Evaluation of Reference Gene Stability in Goat Skeletal Muscle Satellite Cells during Proliferation and Differentiation Phases
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Isolation and Culture of Goat MuSCs
2.3. Selection of Candidate Reference Genes
2.4. RNA Extraction and cDNA Synthesis
2.5. RT-qPCR Analysis
2.6. Stability Analysis of Reference Genes
2.7. Validation of Reference Gene Expression
2.8. Statistical Analysis
3. Results
3.1. Proliferation and Differentiation of Goat MuSCs In Vitro
3.2. RNA Purity, Primer Verification, and Amplification Efficiency
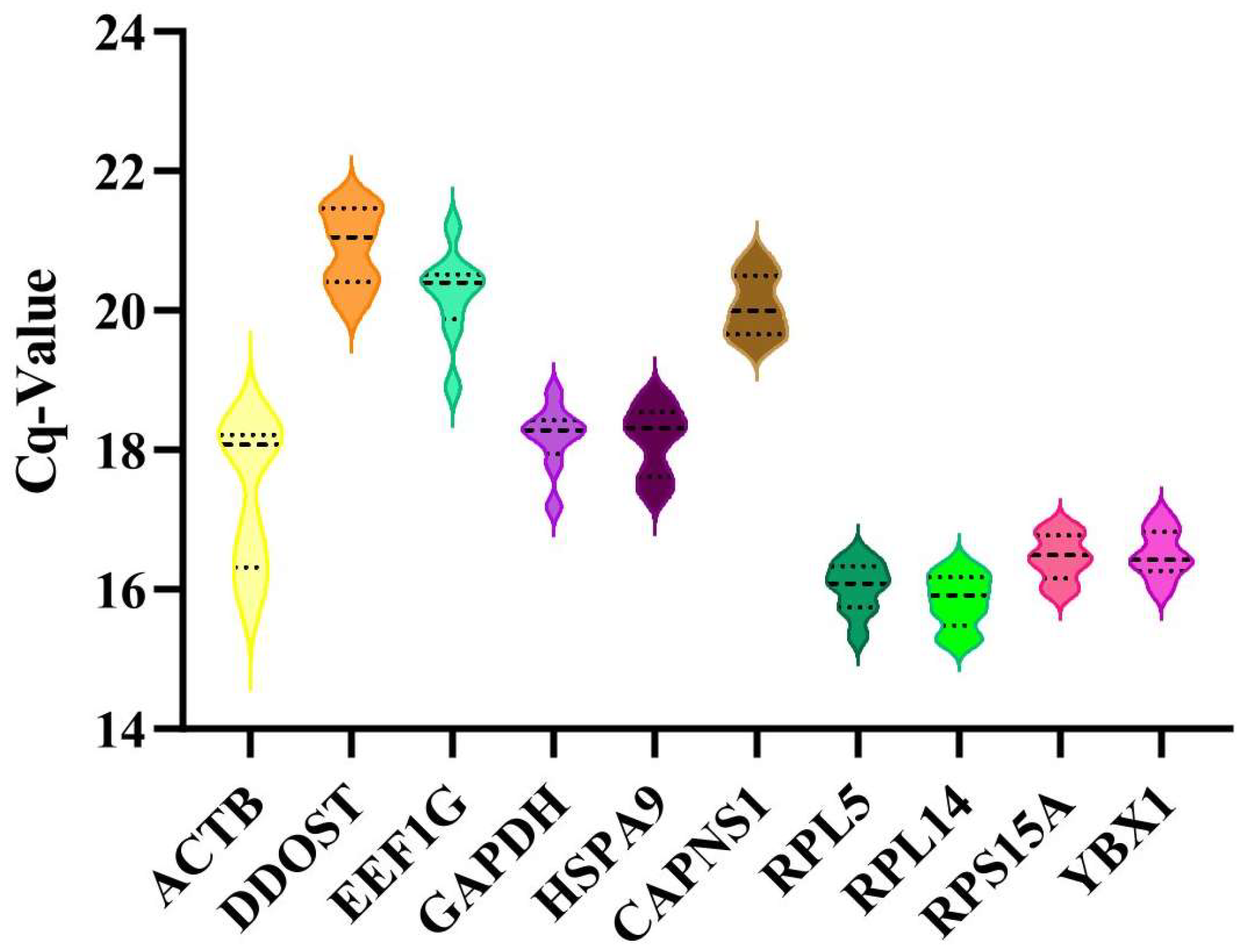
3.3. Analysis of the Expression Levels of the Candidate Reference Genes
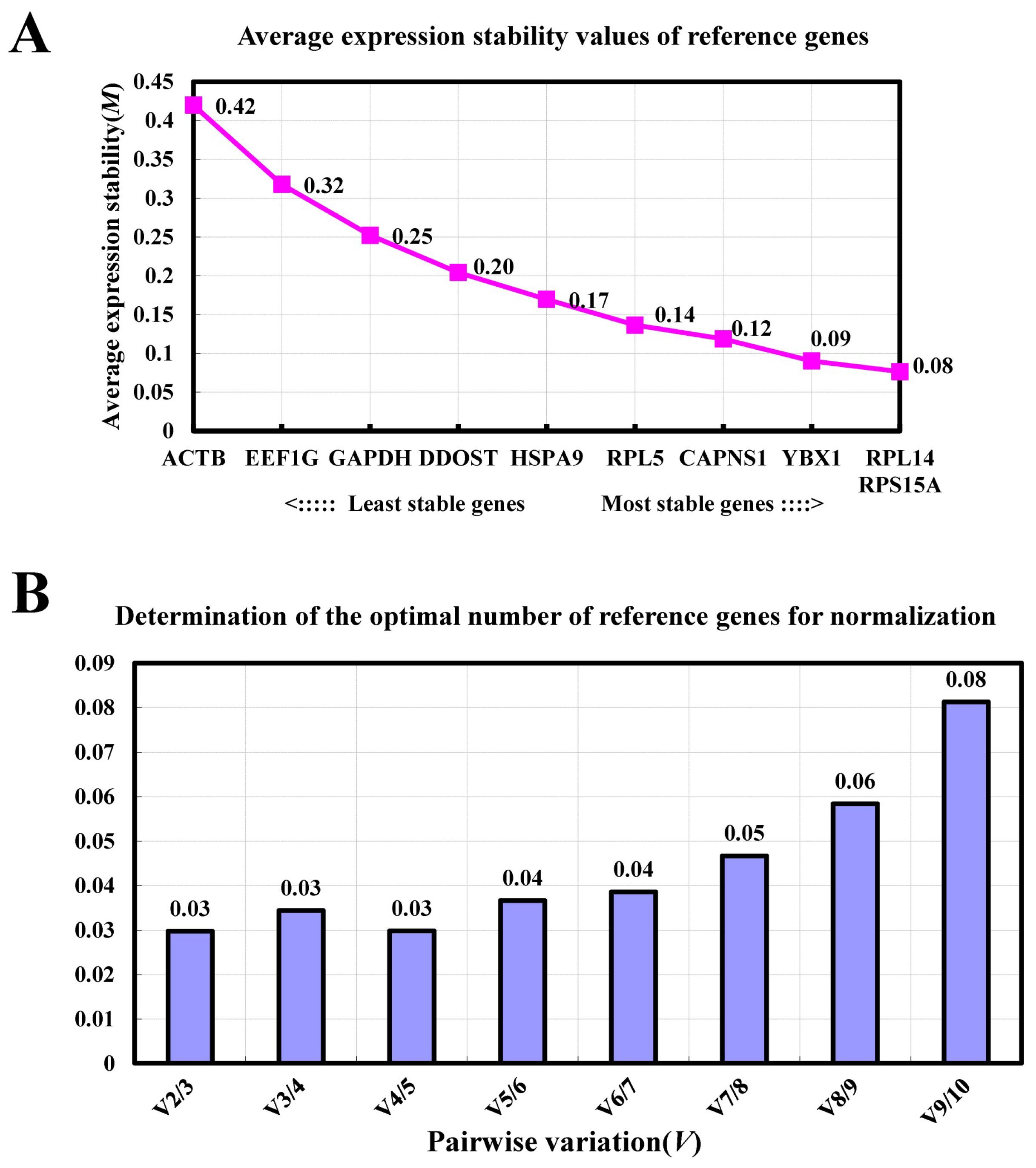
3.4. GeNorm Analysis
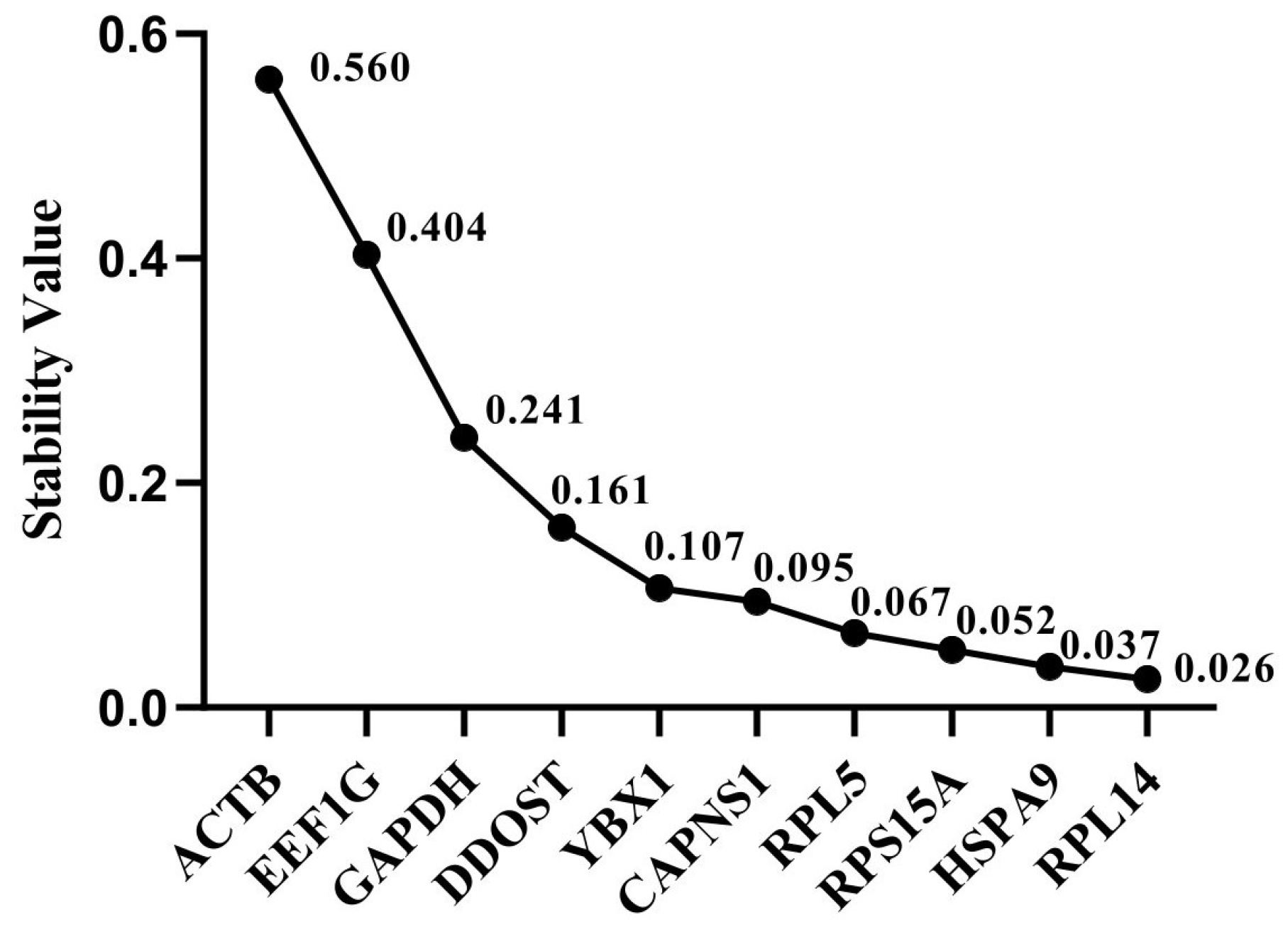
3.5. NormFinder Analysis
3.6. Bestkeeper Analysis
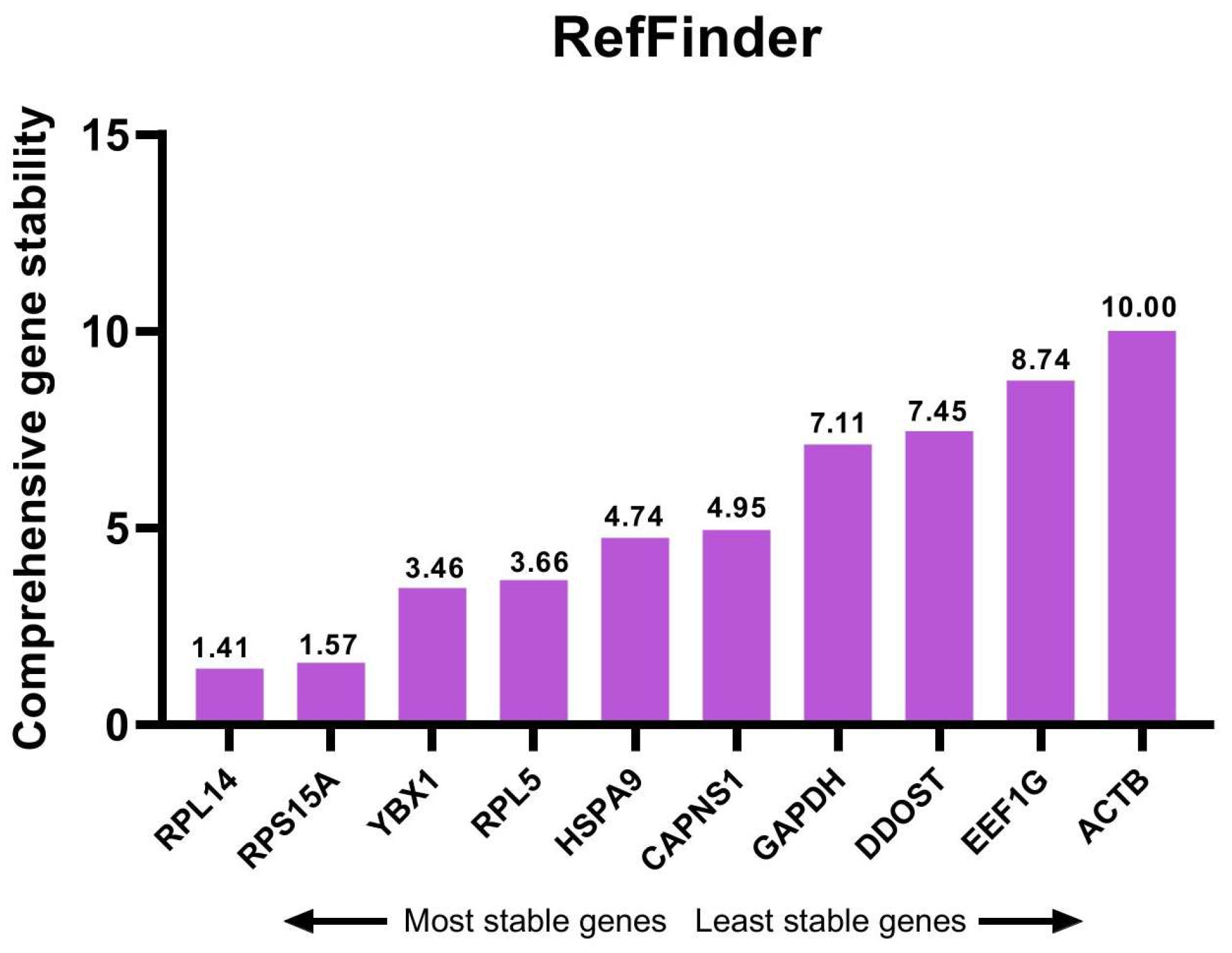
3.7. Comprehensive Analysis of Candidate Reference Genes
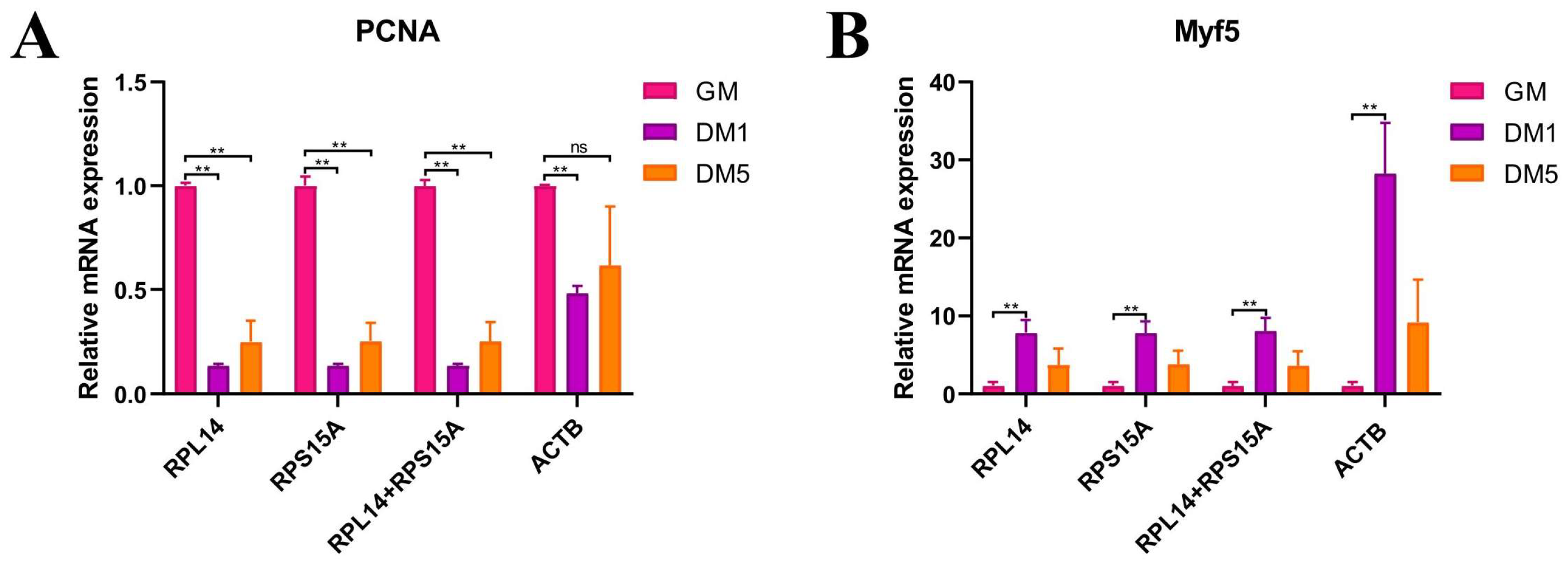
3.8. Expression Validation of Candidate Reference Genes Using Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubeuf, J.P.; Morand-Fehr, P.; Rubino, R. Situation, changes and future of goat industry around the world. Small Rumin. Res. 2004, 51, 165–173. [Google Scholar] [CrossRef]
- Kopantseva, E.E.; Belyavsky, A.V. Key regulators of skeletal myogenesis. Mol. Biol. 2016, 50, 169–192. [Google Scholar] [CrossRef]
