.R.; Ibelli, A.M.G.; Paludo, E.; de Oliveira Peixoto, J.; Jaenisch, F.R.F.; de Córdova Cucco, D.; Ledur, M.C. New Genes Involved in the Bacterial Chondronecrosis with Osteomyelitis in Commercial Broilers. Livest. Sci. 2018, 208, 33–39. [Google Scholar] [CrossRef]
- Rausch, T.; Hsi-Yang Fritz, M.; Korbel, J.O.; Benes, V. Alfred: Interactive Multi-Sample BAM Alignment Statistics, Feature Counting and Feature Annotation for Long- and Short-Read Sequencing. Bioinformatics 2019, 35, 2489–2491. [Google Scholar] [CrossRef]
- Rausch, T.; Fritz, M.H.Y.; Untergasser, A.; Benes, V. Tracy: Basecalling, Alignment, Assembly and Deconvolution of Sanger Chromatogram Trace Files. BMC Genom. 2020, 21, 230. [Google Scholar] [CrossRef]
- Untergasser, A.; Ruijter, J.M.; Benes, V.; van den Hoff, M.J.B. Web-Based LinRegPCR: Application for the Visualization and Analysis of (RT)-QPCR Amplification and Melting Data. BMC Bioinform. 2021, 22, 398. [Google Scholar] [CrossRef]
- GEAR: Genome Analysis Server. Available online: https://www.gear-genomics.com/ (accessed on 3 August 2024).
- EndoGenes—Análise de Genes de Referência. Available online: https://hanielcedraz.shinyapps.io/endoGenes/ (accessed on 3 August 2024).
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Início Gene. NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene (accessed on 3 August 2024).
- Navegador Do Genoma Ensembl 112. Available online: https://www.ensembl.org/index.html (accessed on 3 August 2024).
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Zhang, J.; Kaiser, M.G.; Deist, M.S.; Gallardo, R.A.; Bunn, D.A.; Kelly, T.R.; Dekkers, J.C.M.; Zhou, H.; Lamont, S.J. Transcriptome Analysis in Spleen Reveals Differential Regulation of Response to Newcastle Disease Virus in Two Chicken Lines. Sci. Rep. 2018, 8, 1278. [Google Scholar] [CrossRef] [PubMed]
- Mutryn, M.F.; Brannick, E.M.; Fu, W.; Lee, W.R.; Abasht, B. Characterization of a Novel Chicken Muscle Disorder through Differential Gene Expression and Pathway Analysis Using RNA-Sequencing. BMC Genom. 2015, 16, 399. [Google Scholar] [CrossRef]
- Zambonelli, P.; Zappaterra, M.; Soglia, F.; Petracci, M.; Sirri, F.; Cavani, C.; Davoli, R. Detection of Differentially Expressed Genes in Broiler Pectoralis Major Muscle Affected by White Striping—Wooden Breast Myopathies. Poult. Sci. 2016, 95, 2771–2785. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.W.; Hudson, N.; Seo, D.; Lee, S.; Khatri, B.; Lassiter, K.; Cook, D.; Piekarski, A.; Dridi, S.; Anthony, N.; et al. RNA Sequencing for Global Gene Expression Associated with Muscle Growth in a Single Male Modern Broiler Line Compared to a Foundational Barred Plymouth Rock Chicken Line. BMC Genom. 2017, 18, 82. [Google Scholar] [CrossRef]
- Papah, M.B.; Abasht, B. Dysregulation of Lipid Metabolism and Appearance of Slow Myofiber-Specific Isoforms Accompany the Development of Wooden Breast Myopathy in Modern Broiler Chickens. Sci. Rep. 2019, 9, 17170. [Google Scholar] [CrossRef]
- Zhang, Z.; Du, H.; Yang, C.; Li, Q.; Qiu, M.; Song, X.; Yu, C.; Jiang, X.; Liu, L.; Hu, C.; et al. Comparative Transcriptome Analysis Reveals Regulators Mediating Breast Muscle Growth and Development in Three Chicken Breeds. Anim. Biotechnol. 2019, 30, 233–241. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.W.; Sadayappan, S. Skeletal Myosin Binding Protein-C: An Increasingly Important Regulator of Striated Muscle Physiology. Arch. Biochem. Biophys. 2018, 660, 121–128. [Google Scholar] [CrossRef]
