The Effect of the Species Source of Muscle and/or Digestive Enzymes on the Utilization of Fish Protein Hydrolysates as a Dietary Protein Source in First Feed for Larval Walleye (Sander vitreus)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Conditions
2.2. Muscle Hydrolysis
2.3. Diets
2.4. Diet Analysis
2.5. Experimental Design
2.6. Sampling and Measuring
2.7. PepT1 Analysis
2.8. Free Amino Acid Analysis
2.9. SDS-PAGE
2.10. Statistical Analysis
3. Results
3.1. SDS-PAGE
3.2. Growth, Survival, and Skeletal Deformities
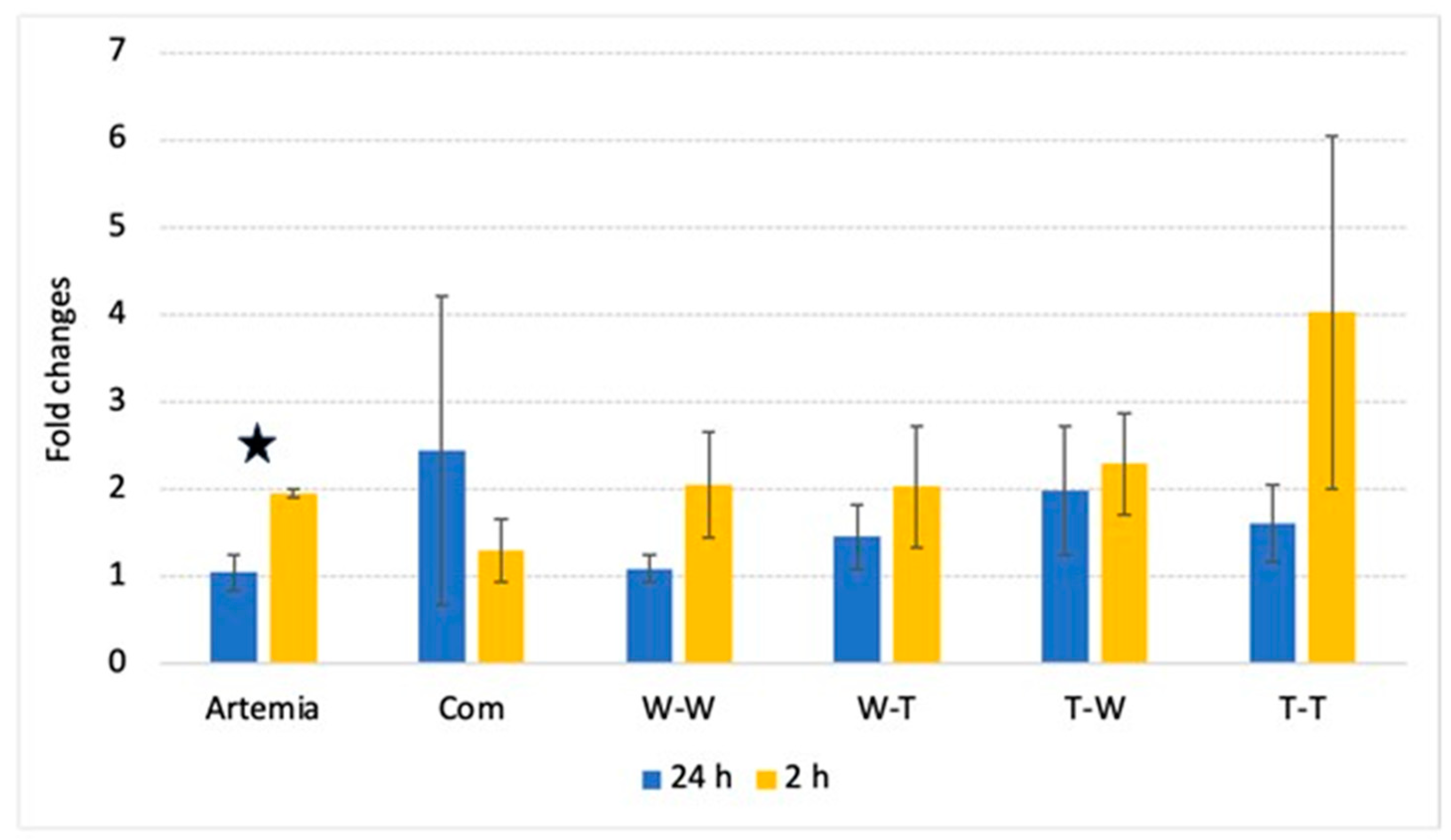
3.3. PepT1 Expression
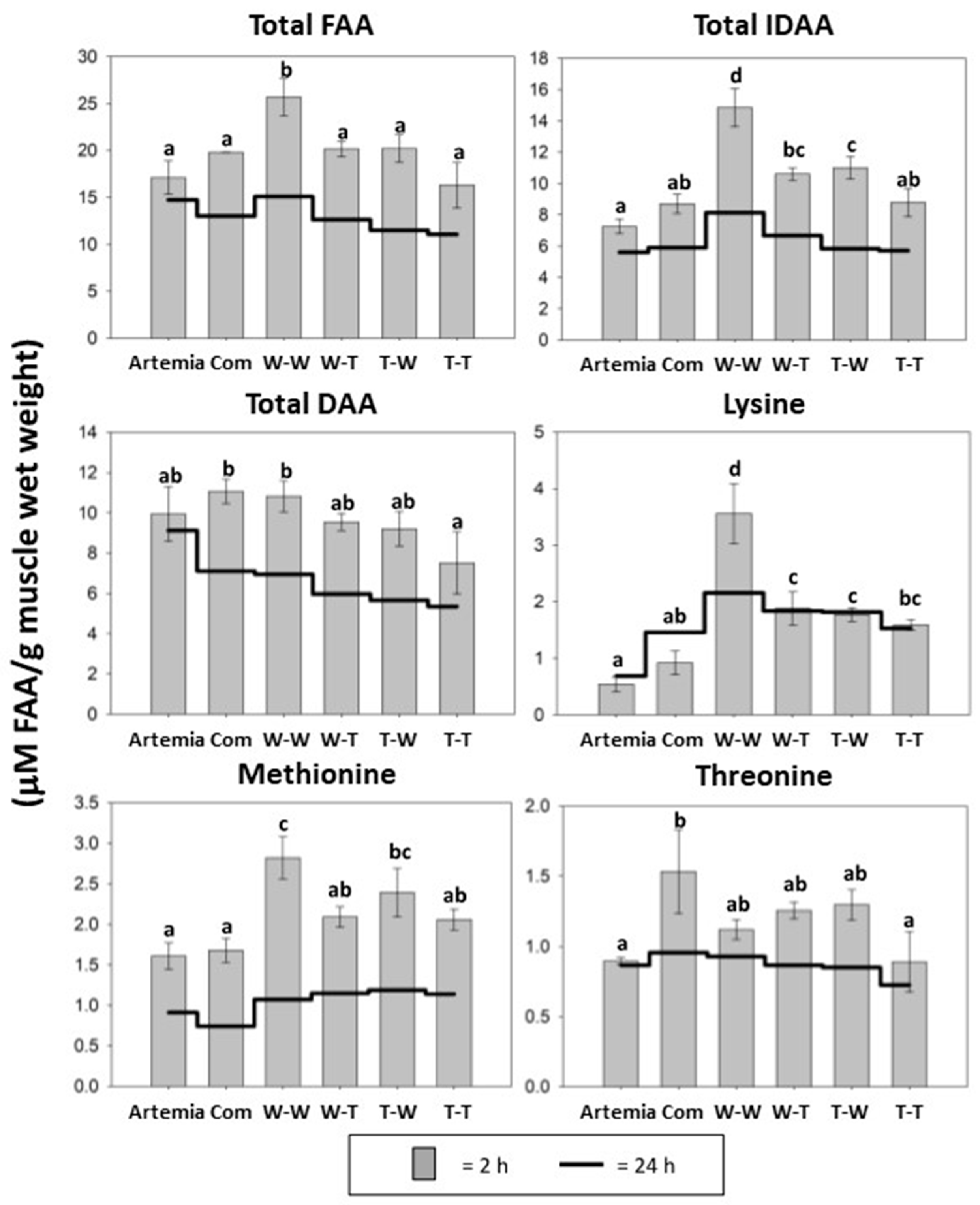
3.4. Muscle Free Amino Acid Composition
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cahu, C.L.; Infante, J.Z. Effect of the molecular form of dietary nitrogen supply in sea bass larvae: Response of pancreatic enzymes and intestinal peptidases. Fish Physiol. Biochem. 1995, 14, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.P.; Escaffre, A.M.; Oliva Teles, A.; Bergot, P. First feeding of common carp larvae on diets with high levels of protein hydrolysates. Aquac. Int. 1997, 5, 361–367. [Google Scholar] [CrossRef]
- Tonheim, S.K.; Espe, M.; Hamre, K.; Rønnestad, I. Pre-hydrolysis improves utilisation of dietary protein in the larval teleost Atlantic halibut (Hippoglossus hippoglossus L.). J. Exp. Mar. Biol. Ecol. 2005, 321, 19–34. [Google Scholar] [CrossRef]
- Kotzamanis, Y.P.; Gisbert, E.; Gatesoupe, F.J.; Infante, J.Z.; Cahu, C. Effects of different dietary levels of fish protein hydrolysates on growth, digestive enzymes, gut microbiota, and resistance to Vibrio anguillarum in European sea bass (Dicentrarchus labrax) larvae. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 147, 205–214. [Google Scholar] [CrossRef]
- Kvåle, A.; Harboe, T.; Mangor-Jensen, A.; Hamre, K. Effects of protein hydrolysate in weaning diets for Atlantic cod (Gadus morhua L.) and Atlantic halibut (Hippoglossus hippoglossus L.). Aquac. Nutr. 2009, 15, 218–227. [Google Scholar] [CrossRef]
- Srichanun, M.; Tantikitti, C.; Kortner, T.M.; Krogdahl, Å.; Chotikachinda, R. Effects of different protein hydrolysate products and levels on growth, survival rate and digestive capacity in Asian seabass (Lates calcarifer Bloch) larvae. Aquaculture 2014, 428, 195–202. [Google Scholar] [CrossRef]
- Kasumyan, A.O.; DÖving, K.B. Taste preferences in fishes. Fish Fish. 2003, 4, 289–347. [Google Scholar] [CrossRef]
- Refstie, S.; Olli, J.J.; Standal, H. Feed intake, growth, and protein utilisation by post-smolt Atlantic salmon (Salmo salar) in response to graded levels of fish protein hydrolysate in the diet. Aquaculture 2004, 239, 331–349. [Google Scholar] [CrossRef]
- Bui, H.T.D.; Khosravi, S.; Fournier, V.; Herault, M.; Lee, K.J. Growth performance, feed utilization, innate immunity, digestibility and disease resistance of juvenile red seabream (Pagrus major) fed diets supplemented with protein hydrolysates. Aquaculture 2014, 418, 11–16. [Google Scholar] [CrossRef]
- Pyz-Łukasik, R.; Paszkiewicz, W. Species variations in the proximate composition, amino acid profile, and protein quality of the muscle tissue of grass carp, bighead carp, siberian sturgeon, and wels catfish. J. Food Qual. 2018, 2018, 2625401. [Google Scholar] [CrossRef]
- Ryu, B.; Shin, K.H.; Kim, S.K. Muscle protein hydrolysates and amino acid composition in fish. Mar. Drugs 2021, 19, 377. [Google Scholar] [CrossRef] [PubMed]
- Clemente, A. Enzymatic protein hydrolysates in human nutrition. Trends Food Sci. Technol. 2000, 11, 254–262. [Google Scholar] [CrossRef]
