Genome-Wide Analysis of Genetic Diversity and Selection Signatures in Zaobei Beef Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. Whole-Genome Sequencing Data Alignment and Variant Calling
2.3. Population Structure and Genetic Analysis
2.4. Detection of Selection Signatures
2.5. Functional Annotation of Selection Signatures and Enrichment Analysis
3. Results
3.1. Whole-Genome Resequence and SNP Identification
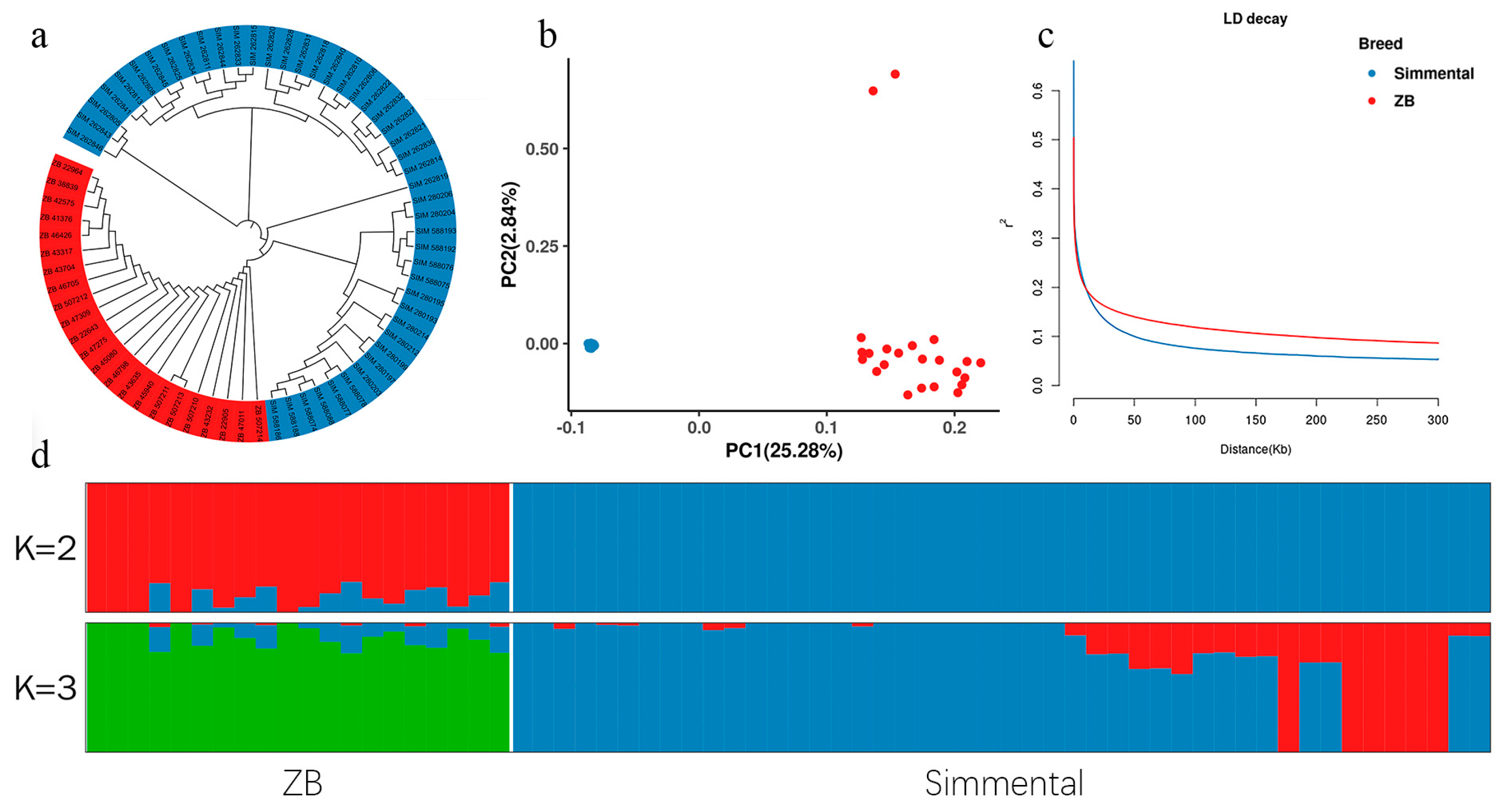
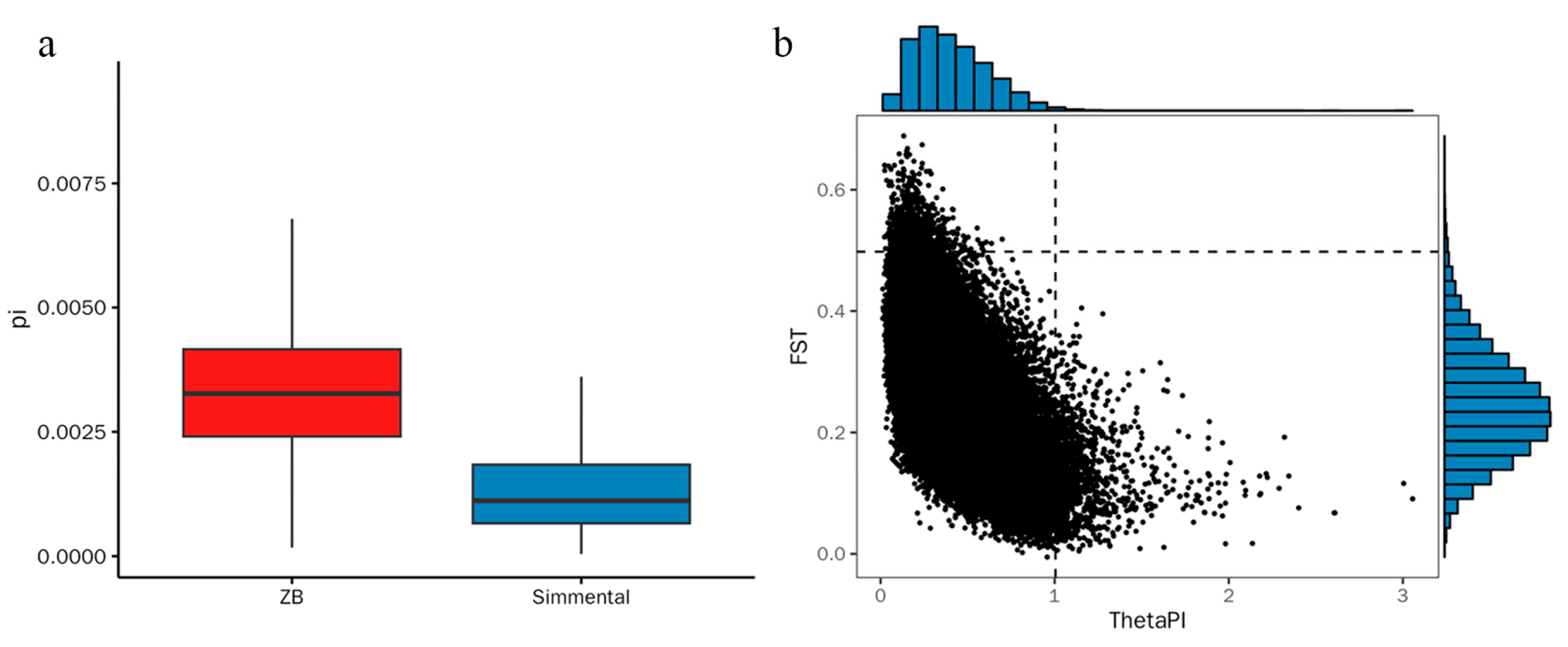
3.2. Population Structure and Linkage Disequilibrium Analysis
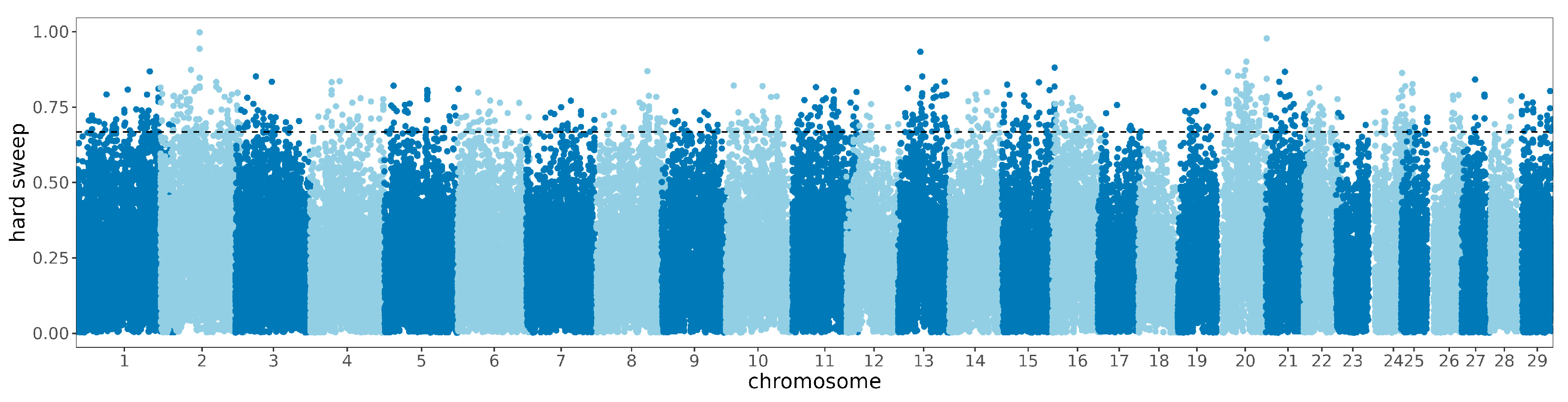
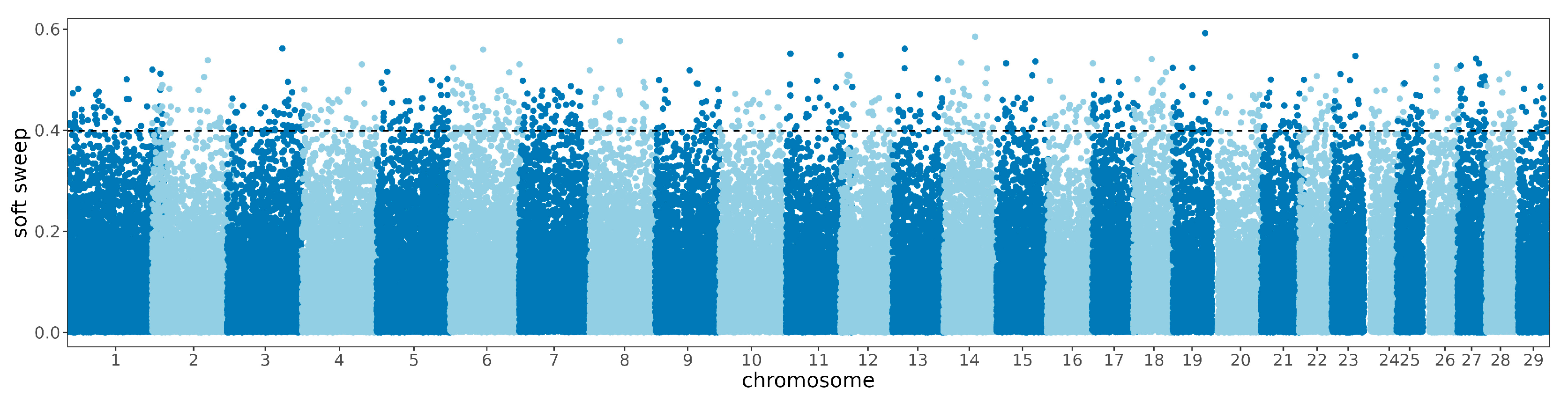
3.3. Signature of Detection in the Zaobei Cattle and Gene Annotationn
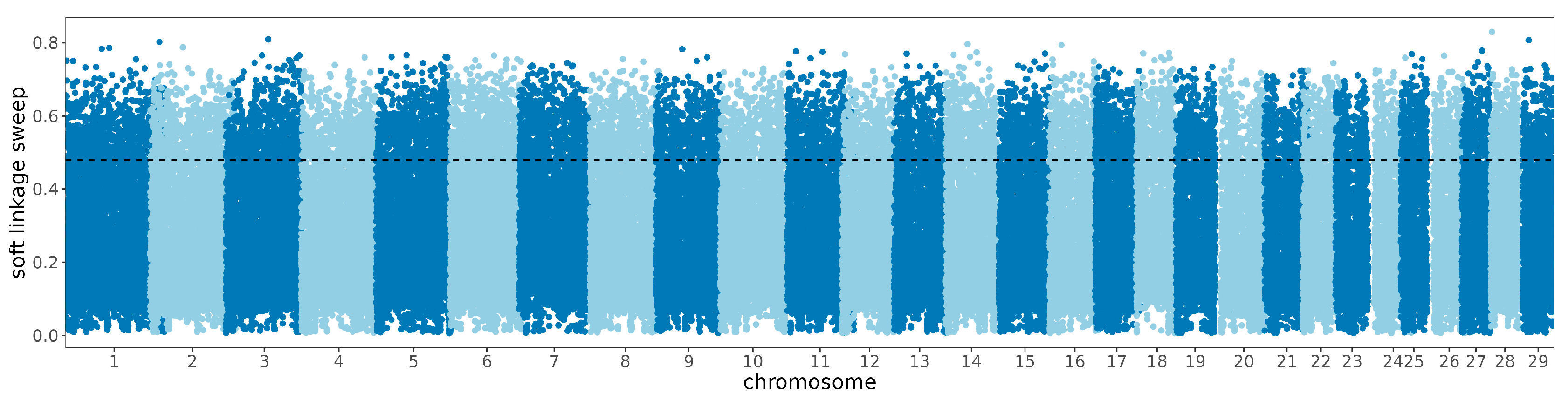
3.4. Selective Sweep and Enrichment Analysis between Breeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Averdunk, G. DAIRY ANIMALS|Minor and Dual-Purpose Bos taurus Breeds. In Encyclopedia of Dairy Sciences; Roginski, H., Ed.; Elsevier: Oxford, UK, 2002; pp. 568–576. [Google Scholar] [CrossRef]