- Wu, J.Y.; Yue, B.L. Regulation of myogenic cell proliferation and differentiation during mammalian skeletal myogenesis. Biomed. Pharmacother. 2024, 174, 116563. [Google Scholar] [CrossRef] [PubMed]
- Nejad, F.M.; Mohammadabadi, M.; Roudbari, Z.; Gorji, A.E.; Sadkowski, T. Network visualization of genes involved in skeletal muscle myogenesis in livestock animals. Bmc Genom. 2024, 25, 294. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR(RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004, 10, 190–212. [Google Scholar] [CrossRef]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N., Jr. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef]
- Bai, W.L.; Yin, R.H.; Zhao, S.J.; Jiang, W.Q.; Yin, R.L.; Ma, Z.J.; Wang, Z.Y.; Zhu, Y.B.; Luo, G.B.; Yang, R.J.; et al. Technical note: Selection of suitable reference genes for studying gene expression in milk somatic cell of yak (Bos grunniens) during the lactation cycle. J. Dairy Sci. 2014, 97, 902–910. [Google Scholar] [CrossRef]
- Kishore, A.; Sodhi, M.; Khate, K.; Kapila, N.; Kumari, P.; Mukesh, M. Selection of stable reference genes in heat stressed peripheral blood mononuclear cells of tropically adapted Indian cattle and buffaloes. Mol. Cell. Probes 2013, 27, 140–144. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- van Rijn, S.J.; Riemers, F.M.; van den Heuvel, D.; Wolfswinkel, J.; Hofland, L.; Meij, B.P.; Penning, L.C. Expression Stability of Reference Genes for Quantitative RT-PCR of Healthy and Diseased Pituitary Tissue Samples Varies Between Humans, Mice, and Dogs. Mol. Neurobiol. 