- Zamora-Sillero, J.; Gharsallaoui, A.; Prentice, C. Peptides from fish by-product protein hydrolysates and its functional properties: An overview. Mar. Biotechnol. 2018, 20, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Sandbakken, I.S.; Five, K.K.; Bardal, T.; Knapp, J.L.; Olsen, R.E. Salmon hydrolysate as a protein source for Atlantic salmon; prion content and effects on growth, digestibility and gut health. Aquaculture 2023, 576, 739863. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; Rasco, B.A. Fish protein hydrolysates: Production, biochemical, and functional properties. Crit. Rev. Food Sci. Nutr. 2000, 40, 43–81. [Google Scholar] [CrossRef]
- Aspmo, S.I.; Horn, S.J.; Eijsink, V.G. Enzymatic hydrolysis of Atlantic cod (Gadus morhua L.) viscera. Process Biochem. 2005, 40, 1957–1966. [Google Scholar] [CrossRef]
- Hathwar, S.C.; Bijinu, B.; Rai, A.K.; Narayan, B. Simultaneous recovery of lipids and proteins by enzymatic hydrolysis of fish industry waste using different commercial proteases. Appl. Biochem. Biotechnol. 2011, 164, 115–124. [Google Scholar] [CrossRef]
- Saidi, S.; Belleville, M.P.; Deratani, A.; Amar, R.B. Optimization of peptide production by enzymatic hydrolysis of tuna dark muscle by-product using commercial proteases. Afr. J. Biotechnol. 2013, 12, 1533–1547. [Google Scholar]
- Elavarasan, K.; Naveen Kumar, V.; Shamasundar, B.A. Antioxidant and functional properties of fish protein hydrolysates from freshwater carp (Catla catla) as influenced by the nature of enzyme. J. Food Process. Preserv. 2014, 38, 1207–1214. [Google Scholar] [CrossRef]
- Herpandi, N.H.; Rosma, A.; Wan Nadiah, W.A. The tuna fishing industry: A new outlook on fish protein hydrolysates. Compr. Rev. Food Sci. Food Saf. 2011, 10, 195–207. [Google Scholar] [CrossRef]
- Silva, J.F.X.; Ribeiro, K.; Silva, J.F.; Cahú, T.B.; Bezerra, R.S. Utilization of tilapia processing waste for the production of fish protein hydrolysate. Anim. Feed. Sci. Technol. 2014, 196, 96–106. [Google Scholar] [CrossRef]
- Kwasek, K.; Gonzalez, C.; Wick, M.; Molinari, G.S.; Wojno, M. Fish muscle hydrolysate obtained using largemouth bass Micropterus salmoides digestive enzymes improves largemouth bass performance in its larval stages. PLoS ONE 2021, 16, e0261847. [Google Scholar] [CrossRef] [PubMed]
- Molinari, G.S.; Wojno, M.; Terova, G.; Wick, M.; Riley, H.; Caminiti, J.T.; Kwasek, K. A Novel Approach in the Development of Larval Largemouth Bass Micropterus salmoides Diets Using Largemouth Bass Muscle Hydrolysates as the Protein Source. Animals 2023, 13, 373. [Google Scholar] [CrossRef]
- Buddington, R.K.; Krogdahl, A.; Bakke-McKellep, A.M. The intestines of carnivorous fish: Structure and functions and the relations with diet. Acta Physiologica Scandinavica. Supplementum 1997, 638, 67–80. [Google Scholar]
- Kramer, D.L.; Bryant, M.J. Intestine length in the fishes of a tropical stream: 2. Relationships to diet—The long and short of a convoluted issue. Environ. Biol. Fishes 1995, 42, 129–141. [Google Scholar] [CrossRef]
- German, D.P.; Horn, M.H.; Gawlicka, A. Digestive enzyme activities in herbivorous and carnivorous prickleback fishes (Teleostei: Stichaeidae): Ontogenetic, dietary, and phylogenetic effects. Physiol. Biochem. Zool. 2004, 77, 789–804. [Google Scholar] [CrossRef]
- Johnson, J.A.; Summerfelt, R.C.; Clayton, R.D. Evaluation of replacement feeds for the fry feed Kyowa for larval walleyes. North Am. J. Aquac. 2008, 70, 446–451. [Google Scholar] [CrossRef]