- Qaisar, R.; Bhaskaran, S.; Van Remmen, H. Muscle Fiber Type Diversification during Exercise and Regeneration. Free Radic. Biol. Med. 2016, 98, 56–67. [Google Scholar] [CrossRef]
- Frey, N.; Barrientos, T.; Shelton, J.M.; Frank, D.; Rütten, H.; Gehring, D.; Kuhn, C.; Lutz, M.; Rothermel, B.; Bassel-Duby, R.; et al. Mice Lacking Calsarcin-1 Are Sensitized to Calcineurin Signaling and Show Accelerated Cardiomyopathy in Response to Pathological Biomechanical Stress. Nat. Med. 2004, 10, 1336–1343. [Google Scholar] [CrossRef]
- Schulz, R.A.; Yutzey, K.E. Calcineurin Signaling and NFAT Activation in Cardiovascular and Skeletal Muscle Development. Dev. Biol. 2004, 266, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Jin, J.P. TNNT1, TNNT2, and TNNT3: Isoform Genes, Regulation, and Structure–Function Relationships. Gene 2016, 582, 1–13. [Google Scholar] [CrossRef]
- Yang, J.; Gong, Y.; Cai, J.; Liu, Q.; Zhang, Z. Lnc-3215 Suppression Leads to Calcium Overload in Selenium Deficiency-Induced Chicken Heart Lesion via the Lnc-3215-MiR-1594-TNN2 Pathway. Mol. Ther. Nucleic Acids. 2019, 18, 1–15. [Google Scholar] [CrossRef]
- Cowling, B.S.; McGrath, M.J.; Nguyen, M.A.; Cottle, D.L.; Kee, A.J.; Brown, S.; Schessl, J.; Zou, Y.; Joya, J.; Bönnemann, C.G.; et al. Identification of FHL1 as a Regulator of Skeletal Muscle Mass: Implications for Human Myopathy. J. Cell Biol. 2008, 183, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.; Owens, C.; Bottje, W.; Shakeri, M.; Choi, J.; Zhuang, H.; Bowker, B. Proteomic Analyses on Chicken Breast Meat with White Striping Myopathy. Poult. Sci. 2024, 103, 103682. [Google Scholar] [CrossRef]
- Zhang, X.; Antonelo, D.; Hendrix, J.; To, V.; Campbell, Y.; Von Staden, M.; Li, S.; Suman, S.P.; Zhai, W.; Chen, J.; et al. Proteomic Characterization of Normal and Woody Breast Meat from Broilers of Five Genetic Strains. Meat Muscle Biol. 2020, 4, 1–17. [Google Scholar] [CrossRef]
- Dowling, P.; Gargan, S.; Zweyer, M.; Sabir, H.; Swandulla, D.; Ohlendieck, K. Proteomic Profiling of Carbonic Anhydrase CA3 in Skeletal Muscle. Expert Rev. Proteom. 2021, 18, 1073–1086. [Google Scholar] [CrossRef]
- Qian, S.H.; Chen, L.; Xiong, Y.L.; Chen, Z.X. Evolution and Function of Developmentally Dynamic Pseudogenes in Mammals. Genome Biol. 2022, 23, 235. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, B.; Ratliff, A.C.; Arlington, J.; Chen, J.; Xiong, Y.; Yue, F.; Nie, Y.; Hu, K.; Jin, W.; et al. Methyltransferase-like 21e Inhibits 26S Proteasome Activity to Facilitate Hypertrophy of Type IIb Myofibers. FASEB J. 2019, 33, 9672. [Google Scholar] [CrossRef]
- Bordini, M.; Wang, Z.; Soglia, F.; Petracci, M.; Schmidt, C.J.; Abasht, B. RNA-Sequencing Revisited Data Shed New Light on Wooden Breast Myopathy. Poult. Sci. 2024, 103, 103902. [Google Scholar] [CrossRef]
- Oehrl, W.; Cotsiki, M.; Panayotou, G. Differential Regulation of M3/6 (DUSP8) Signaling Complexes in Response to Arsenite-Induced Oxidative Stress. Cell Signal. 2013, 25, 429–438. [Google Scholar] [CrossRef]