2013, 49, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Xiong, Y.; Qian, H.; Lei, M.; Xu, D.; Ren, Z. Selection of reference genes for gene expression studies in porcine skeletal muscle using SYBR green qPCR. J. Biotechnol. 2010, 150, 288–293. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-D.; Liu, X.; Li, Y.-S.; Ding, J.-P.; Zhang, X.-R.; Zhang, Y.-H. Reference Gene Screening for Analyzing Gene Expression Across Goat Tissue. Asian-Australas. J. Anim. Sci. 2013, 26, 1665–1671. [Google Scholar] [CrossRef] [PubMed]
- Vasu, M.; Ahlawat, S.; Choudhary, V.; Kaur, R.; Arora, R.; Sharma, R.; Sharma, U.; Chhabra, P.; Mir, M.A.; Kumar Singh, M. Identification and validation of stable reference genes for expression profiling of target genes in diverse ovine tissues. Gene 2024, 897, 148067. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yang, H.; Li, X.; Zhou, Y.; Liu, T.; Zhao, Y. Transcriptome-based selection and validation of optimal reference genes in perirenal adipose developing of goat (Capra hircus). Front. Vet. Sci. 2022, 9, 1055866. [Google Scholar] [CrossRef]
- Niu, G.L.; Yang, Y.L.; Zhang, Y.Y.; Hua, C.J.; Wang, Z.S.; Tang, Z.L.; Li, K. Identifying suitable reference genes for gene expression analysis in developing skeletal muscle in pigs. Peerj 2016, 4, e2428. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Xiao, P.; Chen, D.L.; Xu, L.; Zhang, B.H. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Xie, F.L.; Wang, J.Y.; Zhang, B.H. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Chen, L.; Zhong, T.; Wang, L.; Guo, J.; Dong, Y.; Feng, J.; Song, T.; Li, L.; Zhang, H. The differential proliferation and differentiation ability of skeletal muscle satellite cell in Boer and Nanjiang brown goats. Small Rumin. Res. 2018, 169, 99–107. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Nie, L.; Ding, X.; Zhang, X.; Zhao, W.; Xu, X.; Kyei, B.; Dai, D.; Zhan, S.; et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 807–821. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Dollinger, G.; Walsh, P.S.; Griffith, R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology 1992, 10, 413–417. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Herrmann, M.G.; Moss, A.A.; Rasmussen, R.P. Continuous fluorescence monitoring of rapid cycle DNA amplification. 1997. Biotechniques 2013, 54, 314–320. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Biotechnology 1993, 11, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Oliveira, F.; Leandro, J.G.B.; Ausina, P.; Sola-Penna, M.; Majerowicz, D. Reference genes for quantitative PCR in the adipose tissue of mice with metabolic disease. Biomed. Pharmacother. 2017, 88, 948–955. [Google Scholar] [CrossRef]
- Cankorur-Cetinkaya, A.; Dereli, E.; Eraslan, S.; Karabekmez, E.; Dikicioglu, D.; Kirdar, B. A novel strategy for selection and validation of reference genes in dynamic multidimensional experimental design in yeast. PLoS ONE 2012, 7, e38351. [Google Scholar] [CrossRef] [PubMed]