- Morrison, C.M.; Wright, J.R., Jr. A study of the histology of the digestive tract of the Nile tilapia. J. Fish Biol. 1999, 54, 597–606. [Google Scholar] [CrossRef]
- Dabrowski, K.; Miller, M. Contested paradigm in raising zebrafish (Danio rerio). Zebrafish 2018, 15, 295–309. [Google Scholar] [CrossRef]
- Clayton, R.D.; Summerfelt, R.C. Gas bladder inflation in walleye fry cultured in turbid water with and without a surface spray. North Am. J. Aquac. 2010, 72, 338–342. [Google Scholar] [CrossRef]
- Summerfelt, R.C.; Johnson, J.A. Intensive culture of Walleye from egg incubation to juvenile. Biol. Cult. Percid Fishes: Princ. Pract. 2015, 313–335. [Google Scholar] [CrossRef]
- Roslan, J.; Yunos, K.F.M.; Abdullah, N.; Kamal, S.M.M. Characterization of fish protein hydrolysate from tilapia (Oreochromis niloticus) by-product. Agric. Agric. Sci. Procedia 2014, 2, 312–319. [Google Scholar] [CrossRef]
- Solovyev, M.; Gisbert, E. Influence of time, storage temperature and freeze/thaw cycles on the activity of digestive enzymes from gilthead sea bream (Sparus aurata). Fish Physiol. Biochem. 2016, 42, 1383–1394. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, H. Standard Norwegian Fishmeal-and Fishoil Process; Heat treatment requirements; Nofima rapportserie; Nofima AS: Ås, Norway, 2010. [Google Scholar]
- Ramluckan, K.; Moodley, K.G.; Bux, F. An evaluation of the efficacy of using selected solvents for the extraction of lipids from algal biomass by the soxhlet extraction method. Fuel 2014, 116, 103–108. [Google Scholar] [CrossRef]
- National Academies Press. National Research Council Nutrient Requirements of Fish and Shrimp; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Folch, J.; Lees, M.; Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Cahu, C.; Infante, J.Z.; Takeuchi, T. Nutritional components affecting skeletal development in fish larvae. Aquaculture 2003, 227, 245–258. [Google Scholar] [CrossRef]
- Ng, W.K.; Hung, S.S.O. Estimating the ideal dietary indispensable amino acid pattern for growth of white sturgeon, Acipenser transmontanus (Richardson). Aquac. Nutr. 1995, 1, 85–94. [Google Scholar] [CrossRef]
- Terova, G.; Corà, S.; Verri, T.; Rimoldi, S.; Bernardini, G.; Saroglia, M. Impact of feed availability on PepT1 mRNA expression levels in sea bass (Dicentrarchus labrax). Aquaculture 2009, 294, 288–299. [Google Scholar] [CrossRef]
- Updike, M.S.; Zerby, H.; Utrata, K.L.; Lilburn, M.; Kaletunc, G.; Wick, M. Proteins associated with thermally induced gelation of turkey breast meat. J. Food Sci. 2006, 71, E398–E402. [Google Scholar] [CrossRef]
- Khosravi, S.; Bui, H.T.D.; Rahimnejad, S.; Herault, M.; Fournier, V.; Kim, S.S.; Jeong, J.B.; Lee, K.J. Dietary supplementation of marine protein hydrolysates in fish-meal based diets for red sea bream (Pagrus major) and olive flounder (Paralichthys olivaceus). Aquaculture 2015, 435, 371–376. [Google Scholar] [CrossRef]
- Nurdiani, R.; Dissanayake, M.; Street, W.E.; Donkor, O.N.; Singh, T.K.; Vasiljevic, T. In vitro study of selected physiological and physicochemical properties of fish protein hydrolysates from 4 Australian fish species. Int. Food Res. J. 2016, 23, 2029–2040. [Google Scholar]
- Pastoriza, L.; Sampedro, G.; Cabo, M.L.; Herrera, J.J.; Bernárdez, M. Solubilisation of proteins from rayfish residues by endogenous and commercial enzymes. J. Sci. Food Agric. 2004, 84, 83–88. [Google Scholar] [CrossRef]
- Patterson, J.; Ohs, C.; O’Malley, P.; Palau, A.; D’Abramo, L.; Reigh, R.; Green, C. Feeding larval gulf killifish: Total replacement of Artemia nauplii and co-feeding from hatch. N. Am. J. Aquac. 2016, 78, 396–404. [Google Scholar] [CrossRef]