- Boyer, J.G.; Prasad, V.; Song, T.; Lee, D.; Fu, X.; Grimes, K.M.; Sargent, M.A.; Sadayappan, S.; Molkentin, J.D. ERK1/2 Signaling Induces Skeletal Muscle Slow Fiber-Type Switching and Reduces Muscular Dystrophy Disease Severity. JCI Insight 2019, 4, e127356. [Google Scholar] [CrossRef]
- Almasi, S.; Crawford Parks, T.E.; Ravel-Chapuis, A.; MacKenzie, A.; Côté, J.; Cowan, K.N.; Jasmin, B.J. Differential Regulation of Autophagy by STAU1 in Alveolar Rhabdomyosarcoma and Non-transformed Skeletal Muscle Cells. Cell. Oncol. 2021, 44, 851–870. [Google Scholar] [CrossRef]
- Zhang, X.; Nan, H.; Li, S. Dual Specificity Phosphatases 8 Directly Interacts with Mitogen-Activated Protein Kinase 1 to Regulate the Proliferation and Apoptosis of Rheumatoid Arthritis Fibroblast-like Synovial Cells. Chin. J. Rheumatol. 2022, 433–438, C7-1. [Google Scholar] [CrossRef]
- Xie, P.; Guo, S.; Fan, Y.; Zhang, H.; Gu, D.; Li, H. Atrogin-1/MAFbx Enhances Simulated Ischemia/Reperfusion-Induced Apoptosis in Cardiomyocytes through Degradation of MAPK Phosphatase-1 and Sustained JNK Activation. J. Biol. Chem. 2009, 284, 5488–5496. [Google Scholar] [CrossRef] [PubMed]
- Tintignac, L.A.; Lagirand, J.; Batonnet, S.; Sirri, V.; Leibovitch, M.P.; Leibovitch, S.A. Degradation of MyoD Mediated by the SCF (MAFbx) Ubiquitin Ligase. J. Biol. Chem. 2005, 280, 2847–2856. [Google Scholar] [CrossRef]
- Li, J.; Hu, Y.; Lan, H.; Li, L.; Hu, X.; Li, N. P3014 The Study on the Genetic Mechanism of Varied Atrogin-1 Expression in Different Chicken Lines. J. Anim. Sci. 2016, 94, 58–59. [Google Scholar] [CrossRef]
- Balamurugan, K.; Sterneck, E. The Many Faces of C/EBPδ and Their Relevance for Inflammation and Cancer. Int. J. Biol. Sci. 2013, 9, 917–933. [Google Scholar] [CrossRef]
- Yu, Y.J.; Xu, Y.Y.; Lan, X.O.; Liu, X.Y.; Zhang, X.L.; Gao, X.H.; Geng, L. Shikonin Induces Apoptosis and Suppresses Growth in Keratinocytes via CEBP-δ Upregulation. Int. Immunopharmacol. 2019, 72, 511–521. [Google Scholar] [CrossRef]
- Mao, X.G.; Xue, X.Y.; Lv, R.; Ji, A.; Shi, T.Y.; Chen, X.Y.; Jiang, X.F.; Zhang, X. CEBPD Is a Master Transcriptional Factor for Hypoxia Regulated Proteins in Glioblastoma and Augments Hypoxia Induced Invasion through Extracellular Matrix-Integrin Mediated EGFR/PI3K Pathway. Cell Death Dis. 2023, 14, 269. [Google Scholar] [CrossRef]
- Ko, C.Y.; Chang, W.C.; Wang, J.M. Biological Roles of CCAAT/Enhancer-Binding Protein Delta during Inflammation. J. Biomed. Sci. 2015, 22, 6. [Google Scholar] [CrossRef] [PubMed]
- Bello, N.F.; Lamsoul, I.; Heuzé, M.L.; Métais, A.; Moreaux, G.; Calderwood, D.A.; Duprez, D.; Moog-Lutz, C.; Lutz, P.G. The E3 Ubiquitin Ligase Specificity Subunit ASB2β Is a Novel Regulator of Muscle Differentiation That Targets Filamin B to Proteasomal Degradation. Cell Death Differ. 2009, 16, 921. [Google Scholar] [CrossRef]
- Lu, P.Y.; Taylor, M.; Jia, H.T.; Ni, J.H. Muscle LIM Protein Promotes Expression of the Acetylcholine Receptor γ-Subunit Gene Cooperatively with the Myogenin-E12 Complex. Cell. Mol. Life Sci. 2004, 61, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, D.; Zhang, X.; Li, Y.; Fu, X.; He, W.; Li, M.; Chen, P.; Zeng, G.; DiSanto, M.E.; et al. NELL2 Modulates Cell Proliferation and Apoptosis via ERK Pathway in the Development of Benign Prostatic Hyperplasia. Clin. Sci. 2021, 135, 1591–1608. [Google Scholar] [CrossRef] [PubMed]