- Lallemant, B.; Evrard, A.; Combescure, C.; Chapuis, H.; Chambon, G.; Raynal, C.; Reynaud, C.; Sabra, O.; Joubert, D.; Hollande, F.; et al. Reference gene selection for head and neck squamous cell carcinoma gene expression studies. BMC Mol. Biol. 2009, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Jeon, R.H.; Lee, W.J.; Son, Y.B.; Bharti, D.; Shivakumar, S.B.; Lee, S.L.; Rho, G.J. PPIA, HPRT1, and YWHAZ Genes Are Suitable for Normalization of mRNA Expression in Long-Term Expanded Human Mesenchymal Stem Cells. Biomed. Res. Int. 2019, 2019, 3093545. [Google Scholar] [CrossRef]
- Palombella, S.; Pirrone, C.; Cherubino, M.; Valdatta, L.; Bernardini, G.; Gornati, R. Identification of reference genes for qPCR analysis during hASC long culture maintenance. PLoS ONE 2017, 12, e0170918. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques 2018, 37, 112–119. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, L.; Liu, S. Determination of reference genes for ovine pulmonary adenocarcinoma infected lung tissues using RNA-seq transcriptome profiling. J. Virol. Methods 2020, 284, 113923. [Google Scholar] [CrossRef]
- Niemann, H.; Najafpanah, M.J.; Sadeghi, M.; Bakhtiarizadeh, M.R. Reference Genes Selection for Quantitative Real-Time PCR Using RankAggreg Method in Different Tissues of Capra hircus. PLoS ONE 2013, 8, e83041. [Google Scholar]