- Lipscomb, T.N.; Patterson, J.T.; Wood, A.L.; Watson, C.A.; DiMaggio, M.A. Larval growth, survival, and partial budget analysis related to replacing Artemia in larval culture of six freshwater ornamental fishes. J. World Aquac. Soc. 2020, 51, 1132–1144. [Google Scholar] [CrossRef]
- Cole, D.J.A. The amino acid requirements of pigs-the concept of an ideal protein. Pig News Inf. 1980, 1, 201–205. [Google Scholar]
- Wilson, R.P.; Cowey, C.B. Amino acid composition of whole body tissue of rainbow trout and Atlantic salmon. Aquaculture 1985, 48, 373–376. [Google Scholar] [CrossRef]
- Chevalier, J.R. Cannibalism as a factor in first year survival of walleye in Oneida Lake. Trans. Am. Fish. Soc. 1973, 102, 739–744. [Google Scholar] [CrossRef]
- Hecht, T.; Pienaar, A.G. A review of cannibalism and its implications in fish larviculture. J. World Aquac. Soc. 1993, 24, 246–261. [Google Scholar] [CrossRef]
- Baras, E.; Jobling, M. Dynamics of intracohort cannibalism in cultured fish. Aquac. Res. 2002, 33, 461–479. [Google Scholar] [CrossRef]
- Duk, K.; Pajdak, J.; Terech-Majewska, E.; Szarek, J. Intracohort cannibalism and methods for its mitigation in cultured freshwater fish. Rev. Fish Biol. Fish. 2017, 27, 193–208. [Google Scholar] [CrossRef]
- Kolkovski, S. Digestive enzymes in fish larvae and juveniles—Implications and applications to formulated diets. Aquaculture 2001, 200, 181–201. [Google Scholar] [CrossRef]
- Hansen, J.M.; Lazo, J.P.; Kling, L.J. A method to determine protein digestibility of microdiets for larval and early juvenile fish. Aquac. Nutr. 2009, 15, 615–626. [Google Scholar] [CrossRef]
- Tibbetts, S.M.; Verreth, J.A.J. Lall SP In vitro pH-Stat protein hydrolysis of feed ingredients for Atlantic cod, Gadus morhua. 2. In vitro protein digestibility of common and alternative feed ingredients. Aquaculture 2011, 319, 407–416. [Google Scholar] [CrossRef]
- Yasumaru, F.; Lemos, D. Species specific in vitro protein digestion (pH-stat) for fish: Method development and application for juvenile rainbow trout (Oncorhynchus mykiss), cobia (Rachycentron canadum), and Nile tilapia (Oreochromis niloticus). Aquaculture 2014, 426, 74–84. [Google Scholar] [CrossRef]
- Kwasek, K.; Terova, G.; Wojno, M.; Dabrowski, K.; Wick, M. The effect of dietary dipeptide lysine–glycine on growth, muscle proteins, and intestine PepT1 gene expression in juvenile yellow perch. Rev. Fish Biol. Fish. 2012, 22, 797–812. [Google Scholar] [CrossRef]
- Terova, G.; Robaina, L.; Izquierdo, M.; Cattaneo, A.; Molinari, S.; Bernardini, G.; Saroglia, M. PepT1 mRNA expression levels in sea bream (Sparus aurata) fed different plant protein sources. SpringerPlus 2013, 2, 1–14. [Google Scholar] [CrossRef]
- Gisbert, E.; Skalli, A.; Fernández, I.; Kotzamanis, Y.; Zambonino-Infante, J.L.; Fabregat, R. Protein hydrolysates from yeast and pig blood as alternative raw materials in microdiets for gilthead sea bream (Sparus aurata) larvae. Aquaculture 2012, 338, 96–104. [Google Scholar] [CrossRef]
- Peruzzi, S.; Puvanendran, V.; Riesen, G.; Seim, R.R.; Hagen, Ø.; Martinez-Llorens, S.; Falk-Petersen, I.B.; Fernandes, J.M.; Jobling, M. Growth and development of skeletal anomalies in diploid and triploid Atlantic salmon (Salmo salar) fed phosphorus-rich diets with fish meal and hydrolyzed fish protein. PLoS ONE 2018, 13, e0194340. [Google Scholar] [CrossRef]
- Printzi, A.; Koumoundouros, G.; Fournier, V.; Madec, L.; Zambonino-Infante, J.L.; Mazurais, D. Effect of Early Peptide Diets on Zebrafish Skeletal Development. Biomolecules 2023, 13, 659. [Google Scholar] [CrossRef]