- Gordon, B.S.; Rossetti, M.L.; Kimball, S.R. Alpha Arrestin Domain Containing 2 and Alpha Arrestin Domain Containing 3 Expression Is Reduced in Skeletal Muscle by Anabolic Stimuli and Increased by Catabolic Stimuli. FASEB J. 2019, 33, 700.10. [Google Scholar] [CrossRef]
- Xia, H.; Zhang, Q.; Shen, Y.; Bai, Y.; Ma, X.; Zhang, B.; Qi, Y.; Zhang, J.; Hu, Q.; Du, W.; et al. Ube3d, a New Gene Associated with Age-Related Macular Degeneration, Induces Functional Changes in Both In Vivo and In Vitro Studies. Mol. Ther. Nucleic Acids 2020, 20, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Lilburn, M.S.; Griffin, J.R.; Wick, M. From Muscle to Food: Oxidative Challenges and Developmental Anomalies in Poultry Breast Muscle. Poult. Sci. 2019, 98, 4255–4260. [Google Scholar] [CrossRef] [PubMed]
- Pette, D.; Staron, R.S. Transitions of Muscle Fiber Phenotypic Profiles. Histochem. Cell Biol. 2001, 115, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Abasht, B.; Papah, M.B.; Qiu, J. Evidence of Vascular Endothelial Dysfunction in Wooden Breast Disorder in Chickens: Insights through Gene Expression Analysis, Ultra-Structural Evaluation and Supervised Machine Learning Methods. PLoS ONE 2021, 16, e0243983. [Google Scholar] [CrossRef] [PubMed]
- Lake, J.A.; Abasht, B. Glucolipotoxicity: A Proposed Etiology for Wooden Breast and Related Myopathies in Commercial Broiler Chickens. Front. Physiol. 2020, 11, 528245. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, E.J.; Kim, D.K.; Lee, J.M.; Park, S.B.; Lee, I.K.; Harris, R.A.; Lee, M.O.; Choi, H.S. Hypoxia Induces PDK4 Gene Expression through Induction of the Orphan Nuclear Receptor ERRγ. PLoS ONE 2012, 7, e46324. [Google Scholar] [CrossRef]
- Wang, X.; Shen, X.; Yan, Y.; Li, H. Pyruvate Dehydrogenase Kinases (PDKs): An Overview toward Clinical Applications. Biosci. Rep. 2021, 41, 20204402. [Google Scholar] [CrossRef]
- Pettersen, I.K.N.; Tusubira, D.; Ashrafi, H.; Dyrstad, S.E.; Hansen, L.; Liu, X.Z.; Nilsson, L.I.H.; Løvsletten, N.G.; Berge, K.; Wergedahl, H.; et al. Upregulated PDK4 Expression Is a Sensitive Marker of Increased Fatty Acid Oxidation. Mitochondrion 2019, 49, 97–110. [Google Scholar] [CrossRef]
- Kuttappan, V.A.; Brewer, V.B.; Mauromoustakos, A.; McKee, S.R.; Emmert, J.L.; Meullenet, J.F.; Owens, C.M. Estimation of Factors Associated with the Occurrence of White Striping in Broiler Breast Fillets. Poult. Sci. 2013, 92, 811–819. [Google Scholar] [CrossRef]
- Koves, T.R.; Ussher, J.R.; Noland, R.C.; Slentz, D.; Mosedale, M.; Ilkayeva, O.; Bain, J.; Stevens, R.; Dyck, J.R.B.; Newgard, C.B.; et al. Mitochondrial Overload and Incomplete Fatty Acid Oxidation Contribute to Skeletal Muscle Insulin Resistance. Cell Metab. 2008, 7, 45–56. [Google Scholar] [CrossRef]
- Bezaire, V.; Spriet, L.L.; Campbell, S.; Sabet, N.; Gerrits, M.; Bonen, A.; Harper, M.-E.; Harper, M.-E. Constitutive UCP3 Overexpression at Physiological Levels Increases Mouse Skeletal Muscle Capacity for Fatty Acid Transport and Oxidation. FASEB J. 2005, 19, 977–979. [Google Scholar] [CrossRef]