- Zhao, F.; Xie, R.; Fang, L.; Xiang, R.; Yuan, Z.; Liu, Y.; Wang, L. Analysis of 206 whole-genome resequencing reveals selection signatures associated with breed-specific traits in Hu sheep. Evol. Appl. 2024, 17, e13697. [Google Scholar] [CrossRef] [PubMed]
- Sarviaho, K.; Uimari, P.; Martikainen, K. Signatures of positive selection after the introduction of genomic selection in the Finnish Ayrshire population. J. Dairy Sci. 2024, 107, 4822–4832. [Google Scholar] [CrossRef]
- Tong, X.; Chen, D.; Hu, J.; Lin, S.; Ling, Z.; Ai, H.; Zhang, Z.; Huang, L. Accurate haplotype construction and detection of selection signatures enabled by high quality pig genome sequences. Nat. Commun. 2023, 14, 5126. [Google Scholar] [CrossRef]
- Qanbari, S.; Simianer, H. Mapping signatures of positive selection in the genome of livestock. Livest. Sci. 2014, 166, 133–143. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Luo, H.; Zhang, P.; Zhang, W.; Zheng, Y.; Hao, D.; Shi, Y.; Niu, Y.; Song, T.; Li, Y.; Zhao, S.; et al. Recent positive selection signatures reveal phenotypic evolution in the Han Chinese population. Sci. Bull. 2023, 68, 2391–2404. [Google Scholar] [CrossRef]