- Fernández, I.; Hontoria, F.; Ortiz-Delgado, J.B.; Kotzamanis, Y.; Estévez, A.; Zambonino-Infante, J.L.; Gisbert, E. Larval performance and skeletal deformities in farmed gilthead sea bream (Sparus aurata) fed with graded levels of Vitamin A enriched rotifers (Brachionus plicatilis). Aquaculture 2008, 283, 102–115. [Google Scholar] [CrossRef]
- Shefat, S.H.T.; Karim, M.A. Nutritional diseases of fish in aquaculture and their management: A review. Acta Sci. Pharm. Sci. 2018, 2, 50–58. [Google Scholar]
- Eissa, A.E.; Abu-Seida, A.M.; Ismail, M.M.; Abu-Elala, N.M.; Abdelsalam, M. A comprehensive overview of the most common skeletal deformities in fish. Aquac. Res. 2021, 52, 2391–2402. [Google Scholar] [CrossRef]
- Darias, M.J.; Mazurais, D.; Koumoundouros, G.; Cahu, C.L.; Zambonino-Infante, J.L. Overview of vitamin D and C requirements in fish and their influence on the skeletal system. Aquaculture 2011, 315, 49–60. [Google Scholar] [CrossRef]
- Silverstone, A.M.; Hammell, L. Spinal deformities in farmed Atlantic salmon. Can. Vet. J. 2002, 43, 782. [Google Scholar] [PubMed]
- Lall, S.P.; Kaushik, S.J. Nutrition and metabolism of minerals in fish. Animals 2021, 11, 2711. [Google Scholar] [CrossRef]
- Coutteau, P.; Geurden, I.; Camara, M.R.; Bergot, P.; Sorgeloos, P. Review on the dietary effects of phospholipids in fish and crustacean larviculture. Aquaculture 1997, 155, 149–164. [Google Scholar] [CrossRef]
- Akiyama, T.; Murai, T.; Mori, K. Role of tryptophan metabolites in inhibition of spinal deformity of chulm salmon fry caused by tryptophan deficiency. Nippon. Suisan Gakkaishi 1986, 52, 1255–1259. [Google Scholar] [CrossRef]
- Yúfera, M.; Fernández-Díaz, C.; Pascual, E. Food microparticles for larval fish prepared by internal gelation. Aquaculture 2005, 248, 253–262. [Google Scholar] [CrossRef]
- Drossou, A.; Ueberschär, B.; Rosenthal, H.; Herzig, K.H. Ontogenetic development of the proteolytic digestion activities in larvae of Oreochromis niloticus fed with different diets. Aquaculture 2006, 256, 479–488. [Google Scholar] [CrossRef]
- Lian, P.; Lee, C.M.; Bengtson, D.A. Development of a squid-hydrolysate-based larval diet and its feeding performance on summer flounder, Paralichthys dentatus, larvae. J. World Aquac. Soc. 2008, 39, 196–204. [Google Scholar] [CrossRef]
- Cai, Z.; Li, W.; Mai, K.; Xu, W.; Zhang, Y.; Ai, Q. Effects of dietary size-fractionated fish hydrolysates on growth, activities of digestive enzymes and aminotransferases and expression of some protein metabolism related genes in large yellow croaker (Larimichthys crocea) larvae. Aquaculture 2015, 440, 40–47. [Google Scholar] [CrossRef]
- Kvåle, A.; Yúfera, M.; Nygård, E.; Aursland, K.; Harboe, T.; Hamre, K. Leaching properties of three different micropaticulate diets and preference of the diets in cod (Gadus morhua L.) larvae. Aquaculture 2006, 251, 402–415. [Google Scholar] [CrossRef]
- Nordgreen, A.; Tonheim, S.; Hamre, K. Protein quality of larval feed with increased concentration of hydrolysed protein: Effects of heat treatment and leaching. Aquac. Nutr. 2009, 15, 525–536. [Google Scholar] [CrossRef]
- Martínez-Alvarez, O.; Chamorro, S.; Brenes, A. Protein hydrolysates from animal processing by-products as a source of bioactive molecules with interest in animal feeding: A review. Food Res. Int. 2015, 73, 204–212. [Google Scholar] [CrossRef]
- Hardy, R.W.; Barrows, F.T. Diet formulation and manufacture. In Fish Nutrition; Academic Press: Cambridge, MA, USA, 2003; pp. 505–600. [Google Scholar]


| Ingredients (%) | Diet |