- García-Martínez, C.; Sibille, B.; Solanes, G.; Darimont, C.; Macé, K.; Villarroya, F.; Gómez-Foix, A.M. Overexpression of UCP3 in Cultured Human Muscle Lowers Mitochondrial Membrane Potential, Raises ATP/ADP Ratio, and Favors Fatty Acid versus Glucose Oxidation. FASEB J. 2001, 15, 2033–2035. [Google Scholar] [CrossRef]
- Schrauwen, P.; Hinderling, V.; Hesselink, M.K.C.; Schaart, G.; Kornips, E.; Saris, W.H.M.; Westerterp-Plantenga, M.; Langhans, W. Etomoxir-Induced Increase in UCP3 Supports a Role of Uncoupling Protein 3 as a Mitochondrial Fatty Acid Anion Exporter. FASEB J. 2002, 16, 1688–1690. [Google Scholar] [CrossRef] [PubMed]
- Dobrzyn, A.; Dobrzyn, P.; Lee, S.H.; Miyazaki, M.; Cohen, P.; Asilmaz, E.; Hardie, D.G.; Friedman, J.M.; Ntambi, J.M. Stearoyl-CoA Desaturase-1 Deficiency Reduces Ceramide Synthesis by Downregulating Serine Palmitoyltransferase and Increasing β-Oxidation in Skeletal Muscle. Am. J. Physiol. Endocrinol. Metab. 2005, 288, 599–607. [Google Scholar] [CrossRef]
- Bu, S.Y. Role of Acyl-CoA:Diacylglycerol Acyltransferase 2 (DGAT2) in Energy Metabolism in Skeletal Muscle Cells. FASEB J. 2015, 29, LB176. [Google Scholar] [CrossRef]
- Bu, S.Y. Role of Dgat2 in Glucose Uptake and Fatty Acid Metabolism in C2C12 Skeletal Myotubes. J. Microbiol. Biotechnol. 2023, 33, 1563. [Google Scholar] [CrossRef]
- Guerranti, R.; Pagani, R.; Neri, S.; Errico, S.V.; Leoncini, R.; Marinello, E. Inhibition and Regulation of Rat Liver L-Threonine Dehydrogenase by Different Fatty Acids and Their Derivatives. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2001, 1568, 45–52. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, P.; Fan, S.; Zhai, B.; Jin, W.; Li, D.; Li, H.; Sun, G.; Han, R.; Liu, X.; et al. Weighted Gene Co-Expression Network Indicates That the DYNLL2 Is an Important Regulator of Chicken Breast Muscle Development and Is Regulated by MiR-148a-3p. BMC Genom. 2022, 23, 258. [Google Scholar] [CrossRef]
- Li, F.; Luo, Z.; Huang, W.; Lu, Q.; Wilcox, C.S.; Jose, P.A.; Chen, S. Response Gene to Complement 32, a Novel Regulator for Transforming Growth Factor-β-Induced Smooth Muscle Differentiation of Neural Crest Cells. J. Biol. Chem. 2007, 282, 10133–10137. [Google Scholar] [CrossRef]
- Cheng, S.; Wan, X.; Yang, L.; Qin, Y.; Chen, S.; Liu, Y.; Sun, Y.; Qiu, Y.; Huang, L.; Qin, Q.; et al. RGCC-Mediated PLK1 Activity Drives Breast Cancer Lung Metastasis by Phosphorylating AMPKα2 to Activate Oxidative Phosphorylation and Fatty Acid Oxidation. J. Exp. Clin. Cancer Res. 2023, 42, 342. [Google Scholar] [CrossRef]
- Tang, R.; Zhang, G.; Chen, S.Y. Response Gene to Complement 32 Protein Promotes Macrophage Phagocytosis via Activation of Protein Kinase C Pathway. J. Biol. Chem. 2014, 289, 22715–22722. [Google Scholar] [CrossRef]
- Shoshani, T.; Faerman, A.; Mett, I.; Zelin, E.; Tenne, T.; Gorodin, S.; Moshel, Y.; Elbaz, S.; Budanov, A.; Chajut, A.; et al. Identification of a Novel Hypoxia-Inducible Factor 1-Responsive Gene, RTP801, Involved in Apoptosis. Mol. Cell Biol. 2002, 22, 2283. [Google Scholar] [CrossRef]