- Zeng, J.; Xue, A.; Jiang, L.; Lloyd-Jones, L.R.; Wu, Y.; Wang, H.; Zheng, Z.; Yengo, L.; Kemper, K.E.; Goddard, M.E.; et al. Widespread signatures of natural selection across human complex traits and functional genomic categories. Nat. Commun. 2021, 12, 1164. [Google Scholar] [CrossRef]
- Lukic, B.; Curik, I.; Drzaic, I.; Galic, V.; Shihabi, M.; Vostry, L.; Cubric-Curik, V. Genomic signatures of selection, local adaptation and production type characterisation of East Adriatic sheep breeds. J. Anim. Sci. Biotechnol. 2023, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, M.; Cheng, H.; Fan, W.; Yuan, Z.; Gao, Q.; Xu, Y.; Guo, Z.; Zhang, Y.; Hu, J.; et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018, 9, 2648. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Pan, D.; Ren, H.; Fu, J.; Li, J.; Su, G.; Wang, A.; Jiang, L.; Zhang, Q.; Liu, J.F. Identification of selective sweeps reveals divergent selection between Chinese Holstein and Simmental cattle populations. Genet. Sel. Evol. 2016, 48, 76. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Liu, R.; Zhao, D.; He, Z.; Li, W.; Zheng, M.; Li, Q.; Wang, Q.; Liu, D.; Feng, F.; et al. Large-scale genomic and transcriptomic analyses elucidate the genetic basis of high meat yield in chickens. J. Adv. Res. 2024, 55, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Berg, I.V.D.; MacLeod, I.M.; Hayes, B.J.; Prowse-Wilkins, C.P.; Wang, M.; Bolormaa, S.; Liu, Z.; Rochfort, S.J.; Reich, C.M.; et al. Quantifying the contribution of sequence variants with regulatory and evolutionary significance to 34 bovine complex traits. Proc. Natl. Acad. Sci. USA 2019, 116, 19398–19408. [Google Scholar] [CrossRef]
- Na, W.; Yu, J.Q.; Xu, Z.C.; Zhang, X.Y.; Yang, L.L.; Cao, Z.P.; Li, H.; Zhang, H. Important candidate genes for abdominal fat content identified by linkage disequilibrium and fixation index information. Poult. Sci. 2019, 98, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006, 4, e72. [Google Scholar] [CrossRef]
- Pavlidis, P.; Alachiotis, N. A survey of methods and tools to detect recent and strong positive selection. J. Biol. Res. 2017, 24, 7. [Google Scholar] [CrossRef]
- Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005, 39, 197–218. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Reich, D.E.; Higgins, J.M.; Levine, H.Z.; Richter, D.J.; Schaffner, S.F.; Gabriel, S.B.; Platko, J.V.; Patterson, N.J.; McDonald, G.J.; et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, K.; Cheng, G.; Mei, C.; Wang, H.; Zan, L. Genome-wide analysis reveals genomic diversity and signatures of selection in Qinchuan beef cattle. BMC Genom. 2024, 25, 558. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, F.; Li, S.; Luo, X.; Peng, L.; Dong, Z.; Pausch, H.; Leonard, A.S.; Crysnanto, D.; Wang, S.; et al. Structural variation and introgression from wild populations in East Asian cattle genomes confer adaptation to local environment. Genome Biol. 2023, 24, 211. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Picard Toolkit. Broad Institute, GitHub Repository 2019. Available online: https://broadinstitute.github.io/picard/ (accessed on 19 July 2024).