|---|---|
| Unhydrolyzed Protein a | 37.00 |
| Protein Hydrolysate b | 37.00 |
| Krill Meal c | 5.00 |
| Fish oil d | 9.00 |
| Lecithin e | 4.00 |
| Mineral mix f | 3.00 |
| Vitamin mix g | 3.00 |
| CaHPO4 | 1.00 |
| Taurine | 1.00 |
| Choline chloride | 0.10 |
| Vitamin C h | 0.05 |
| Sum | 100 |
| Analyzed Composition (g/100g) Dry Matter | W-W | W-T | T-W | T-T |
|---|---|---|---|---|
| Crude Protein (N × 6.25) | 60.23 (±0.09) | 60.20 (±0.15) | 59.92 (±0.25) | 59.93 (±0.05) |
| Crude Lipids | 15.30 (±0.05) | 15.47 (±0.02) | 15.66 (±0.02) | 15.08 (±0.08) |
| Ash | 13.48 (±0.21) | 13.45 (±0.09) | 13.82 (±0.09) | 13.74 (±0.45) |
| Alanine | 3.70 (±0.02) | 3.86 (±0.13) | 3.73 (±0.01) | 3.77 (±0.09) |
| Arginine | 3.40 (±0.03) | 3.08 (±0.12) | 3.57 (±0.01) | 3.17 (±0.09) |
| Aspartic Acid | 5.58 (±0.02) | 5.63 (±0.04) | 5.30 (±0.07) | 5.45 (±0.09) |
| Cysteine | 0.59 (±0.02) | 0.63 (±0.00) | 0.53 (±0.01) | 0.58 (±0.02) |
| Glutamic Acid | 7.97 (±0.24) | 7.81 (±0.10) | 7.71 (±0.03) | 7.97 (±0.09) |
| Glycine | 3.86 (±0.04) | 4.20 (±0.28) | 4.51 (±0.00) | 4.35 (±0.32) |
| Histidine | 1.49 (±0.02) | 1.43 (±0.03) | 1.27 (±0.01) | 1.36 (±0.03) |
| Hydroxyproline | 0.60 (±0.02) | 0.65 (±0.01) | 1.13 (±0.11) | 0.94 (±0.14) |
| Isoleucine | 2.65 (±0.03) | 2.66 (±0.04) | 2.49 (±0.06) | 2.59 (±0.08) |
| Leucine | 4.31 (±0.03) | 4.31 (±0.05) | 4.07 (±0.02) | 4.23 (±0.09) |
| Lysine | 5.04 (±0.03) | 4.61 (±0.07) | 4.52 (±0.01) | 4.28 (±0.08) |
| Methionine | 1.69 (±0.01) | 1.70 (±0.00) | 1.45 (±0.03) | 1.51 (±0.02) |
| Phenylalanine | 2.32 (±0.02) | 2.34 (±0.05) | 2.17 (±0.03) | 2.28 (±0.04) |
| Proline | 2.34 (±0.03) | 2.50 (±0.08) | 2.70 (±0.04) | 2.63 (±0.14) |
| Serine | 2.26 (±0.11) | 2.24 (±0.05) | 2.02 (±0.06) | 2.00 (±0.02) |
| Taurine | 1.53 (±0.01) | 1.54 (±0.03) | 1.87 (±0.03) | 1.86 (±0.13) |
| Threonine | 2.51 (±0.00) | 2.56 (±0.01) | 2.41 (±0.03) | 2.47 (±0.03) |
| Tryptophan | 0.61 (±0.03) | 0.64 (±0.03) | 0.54 (±0.01) | 0.61 (±0.03) |
| Tyrosine | 1.79 (±0.01) | 1.80 (±0.05) | 1.62 (±0.11) | 1.73 (±0.04) |
| Valine | 2.83 (±0.04) | 2.91 (±0.02) | 2.73 (±0.04) | 2.84 (±0.05) |
| IDAA | 26.85 (±0.11) | 26.24 (±0.36) | 25.22 (±0.19) | 25.34 (±0.34) |
| DAA | 30.21 (±0.35) | 30.87 (±0.44) | 31.12 (±0.13) | 31.29 (±0.50) |
| Total | 57.06 (±0.24) | 57.11 (±0.20) | 56.33 (±0.27) | 56.63 (±0.41) |
| Gene | Primer | Nucleotide Sequence (5′–3′) |
|---|---|---|
| PepT1 | Forward | CACACCCAGCAGAAGTGCTACT |
| Reverse | ACAATCAGAGCTACCACCATGAGA | |
| Probe | FAM-ACTGGCCTTTGGTGTCCCCGC-NFQ | |
| eEF1a1 | Forward | GGAAATCCGTCGTGGATATGTG |
| Reverse | TGACCTGGGCGTTGAAGTTG | |
| Probe | FAM-CTGGCGACAGCAAGAACGACCCACC-NFQ | |
| α-tubulin | Forward | ACCAACCTCAACAGGCTAATTGG |
| Reverse | GAGGGCACCATCGAAACGA | |
| Probe | FAM-CAGATTGTGTCCTCCATCACTGCCTCCC-NFQ | |
| β-actin | Forward | CCCTCTTCCAGCCTTCCTT |
| Reverse | GTAGGTGGTCTCGTGGATTCC | |
| Probe | FAM-CCTCGGTATGGAGTCCTG-NFQ |
| Group | Avg. Weight (mg) | Survival (%) |
|---|---|---|
| Artemia | 61.01 a (±12.47) | 53.63 b (±11.43) |
| Com | 57.35 a (±6.18) | 39.41 b (±1.39) |
| W-W | 89.84 b (±6.09) | 16.87 a (±5.71) |
| W-T | 62.83 a (±4.03) | 13.95 a (±2.41) |
| T-W | 61.22 a (±9.61) | 20.98 a (±3.16) |
| T-T | 54.70 a (±8.62) | 21.69 a (±4.80) |
| Parameter | Factor | p-Value |
|---|---|---|
| Avg. Weight | Muscle Source | - |
| Enzyme Source | - | |
| Interaction * | 0.0437 | |
| Survival | Muscle Source * | 0.0344 |
| Enzyme Source | 0.6553 | |
| Interaction | NS | |
| Total Occurrence of Skeletal Deformities | Muscle Source | 0.2126 |
| Enzyme Source | 0.3406 | |
| Interaction | NS | |