- Brugarolas, J.; Lei, K.; Hurley, R.L.; Manning, B.D.; Reiling, J.H.; Hafen, E.; Witters, L.A.; Ellisen, L.W.; Kaelin, W.G. Regulation of MTOR Function in Response to Hypoxia by REDD1 and the TSC1/TSC2 Tumor Suppressor Complex. Genes Dev. 2004, 18, 2893. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. MTOR Signaling at a Glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Ayansola, H.; Liao, C.; Dong, Y.; Yu, X.; Zhang, B.; Wang, B. Prospect of Early Vascular Tone and Satellite Cell Modulations on White Striping Muscle Myopathy. Poult. Sci. 2021, 100, 100945. [Google Scholar] [CrossRef]
- Obach, M.; Navarro-Sabaté, À.; Caro, J.; Kong, X.; Duran, J.; Gómez, M.; Perales, J.C.; Ventura, F.; Rosa, J.L.; Bartrons, R. 6-Phosphofructo-2-Kinase (Pfkfb3) Gene Promoter Contains Hypoxia-Inducible Factor-1 Binding Sites Necessary for Transactivation in Response to Hypoxia. J. Biol. Chem. 2004, 279, 53562–53570. [Google Scholar] [CrossRef]
- Yalcin, A.; Clem, B.F.; Simmons, A.; Lane, A.; Nelson, K.; Clem, A.L.; Brock, E.; Siow, D.; Wattenberg, B.; Telang, S.; et al. Nuclear Targeting of 6-Phosphofructo-2-Kinase (PFKFB3) Increases Proliferation via Cyclin-Dependent Kinases. J. Biol. Chem. 2009, 284, 24223. [Google Scholar] [CrossRef]
- Yalcin, A.; Clem, B.F.; Imbert-Fernandez, Y.; Ozcan, S.C.; Peker, S.; O’Neal, J.; Klarer, A.C.; Clem, A.L.; Telang, T.; Chesney, J. 6-Phosphofructo-2-Kinase (PFKFB3) Promotes Cell Cycle Progression and Suppresses Apoptosis via Cdk1-Mediated Phosphorylation of P27. Cell Death Dis. 2014, 5, e1337. [Google Scholar] [CrossRef]
- Gassen, N.C.; Hartmann, J.; Schmidt, M.V.; Rein, T. FKBP5/FKBP51 Enhances Autophagy to Synergize with Antidepressant Action. Autophagy 2015, 11, 578–580. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.; Wang, Y.; Zhuang, J.; Peng, Y.; Mo, X.; Chen, J.; Shi, Y.; Yu, M.; Cai, W.; et al. FKBP51 Induces P53-Dependent Apoptosis and Enhances Drug Sensitivity of Human Non-Small-Cell Lung Cancer Cells. Exp. Ther. Med. 2020, 19, 2236–2242. [Google Scholar] [CrossRef]
- Gao, S.; Huang, S.; Zhang, Y.; Fang, G.; Liu, Y.; Zhang, C.; Li, Y.; Du, J. The Transcriptional Regulator KLF15 Is Necessary for Myoblast Differentiation and Muscle Regeneration by Activating FKBP5. J. Biol. Chem. 2023, 299, 105226. [Google Scholar] [CrossRef]
- Lee, J.H.; Budanov, A.V.; Karin, M. Sestrins Orchestrate Cellular Metabolism to Attenuate Aging. Cell Metab. 2013, 18, 792–801. [Google Scholar] [CrossRef]
- Peeters, H.; Debeer, P.; Bairoch, A.; Wilquet, V.; Huysmans, C.; Parthoens, E.; Fryns, J.P.; Gewillig, M.; Nakamura, Y.; Niikawa, N.; et al. PA26 Is a Candidate Gene for Heterotaxia in Humans: Identification of a Novel PA26-Related Gene Family in Human and Mouse. Hum. Genet. 2003, 112, 573–580. [Google Scholar] [CrossRef]
- Cai, B.; Ma, M.; Chen, B.; Li, Z.; Abdalla, B.A.; Nie, Q.; Zhang, X. MiR-16-5p Targets SESN1 to Regulate the P53 Signaling Pathway, Affecting Myoblast Proliferation and Apoptosis, and Is Involved in Myoblast Differentiation. Cell Death Dis. 2018, 9, 367. [Google Scholar] [CrossRef]
- Thomas, T.M.; Miyaguchi, K.; Edwards, L.A.; Wang, H.; Wollebo, H.; Aiguo, L.; Murali, R.; Wang, Y.; Braas, D.; Michael, J.S.; et al. Elevated Asparagine Biosynthesis Drives Brain Tumor Stem Cell Metabolic Plasticity and Resistance to Oxidative Stress. Mol. Cancer Res. 2021, 19, 1375–1388. [Google Scholar] [CrossRef]