- Saremi, B.; Sauerwein, H.; Danicke, S.; Mielenz, M. Technical note: Identification of reference genes for gene expression studies in different bovine tissues focusing on different fat depots. J. Dairy Sci. 2012, 95, 3131–3138. [Google Scholar] [CrossRef]
- Thomas, K.C.; Zheng, X.F.; Garces Suarez, F.; Raftery, J.M.; Quinlan, K.G.; Yang, N.; North, K.N.; Houweling, P.J. Evidence based selection of commonly used RT-qPCR reference genes for the analysis of mouse skeletal muscle. PLoS ONE 2014, 9, e88653. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Petibon, C.; Malik Ghulam, M.; Catala, M.; Abou Elela, S. Regulation of ribosomal protein genes: An ordered anarchy. Wiley Interdiscip. Rev. RNA 2021, 12, e1632. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, C.; Zhang, L.; Lei, A.A.; Wang, L.J.; Niu, L.L.; Zhan, S.Y.; Guo, J.Z.; Cao, J.X.; Li, L.; et al. Genome-Wide Identification of Reference Genes for Reverse-Transcription Quantitative PCR in Goat Rumen. Animals 2021, 11, 3137. [Google Scholar] [CrossRef]
- Wang, G.H.; Liang, C.C.; Li, B.Z.; Du, X.Z.; Zhang, W.Z.; Cheng, G.; Zan, L.S. Screening and validation of reference genes for qRT-PCR of bovine skeletal muscle-derived satellite cells. Sci. Rep. 2022, 12, 5653. [Google Scholar] [CrossRef]
- Bonnet, M.; Bernard, L.; Bes, S.; Leroux, C. Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal 2013, 7, 1344–1353. [Google Scholar] [CrossRef]