| Postprandial (2 h) PepT1 Expression | Muscle Source | 0.3442 |
| Enzyme Source | 0.4652 | |
| Interaction | NS | |
| Postprandial (2 h) Total FAA Level in Muscle | Muscle Source * | 0.0013 |
| Enzyme Source * | 0.0012 | |
| Interaction | NS | |
| Postprandial (2 h) Total IDAA Level in Muscle | Muscle Source * | 0.0008 |
| Enzyme Source * | 0.0003 | |
| Interaction | NS | |
| Postprandial (2 h) Total DAA Level in Muscle | Muscle Source * | 0.0085 |
| Enzyme Source * | 0.0243 | |
| Interaction | NS |
| Amino Acid (μM/g) | Artemia | Com | W-W | W-T | T-W | T-T |
|---|---|---|---|---|---|---|
| Aspartic Acid | 1.00 (±0.16) | 1.41 (±0.25) | 1.03 (±0.10) | 1.16 (±0.04) | 1.09 (±0.14) | 1.00 (±0.19) |
| Glutamic Acid | 1.78 ab (±0.18) | 2.14 b (±0.28) | 1.84 ab (±0.15) | 1.86 ab (±0.03) | 1.80 ab (±0.19) | 1.45 a (±0.21) |
| Asparagine | 0.44 c (±0.12) | 0.36 bc (±0.09) | 0.20 ab (±0.01) | 0.17 a (±0.02) | 0.19 ab (±0.03) | 0.15 a (±0.06) |
| Serine | 1.55 b (±0.50) | 1.20 ab (±0.33) | 0.68 a (±0.04) | 0.84 ab (±0.06) | 0.70 a (±0.11) | 0.53 a (±0.18) |
| Glutamine | 0.50 ab (±0.02) | 0.58 b (±0.04) | 0.63 b (±0.03) | 0.52 ab (±0.03) | 0.59 b (±0.08) | 0.38 a (±0.08) |
| Histidine | 1.77 a (±0.48) | 1.73 a (±0.28) | 2.66b (±0.04) | 1.53 a (±0.11) | 1.63 a (±0.28) | 1.07 a (±0.31) |
| Glycine | 1.96 a (±0.67) | 1.75 a (±0.44) | 3.36 b (±0.05) | 1.95 a (±0.12) | 1.91a (±0.24) | 1.86a (±0.41) |
| Threonine | 0.90 a (±0.03) | 1.53 b (±0.30) | 1.21 ab (±0.07) | 1.26 ab (±0.06) | 1.30 ab (±0.11) | 0.89 a (±0.22) |
| Arginine | 0.29 a (±0.03) | 0.45 ab (±0.08) | 1.04 c (±0.17) | 0.53 b (±0.03) | 0.63 b (±0.02) | 0.44 ab (±0.08) |
| Alanine | 1.80 bc (±0.51) | 2.43 c (±0.27) | 1.49 ab (±0.16) | 1.64 ab (±0.17) | 1.37 ab (±0.12) | 0.89 a (±0.25) |
| Tyrosine | 0.45 a (±0.02) | 0.46 a (±0.08) | 0.79 b (±0.15) | 0.62 ab (±0.06) | 0.55 ab (±0.03) | 0.54 ab (±0.14) |
| Lysine | 0.55 a (±0.14) | 0.93 ab (±0.22) | 3.57d (±0.53) | 1.88 c (±0.30) | 1.77 c (±0.12) | 1.59 bc (±0.09) |
| Methionine | 1.61 a (±0.16) | 1.68 a (±0.15) | 2.83 c (±0.26) | 2.10 ab (±0.12) | 2.40 bc (±0.30) | 2.06 ab (±0.13) |
| Valine | 1.44 a (±0.07) | 1.49 a (±0.09) | 2.40 c (±0.07) | 2.03 b (±0.10) | 2.39 c (±0.12) | 2.04 b (±0.11) |
| Cysteine * Not detected | - | - | - | - | - | - |
| Tryptophan | 0.03 ab (±0.01) | 0.02 ab (±0.01) | 0.04 b (±0.00) | 0.04 b (±0.00) | 0.03 a b (±0.00) | 0.02 a (±0.01) |
| Phenylalanine | 0.19 a (±0.03) | 0.17 a (±0.02) | 0.25 ab (±0.03) | 0.27 b (±0.02) | 0.19 ab (±0.03) | 0.20 ab (±0.04) |
| Isoleucine | 0.18 ab (±0.02) | 0.23 bc (±0.04) | 0.28 cd (±0.03) | 0.32 d (±0.00) | 0.21 ac (±0.02) | 0.14 a (±0.03) |
| Leucine | 0.32 a (±0.07) | 0.47 ab (±0.10) | 0.59 bc (±0.05) | 0.66 c (±0.00) | 0.47 ab (±0.05) | 0.34 a (±0.07) |
| Proline | 0.46 a (±0.06) | 0.74 ac (±0.11) | 0.80 bc (±0.10) | 0.80 b c (±0.02) | 1.02 c (±0.19) | 0.71 ab (±0.10) |
| IDAA | 7.27 a (±0.47) | 8.71 ab (±0.63) | 14.87d (±1.23) | 10.61 bc (±0.41) | 11.02 c (±0.72) | 8.78 ab (±0.90) |
| DAA | 9.94 ab (±1.36) | 11.09 b (±0.60) | 10.82 b (±0.77) | 9.56 ab (±0.43) | 9.20 ab (±0.87) | 7.52 a (±1.56) |
| TFAA | 17.21 a (±1.79) | 19.80 a (±0.03) | 25.69 b (±2.00) | 20.17 a (±0.84) | 20.22 a (±1.49) | 16.30 a (±2.43) |
| Amino Acid (μM/g) | Artemia | Com | W-W | W-T | T-W | T-T |
|---|---|---|---|---|---|---|
| Aspartic Acid | 0.68 (±0.02) | 1.29 (±0.30) | 0.75 (±0.06) | 0.87 (±0.02) | 0.74 (±0.02) | 0.93 (±0.05) |
| Glutamic Acid | 1.38 (±0.03) | 1.74 (±0.26) | 1.57 (±0.10) | 1.53 (±0.10) | 1.18 (±0.03) | 1.36 (±0.05) |