- Aung, H.H.; Lame, M.W.; Gohil, K.; An, C.I.; Wilson, D.W.; Rutledge, J.C. Induction of ATF3 Gene Network by Triglyceride-Rich Lipoprotein Lipolysis Products Increases Vascular Apoptosis and Inflammation. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2088–2096. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, Y.; Jin, X.; Gong, R.; Xia, Z. ATF3 Regulates Oxidative Stress and Extracellular Matrix Degradation via P38/Nrf2 Signaling Pathway in Pelvic Organ Prolapse. Tissue Cell 2021, 73, 101660. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, K.J.; Hwang, J.Y.; Kim, G.H.; Lee, D.Y.; Lee, Y.J.; Song, E.H.; Yoo, M.G.; Kim, B.J.; Suh, Y.H.; et al. Activating Transcription Factor 3 Is a Target Molecule Linking Hepatic Steatosis to Impaired Glucose Homeostasis. J. Hepatol. 2017, 67, 349–359. [Google Scholar] [CrossRef]
- Bordini, M.; Zappaterra, M.; Soglia, F.; Petracci, M.; Davoli, R. Weighted Gene Co-Expression Network Analysis Identifies Molecular Pathways and Hub Genes Involved in Broiler White Striping and Wooden Breast Myopathies. Sci. Rep. 2021, 11, 1776. [Google Scholar] [CrossRef]
- Lee, B.; Kim, J.Y.; Ryu, Y.C.; Lee, K.; Choi, Y.M. Research Note: Expression Levels of Collagen-Related Genes in PSE Conditions and White Striping Features of Broiler Pectoralis Major Muscle. Poult. Sci. 2023, 102, 102471. [Google Scholar] [CrossRef]
- Malila, Y.; U-Chupaj, J.; Srimarut, Y.; Chaiwiwattrakul, P.; Uengwetwanit, T.; Arayamethakorn, S.; Punyapornwithaya, V.; Sansamur, C.; Kirschke, C.P.; Huang, L.; et al. Monitoring of White Striping and Wooden Breast Cases and Impacts on Quality of Breast Meat Collected from Commercial Broilers (Gallus gallus). Asian-Australas J. Anim. Sci. 2018, 31, 1807. [Google Scholar] [CrossRef]
- Petracci, M.; Mudalal, S.; Babini, E.; Cavani, C. Effect of White Striping on Chemical Composition and Nutritional Value of Chicken Breast Meat. Ital. J. Anim. Sci. 2014, 13, 179–183. [Google Scholar] [CrossRef]
- Wang, D.; Mohammad, M.; Wang, Y.; Tan, R.; Murray, L.S.; Ricardo, S.; Dagher, H.; van Agtmael, T.; Savige, J. The Chemical Chaperone, PBA, Reduces ER Stress and Autophagy and Increases Collagen IV A5 Expression in Cultured Fibroblasts from Men With X-Linked Alport Syndrome and Missense Mutations. Kidney Int. Rep. 2017, 2, 739–748. [Google Scholar] [CrossRef]
- Valentine, B.A. Skeletal Muscle. In Pathologic Basis of Veterinary Disease, 6th ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 908–953. [Google Scholar] [CrossRef]



| Upregulated | |||||
| Chromosome | Associated Gene Name | Gene Description | Ensembl Gene Id | logFC | FDR |
| 2 | PDK4 | pyruvate dehydrogenase kinase 4 | ENSGALG00010002741 | 3.41421 | 1.571 × 10−24 |
| 1 | MYBPC1 | myosin binding protein C1 | ENSGALG00010011558 | 2.75445 | 8.349 × 10−17 |
| 26 | FKBP5 | FK506 binding protein 5 | ENSGALG00010026893 | 2.40214 | 1.345 × 10−12 |
| 2 | CEBPD | CCAAT enhancer binding protein delta | ENSGALG00010005405 | 2.01735 | 7.776 × 10−9 |
| 2 | CA3A | carbonic anhydrase 3A | ENSGALG00010013288 | 2.00895 | 7.776 × 10−9 |
| 26 | TNNT2 | troponin T2, cardiac type | ENSGALG00010026743 | 1.57929 | 3.182 × 10−5 |
| 2 | ASNS | asparagine synthetase (glutamine-hydrolyzing) | ENSGALG00010003382 | 1.52297 | 9.686 × 10−5 |