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Browning, B.L.; Tian, X.; Zhou, Y.; Browning, S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021, 108, 1880–1890. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Schrider, D.R.; Kern, A.D. S/HIC: Robust Identification of Soft and Hard Sweeps Using Machine Learning. PLoS Genet. 2016, 12, e1005928. [Google Scholar] [CrossRef]
- Song, H.; Chu, J.; Li, W.; Li, X.; Fang, L.; Han, J.; Zhao, S.; Ma, Y. A Novel Approach Utilizing Domain Adversarial Neural Networks for the Detection and Classification of Selective Sweeps. Adv. Sci. 2024, 11, e2304842. [Google Scholar] [CrossRef]
- Schrider, D.R.; Kern, A.D. Soft Sweeps Are the Dominant Mode of Adaptation in the Human Genome. Mol. Biol. Evol. 2017, 34, 1863–1877. [Google Scholar] [CrossRef]
- Mughal, M.R.; DeGiorgio, M. Localizing and Classifying Adaptive Targets with Trend Filtered Regression. Mol. Biol. Evol. 2019, 36, 252–270. [Google Scholar] [CrossRef] [PubMed]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Szpiech, Z.A.; Hernandez, R.D. selscan: An efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef]
- Szpiech, Z.A. selscan 2.0: Scanning for sweeps in unphased data. Bioinformatics 2024, 40, btae006. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.-P.; Wei, C.-H.; Zhang, L.; Liu, J.-S.; Wang, G.-K.; Zeng, T.; Du, L.-X. A genome scan of recent positive selection signatures in three sheep populations. J. Integr. Agric. 2016, 15, 162–174. [Google Scholar] [CrossRef]
- Kinsella, R.J.; Kahari, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef] [PubMed]
- Elizarraras, J.M.; Liao, Y.; Shi, Z.; Zhu, Q.; Pico, A.R.; Zhang, B. WebGestalt 2024: Faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 2024, 52, W415–W421. [Google Scholar] [CrossRef]
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Huson, H.J.; Kim, E.S.; Godfrey, R.W.; Olson, T.A.; McClure, M.C.; Chase, C.C.; Rizzi, R.; O‘Brien, A.M.; Van Tassell, C.P.; Garcia, J.F.; et al. Genome-wide association study and ancestral origins of the slick-hair coat in tropically adapted cattle. Front. Genet. 2014, 5, 101. [Google Scholar] [CrossRef]
- Sweett, H.; Fonseca, P.A.S.; Suarez-Vega, A.; Livernois, A.; Miglior, F.; Canovas, A. Genome-wide association study to identify genomic regions and positional candidate genes associated with male fertility in beef cattle. Sci. Rep. 2020, 10, 20102. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yang, B.; Ju, Z.H.; Wang, X.G.; Qi, C.; Zhang, Y.; Wang, C.F.; Liu, H.D.; Feng, M.Y.; Chen, Y.; et al. Alternative splicing, promoter methylation, and functional SNPs of sperm flagella 2 gene in testis and mature spermatozoa of Holstein bulls. Reproduction 2014, 147, 241–252. [Google Scholar] [CrossRef]
- Araujo, A.C.; Carneiro, P.L.S.; Alvarenga, A.B.; Oliveira, H.R.; Miller, S.P.; Retallick, K.; Brito, L.F. Haplotype-Based Single-Step GWAS for Yearling Temperament in American Angus Cattle. Genes 2021, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.; Zhang, S.; Wang, S.; Wu, X.; Zhang, Q.; Liu, L.; Li, Y.; Qiao, L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS ONE 2014, 9, e96186. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, C.; Pan, C.; Feng, X.; Lei, Z.; Huang, J.; Wei, X.; Li, F.; Ma, Y. Identification of key genes and functional enrichment pathways involved in fat deposition in Xinyang buffalo by WGCNA. Gene 2022, 818, 146225. [Google Scholar] [CrossRef] [PubMed]
- Stepanjuk, A.; Koel, M.; Pook, M.; Saare, M.; Jaager, K.; Peters, M.; Krjutskov, K.; Ingerpuu, S.; Salumets, A. MUC20 expression marks the receptive phase of the human endometrium. Reprod. Biomed. Online 2019, 39, 725–736. [Google Scholar] [CrossRef]