- Macabelli, C.H.; Ferreira, R.M.; Gimenes, L.U.; de Carvalho, N.A.; Soares, J.G.; Ayres, H.; Ferraz, M.L.; Watanabe, Y.F.; Watanabe, O.Y.; Sangalli, J.R.; et al. Reference gene selection for gene expression analysis of oocytes collected from dairy cattle and buffaloes during winter and summer. PLoS ONE 2014, 9, e93287. [Google Scholar] [CrossRef]
- Jang, S.J.; Jeon, R.H.; Kim, H.D.; Hwang, J.C.; Lee, H.J.; Bae, S.G.; Lee, S.L.; Rho, G.J.; Kim, S.J.; Lee, W.J. TATA box binding protein and ribosomal protein 4 are suitable reference genes for normalization during quantitative polymerase chain reaction study in bovine mesenchymal stem cells. Asian-Australas. J. Anim. Sci. 2020, 33, 2021–2030. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, X.; Ding, X.; Chu, M.; Liang, C.; Pei, J.; Xiong, L.; Bao, P.; Guo, X.; Yan, P. Reference gene selection and myosin heavy chain (MyHC) isoform expression in muscle tissues of domestic yak (Bos grunniens). PLoS ONE 2020, 15, e0228493. [Google Scholar] [CrossRef]
- Thorrez, L.; Van Deun, K.; Tranchevent, L.C.; Van Lommel, L.; Engelen, K.; Marchal, K.; Moreau, Y.; Van Mechelen, I.; Schuit, F. Using ribosomal protein genes as reference: A tale of caution. PLoS ONE 2008, 3, e1854. [Google Scholar] [CrossRef] [PubMed]

| Gene | Coefficient of Variation (CV) | Standard Deviation (SD) | Rank |
|---|---|---|---|
| RPS15A | 1.58 | 0.26 | 1 |
| YBX1 | 1.66 | 0.27 | 2 |
| RPL5 | 1.76 | 0.28 | 3 |
| RPL14 | 1.98 | 0.31 | 4 |
| GAPDH | 1.79 | 0.33 | 5 |
| CAPNS1 | 1.82 | 0.36 | 6 |