| Asparagine | 0.35 (±0.05) | 0.15 (±0.04) | 0.15 (±0.01) | 0.11 (±0.01) | 0.12 (±0.01) | 0.11 (±0.01) |
| Serine | 1.23 (±0.08) | 0.57 (±0.09) | 0.42 (±0.01) | 0.39 (±0.02) | 0.45 (±0.01) | 0.39 (±0.04) |
| Glutamine | 0.54 (±0.04) | 0.40 (±0.06) | 0.35 (±0.02) | 0.40 (±0.04) | 0.31 (±0.01) | 0.31 (±0.00) |
| Histidine | 1.52 (±0.12) | 0.54 (±0.11) | 1.26 (±0.22) | 0.55 (±0.03) | 0.51 (±0.03) | 0.39 (±0.00) |
| Glycine | 2.32 (±0.17) | 0.76 (±0.18) | 1.97 (±0.08) | 1.04 (±0.01) | 1.30 (±0.29) | 1.04 (±0.06) |
| Threonine | 0.87 (±0.03) | 0.96 (±0.22) | 0.93 (±0.09) | 0.87 (±0.07) | 0.85 (±0.08) | 0.73 (±0.06) |
| Arginine | 0.17 (±0.00) | 0.22 (±0.02) | 0.38 (±0.01) | 0.30 (±0.04) | 0.36 (±0.03) | 0.22 (±0.05) |
| Alanine | 1.66 (±0.08) | 1.32 (±0.26) | 0.83 (±0.10) | 0.92 (±0.07) | 0.87 (±0.03) | 0.73 (±0.06) |
| Tyrosine | 0.44 (±0.08) | 0.40 (±0.04) | 0.56 (±0.01) | 0.47 (±0.02) | 0.60 (±0.01) | 0.28 (±0.26) |
| Lysine | 0.69 (±0.12) | 1.46 (±0.26) | 2.16 (±0.06) | 1.85 (±0.31) | 1.82 (±0.40) | 1.53 (±0.62) |
| Methionine | 0.91 (±0.17) | 0.74 (±0.06) | 1.07 (±0.10) | 1.15 (±0.25) | 1.19 (±0.06) | 1.14 (±0.07) |
| Valine | 1.10 (±0.11) | 1.45 (±0.06) | 1.90 (±0.05) | 1.56 (±0.39) | 0.80 (±0.48) | 1.17 (±0.02) |
| Cysteine * Not detected | - | - | - | - | - | - |
| Tryptophan | 0.02 (±0.00) | 0.03 (±0.00) | 0.02 (±0.00) | 0.03 (±0.02) | 0.04 (±0.00) | 0.06 (±0.01) |
| Phenylalanine | 0.09 (±0.01) | 0.14 (±0.01) | 0.12 (±0.01) | 0.10 (±0.01) | 0.08 (±0.03) | 0.12 (±0.02) |
| Isoleucine | 0.08 (±0.01) | 0.11 (±0.01) | 0.09 (±0.01) | 0.07 (±0.04) | 0.02 (±0.00) | 0.11 (±0.02) |
| Leucine | 0.16 (±0.03) | 0.25 (±0.01) | 0.22 (±0.03) | 0.19 (±0.10) | 0.17 (±0.08) | 0.24 (±0.01) |
| Proline | 0.53 (±0.10) | 0.49 (±0.09) | 0.37 (±0.03) | 0.22 (±0.03) | 0.10 (±0.08) | 0.20 (±0.04) |
| IDAA | 5.61 (±0.30) | 5.90 (±0.68) | 8.14 (±0.16) | 6.67 (±0.57) | 5.84 (±0.92) | 5.71 (±0.86) |
| DAA | 9.12 (±0.41) | 7.12 (±1.22) | 6.95 (±0.14) | 5.97 (±0.29) | 5.66 (±0.37) | 5.35 (±0.25) |
| TFAA | 14.73 (±0.63) | 13.02 (±1.84) | 15.09 (±0.23) | 12.64 (±0.74) | 11.50 (±1.19) | 11.06 (±1.09) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molinari, G.S.; Wojno, M.; Terova, G.; Wick, M.; Riley, H.; Caminiti, J.T.; Kwasek, K. The Effect of the Species Source of Muscle and/or Digestive Enzymes on the Utilization of Fish Protein Hydrolysates as a Dietary Protein Source in First Feed for Larval Walleye (Sander vitreus). Animals 2024, 14, 2493. https://doi.org/10.3390/ani14172493
Molinari GS, Wojno M, Terova G, Wick M, Riley H, Caminiti JT, Kwasek K. The Effect of the Species Source of Muscle and/or Digestive Enzymes on the Utilization of Fish Protein Hydrolysates as a Dietary Protein Source in First Feed for Larval Walleye (Sander vitreus). Animals. 2024; 14(17):2493. https://doi.org/10.3390/ani14172493
Chicago/Turabian StyleMolinari, Giovanni S., Michal Wojno, Genciana Terova, Macdonald Wick, Hayden Riley, Jeffery T. Caminiti, and Karolina Kwasek. 2024. "The Effect of the Species Source of Muscle and/or Digestive Enzymes on the Utilization of Fish Protein Hydrolysates as a Dietary Protein Source in First Feed for Larval Walleye (Sander vitreus)" Animals 14, no. 17: 2493. https://doi.org/10.3390/ani14172493
APA StyleMolinari, G. S., Wojno, M., Terova, G., Wick, M., Riley, H., Caminiti, J. T., & Kwasek, K. (2024). The Effect of the Species Source of Muscle and/or Digestive Enzymes on the Utilization of Fish Protein Hydrolysates as a Dietary Protein Source in First Feed for Larval Walleye (Sander vitreus). Animals, 14(17), 2493. https://doi.org/10.3390/ani14172493