- Martins-de-Souza, D.; Gattaz, W.F.; Schmitt, A.; Novello, J.C.; Marangoni, S.; Turck, C.W.; Dias-Neto, E. Proteome analysis of schizophrenia patients Wernicke’s area reveals an energy metabolism dysregulation. BMC Psychiatry 2009, 9, 17. [Google Scholar] [CrossRef]
- Hammond, E.R.; Stewart, B.; Peek, J.C.; Shelling, A.N.; Cree, L.M. Assessing embryo quality by combining non-invasive markers: Early time-lapse parameters reflect gene expression in associated cumulus cells. Hum. Reprod. 2015, 30, 1850–1860. [Google Scholar] [CrossRef][Green Version]
- Melo, T.P.; Fortes, M.R.S.; Bresolin, T.; Mota, L.F.M.; Albuquerque, L.G.; Carvalheiro, R. Multitrait meta-analysis identified genomic regions associated with sexual precocity in tropical beef cattle. J. Anim. Sci. 2018, 96, 4087–4099. [Google Scholar] [CrossRef]
- Hiver, S.; Shimizu-Mizuno, N.; Ikawa, Y.; Kajikawa, E.; Sai, X.; Nishimura, H.; Takaoka, K.; Nishimura, O.; Kuraku, S.; Tanaka, S.; et al. Gse1, a component of the CoREST complex, is required for placenta development in the mouse. Dev. Biol. 2023, 498, 97–105. [Google Scholar] [CrossRef]
- Abo-Ismail, M.; Miller, S.; Sargolzaei, M.; Grossi, D.; Nayeri, S.; Moore, S.; Plastow, G.; Stothard, P.; Schenkel, F. Genome wide association analyses identify new loci for milking speed and temperament in North American Holsteins. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Volume Genetics of Trait Complexes: Lactation, Vancouver, BC, Canada, 17–22 August 2014; pp. 17–22. [Google Scholar]
- Cheng, R.; Zheng, X.; Wang, Y.; Ma, X.; Liu, X.; Xu, W.; Wang, M.; Gao, Y.; Xing, X.; Zhou, C.; et al. Modification of alternative splicing in bovine somatic cell nuclear transfer embryos using engineered CRISPR-Cas13d. Sci. China Life Sci. 2022, 65, 2257–2268. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Cheng, H.; Liu, Y.; Sun, L.; Chen, N.; Jiang, F.; You, W.; Yang, Z.; Zhang, B.; Song, E.; et al. Assessing Genomic Diversity and Selective Pressures in Bohai Black Cattle Using Whole-Genome Sequencing Data. Animals 2022, 12, 665. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Pickrell, J.K.; Coop, G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010, 20, R208–R215. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Ben Jemaa, S.; Ciani, E.; Sottile, G.; Moscarelli, A.; Boussaha, M.; Montedoro, M.; Pilla, F.; Cassandro, M. Genome-wide detection of signatures of selection in three Valdostana cattle populations. J. Anim. Breed. Genet. 2020, 137, 609–621. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Huang, M.; Tang, J.; Yang, L.; Yu, Z.; Li, D.; Li, G.; Jiang, Y.; Sun, Y.; et al. Whole-genome SNP markers reveal conservation status, signatures of selection, and introgression in Chinese Laiwu pigs. Evol. Appl. 2021, 14, 383–398. [Google Scholar] [CrossRef]
- Laodim, T.; Koonawootrittriron, S.; Elzo, M.A.; Suwanasopee, T.; Jattawa, D.; Sarakul, M. Genetic factors influencing milk and fat yields in tropically adapted dairy cattle: Insights from quantitative trait loci analysis and gene associations. Anim. Biosci. 2024, 37, 576–590. [Google Scholar] [CrossRef]
- Costa, C.; Germena, G.; Martin-Conte, E.L.; Molineris, I.; Bosco, E.; Marengo, S.; Azzolino, O.; Altruda, F.; Ranieri, V.M.; Hirsch, E. The RacGAP ArhGAP15 is a master negative regulator of neutrophil functions. Blood 2011, 118, 1099–1108. [Google Scholar] [CrossRef]
- Wang, S.; Raza, S.H.A.; Zhang, K.; Mei, C.; Alamoudi, M.O.; Aloufi, B.H.; Alshammari, A.M.; Zan, L. Selection signatures of Qinchuan cattle based on whole-genome sequences. Anim. Biotechnol. 2023, 34, 1483–1491. [Google Scholar] [CrossRef]