| 1 | PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | ENSGALG00010006245 | 1.45674 | 0.0002375 |
| 1 | NELL2 | neural EGFL like 2 | ENSGALG00010013749 | 1.43497 | 0.0006832 |
| 2 | FBXO32 | F-box protein 32 | ENSGALG00010008245 | 1.37948 | 0.0006832 |
| 3 | SESN1 | sestrin 1 | ENSGALG00010014244 | 1.34848 | 0.0009661 |
| 3 | TDH | L-treonina desidrogenase | ENSGALG00010011278 | 1.23466 | 0.0064443 |
| 4 | MYOZ2 | myozenin 2 | ENSGALG00010005510 | 1.21752 | 0.0089447 |
| 4 | COL4A6 | collagen type IV alpha 6 chain | ENSGALG00010015733 | 1.20722 | 0.0107838 |
| 4 | FHL1 | four and a half LIM domains 1 | ENSGALG00010014929 | 1.20013 | 0.0088713 |
| 9 | CHRNG | cholinergic receptor nicotinic gamma subunit | ENSGALG00010017878 | 1.18978 | 0.0143605 |
| 5 | ASB2 | ankyrin repeat and SOCS box containing 2 | ENSGALG00010016987 | 1.17026 | 0.0122158 |
| 4 | COL4A5 | collagen type IV alpha 5 chain | ENSGALG00010015632 | 1.14888 | 0.0228397 |
| 5 | MYBPC3 | myosin binding protein C3 | ENSGALG00010022468 | 1.14728 | 0.0190541 |
| 28 | ARRDC2 | arrestin domain containing 2 | ENSGALG00010027232 | 1.14249 | 0.0216413 |
| 1 | UCP3 | uncoupling protein 3 | ENSGALG00010002543 | 1.08505 | 0.0337988 |
| 6 | DDIT4 | DNA damage inducible transcript 4 | ENSGALG00010021509 | 1.0787 | 0.0370386 |
| 1 | RGCC | regulator of cell cycle | ENSGALG00010003192 | 1.07199 | 0.0372903 |
| Downregulated | |||||
| Chromosome | Associated gene name | Gene description | Ensembl gene id | logFC | FDR |
| 1 | METTL21EP | methyltransferase like 21E, pseudogene | ENSGALG00010006037 | −2.2487 | 1.40 × 10−10 |
| 3 | UBE3D | ubiquitin protein ligase E3D | ENSGALG00010003115 | −2.0996 | 4.26 × 10−9 |
| 6 | SCD | stearoyl-CoA desaturase | ENSGALG00010021083 | −1.9290 | 3.38 × 10−8 |
| 1 | DGAT2 | diacylglycerol O-acyltransferase 2 | ENSGALG00010003035 | −1.5278 | 9.69 × 10−5 |
| 5 | DUSP8 | dual specificity phosphatase 8 | ENSGALG00010024637 | −1.3427 | 0.0017 |
| Z | LIPG | lipase G, endothelial type | ENSGALG00010009013 | -1.0878 | 0.0466 |
| 3 | ATF3 | activating transcription factor 3 | ENSGALG00010019307 | −1.0621 | 0.0431 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilha, S.F.; Ibelli, A.M.G.; Peixoto, J.O.; Cantão, M.E.; Moreira, G.C.M.; Fernandes, L.T.; Tavernari, F.C.; Morés, M.A.Z.; Bastos, A.P.A.; Dias, L.T.; et al. Novel Candidate Genes Involved in an Initial Stage of White Striping Development in Broiler Chickens. Animals 2024, 14, 2379. https://doi.org/10.3390/ani14162379
Padilha SF, Ibelli AMG, Peixoto JO, Cantão ME, Moreira GCM, Fernandes LT, Tavernari FC, Morés MAZ, Bastos APA, Dias LT, et al. Novel Candidate Genes Involved in an Initial Stage of White Striping Development in Broiler Chickens. Animals. 2024; 14(16):2379. https://doi.org/10.3390/ani14162379
Chicago/Turabian StylePadilha, Suelen Fernandes, Adriana Mércia Guaratini Ibelli, Jane Oliveira Peixoto, Maurício Egídio Cantão, Gabriel Costa Monteiro Moreira, Lana Teixeira Fernandes, Fernando Castro Tavernari, Marcos Antônio Zanella Morés, Ana Paula Almeida Bastos, Laila Talarico Dias, and et al. 2024. "Novel Candidate Genes Involved in an Initial Stage of White Striping Development in Broiler Chickens" Animals 14, no. 16: 2379. https://doi.org/10.3390/ani14162379