| EEF1G | 2.22 | 0.45 | 7 |
| HSPA9 | 2.60 | 0.47 | 8 |
| DDOST | 2.30 | 0.48 | 9 |
| ACTB | 5.39 | 0.94 | 10 |
| Gene | Program | Mean Rank | Comprehensive Rank | ||
|---|---|---|---|---|---|
| GeNorm | NormFinder | Bestkeeper | |||
| RPL14 | 1 | 1 | 4 | 1.59 | 1 |
| RPS15A | 2 | 3 | 1 | 1.82 | 2 |
| YBX1 | 3 | 6 | 2 | 3.30 | 3 |
| RPL5 | 5 | 4 | 3 | 3.91 | 4 |
| HSPA9 | 6 | 2 | 8 | 4.58 | 5 |
| CAPNS1 | 4 | 5 | 6 | 4.93 | 6 |
| GAPDH | 8 | 8 | 5 | 6.84 | 7 |
| DDOST | 7 | 7 | 9 | 7.61 | 8 |
| EEF1G | 9 | 9 | 7 | 8.28 | 9 |
| ACTB | 10 | 10 | 10 | 10.00 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, S.; Zhang, L.; Zhong, T.; Wang, L.; Guo, J.; Cao, J.; Li, L.; Zhang, H. Evaluation of Reference Gene Stability in Goat Skeletal Muscle Satellite Cells during Proliferation and Differentiation Phases. Animals 2024, 14, 2479. https://doi.org/10.3390/ani14172479
Zhan S, Zhang L, Zhong T, Wang L, Guo J, Cao J, Li L, Zhang H. Evaluation of Reference Gene Stability in Goat Skeletal Muscle Satellite Cells during Proliferation and Differentiation Phases. Animals. 2024; 14(17):2479. https://doi.org/10.3390/ani14172479
Chicago/Turabian StyleZhan, Siyuan, Lufei Zhang, Tao Zhong, Linjie Wang, Jiazhong Guo, Jiaxue Cao, Li Li, and Hongping Zhang. 2024. "Evaluation of Reference Gene Stability in Goat Skeletal Muscle Satellite Cells during Proliferation and Differentiation Phases" Animals 14, no. 17: 2479. https://doi.org/10.3390/ani14172479