- Elati, K.; Tajeri, S.; Obara, I.; Mhadhbi, M.; Zweygarth, E.; Darghouth, M.A.; Nijhof, A.M. Dual RNA-seq to catalogue host and parasite gene expression changes associated with virulence of T. annulata-transformed bovine leukocytes: Towards identification of attenuation biomarkers. Sci. Rep. 2023, 13, 18202. [Google Scholar] [CrossRef]
- Passaro, A.; Miselli, M.A.; Sanz, J.M.; Dalla Nora, E.; Morieri, M.L.; Colonna, R.; Pisot, R.; Zuliani, G. Gene expression regional differences in human subcutaneous adipose tissue. BMC Genom. 2017, 18, 202. [Google Scholar] [CrossRef]
- Wang, B.B.; Hou, L.M.; Zhou, W.D.; Liu, H.; Tao, W.; Wu, W.J.; Niu, P.P.; Zhang, Z.P.; Zhou, J.; Li, Q.; et al. Genome-wide association study reveals a quantitative trait locus and two candidate genes on Sus scrofa chromosome 5 affecting intramuscular fat content in Suhuai pigs. Animal 2021, 15, 100341. [Google Scholar] [CrossRef]
- Zhou, P.; Yin, C.; Wang, Y.; Yin, Z.; Liu, Y. Genomic Association Analysis of Growth and Backfat Traits in Large White Pigs. Genes 2023, 14, 1258. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Stratton, C.J.; Bao, J.; Zheng, H.; Bhetwal, B.P.; Yanagimachi, R.; Yan, W. Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc. Natl. Acad. Sci. USA 2015, 112, E430–E439. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yao, X.; Xie, H.; Deng, M.; Gao, X.; Deng, K.; Bao, Y.; Wang, Q.; Wang, F. Effects of SPATA6 on proliferation, apoptosis and steroidogenesis of Hu sheep Leydig cells in vitro. Theriogenology 2021, 166, 9–20. [Google Scholar] [CrossRef]
- Chen, I.J.; Yang, C.P.; Lin, S.H.; Lai, C.M.; Wong, C.S. The Circadian Hormone Melatonin Inhibits Morphine-Induced Tolerance and Inflammation via the Activation of Antioxidative Enzymes. Antioxidants 2020, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.X.; Korkmaz, A.; Rosales-Corral, S.A. Melatonin and stable circadian rhythms optimize maternal, placental and fetal physiology. Hum. Reprod. Update 2014, 20, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Buitenhuis, B.; Poulsen, N.A.; Gebreyesus, G.; Larsen, L.B. Estimation of genetic parameters and detection of chromosomal regions affecting the major milk proteins and their post translational modifications in Danish Holstein and Danish Jersey cattle. BMC Genet. 2016, 17, 114. [Google Scholar] [CrossRef]
- Mesbah-Uddin, M.; Guldbrandtsen, B.; Iso-Touru, T.; Vilkki, J.; De Koning, D.J.; Boichard, D.; Lund, M.S.; Sahana, G. Genome-wide mapping of large deletions and their population-genetic properties in dairy cattle. DNA Res. 2018, 25, 49–59. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, L.; Zhang, P.; Liu, Q.; Liu, C.; Cheng, L.; Yu, B.; Chen, H. Genome-Wide Analysis of Genetic Diversity and Selection Signatures in Zaobei Beef Cattle. Animals 2024, 14, 2447. https://doi.org/10.3390/ani14162447
Shi L, Zhang P, Liu Q, Liu C, Cheng L, Yu B, Chen H. Genome-Wide Analysis of Genetic Diversity and Selection Signatures in Zaobei Beef Cattle. Animals. 2024; 14(16):2447. https://doi.org/10.3390/ani14162447
Chicago/Turabian StyleShi, Liangyu, Pu Zhang, Qing Liu, Chenhui Liu, Lei Cheng, Bo Yu, and Hongbo Chen. 2024. "Genome-Wide Analysis of Genetic Diversity and Selection Signatures in Zaobei Beef Cattle" Animals 14, no. 16: 2447. https://doi.org/10.3390/ani14162447
APA StyleShi, L., Zhang, P., Liu, Q., Liu, C., Cheng, L., Yu, B., & Chen, H. (2024). Genome-Wide Analysis of Genetic Diversity and Selection Signatures in Zaobei Beef Cattle. Animals, 14(16), 2447. https://doi.org/10.3390/ani14162447





