Association between Gut Microbiota and Metabolic Health and Obesity Status in Cats
Abstract
:Simple Summary
Abstract
1. Introduction
| In Humans (Smith et al., 2019) [7] | In Cats (Okada et al., 2019) [9] | In Cats (In This Study) |
|---|---|---|
| BMI > 30 kg/m2 | BCS > 7/9 | BCS > 8/9 BMI > 30 kg/m2 |
| Meets the above conditions and at least two of the following: | ||
| Waist circumference > 102 cm in men | Low adiponectin (<3 μg/mL) | Low adiponectin (<1.53 μg/mL) |
| Triglycerides > 150 mg/dL | Triglycerides > 165 mg/dL | Triglycerides > 165 mg/dL |
| HDL cholesterol < 40 mg/dL | High SAA (>200 ng/mL) | |
| Blood pressure > 130/85 mmHg | ||
| Fasting glucose > 100 mg/dL | ||
2. Materials and Methods
2.1. Study Design
2.2. MHO and MUO Phenotypes in Patients with Obesity
2.3. Dietary Data and Adaptation Period
2.4. Blood Sampling and Biochemical Data
2.5. Stool Collection, DNA Extraction, PCR Amplification, and Bioinformatic Data Analysis
2.6. Statistical Analyses
3. Results
3.1. Cat Characteristics and Grouping
3.2. Blood Biochemical Data
3.3. Fecal Microbiota Analysis
3.4. Relative Abundance
3.5. Alpha and Beta Diversity Indices
3.6. Principal Component Analysis of Fecal Bacteria and BW, BMI, Adiponectin, TG, TChol, and fSAA Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- U.S. Pet Obesity Rates Plateau and Nutritional Confusion Grows. Available online: https://petobesityprevention.org/2018 (accessed on 23 June 2024).
- Cave, N.J.; Allan, F.J.; Schokkenbroek, S.L.; Metekohy, C.A.; Pfeiffer, D.U. A cross-sectional study to compare changes in the prevalence and risk factors for feline obesity between 1993 and 2007 in New Zealand. Prev. Vet. Med. 2012, 107, 121–133. [Google Scholar] [CrossRef]
- Kipperman, B.S.; German, A.J. The Responsibility of Veterinarians to Address Companion Animal Obesity. Animals 2018, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Anekwe, C.V.; Jarrell, A.R.; Townsend, M.J.; Gaudier, G.I.; Hiserodt, J.M.; Stanford, F.C. Socioeconomics of obesity. Curr. Obes. Rep. 2020, 9, 272–279. [Google Scholar] [CrossRef]
- Blüher, M. Metabolically healthy obesity. Endocr. Rev. 2020, 41, bnaa004. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.I.; Mittendorfer, B.; Klein, S. Metabolically healthy obesity: Facts and fantasies. J. Clin. Investig. 2019, 129, 3978–3989. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, H.; Janssens, G.P.; de Rooster, H.; Polis, I.; Peters, I.; Ducatelle, R.; Nguyen, P.; Buyse, J.; Rochus, K.; Xu, J.; et al. The cat as a model for human obesity: Insights into depot-specific inflammation associated with feline obesity. Br. J. Nutr. 2013, 110, 1326–1335. [Google Scholar] [CrossRef]
- Okada, Y.; Ueno, H.; Mizorogi, T.; Ohara, K.; Kawasumi, K.; Arai, T. Diagnostic criteria for obesity disease in cats. Front. Vet. Sci. 2019, 6, 284. [Google Scholar] [CrossRef]
- Chartrand, D.J.; Murphy-Després, A.; Alméras, N.; Lemieux, I.; Larose, E.; Després, J.P. Overweight, obesity, and CVD risk: A focus on visceral/ectopic fat. Curr. Atheroscler. Rep. 2022, 24, 185–195. [Google Scholar] [CrossRef]
- Ross, R.; Neeland, I.J.; Yamashita, S.; Shai, I.; Seidell, J.; Magni, P.; Santos, R.D.; Arsenault, B.; Cuevas, A.; Hu, F.B.; et al. Waist circumference as a vital sign in clinical practice: A Consensus Statement from the IAS and ICCR Working Group on Visceral Obesity. Nat. Rev. Endocrinol. 2020, 16, 177–189. [Google Scholar] [CrossRef]
- Okada, Y.; Kobayashi, M.; Sawamura, M.; Arai, T. Comparison of visceral fat accumulation and metabolome markers among cats of varying BCS and novel classification of feline obesity and metabolic syndrome. Front. Vet. Sci. 2017, 4, 17. [Google Scholar] [CrossRef]
- Aljuraiban, G.S.; Alfhili, M.A.; Aldhwayan, M.M.; Aljazairy, E.A.; Al-Musharaf, S. Shared and distinct gut microbial profiles in Saudi women with metabolically healthy and unhealthy obesity. Microorganisms 2023, 11, 1430. [Google Scholar] [CrossRef]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef]
- Kieler, I.N.; Osto, M.; Hugentobler, L.; Puetz, L.; Gilbert, M.T.P.; Hansen, T.; Pedersen, O.; Reusch, C.E.; Zini, E.; Lutz, T.A.; et al. Diabetic cats have decreased gut microbial diversity and a lack of butyrate producing bacteria. Sci. Rep. 2019, 9, 4822. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Brinker, E.; Graff, E.C.; Cao, W.; Gross, A.L.; Johnson, A.K.; Zhang, C.; Martin, D.R.; Wang, X. Whole-genome shotgun metagenomic sequencing reveals distinct gut microbiome signatures of obese cats. Microbiol. Spectr. 2022, 10, e0083722. [Google Scholar] [CrossRef]
- Tal, M.; Weese, J.S.; Gomez, D.E.; Hesta, M.; Steiner, J.M.; Verbrugghe, A. Bacterial fecal microbiota is only minimally affected by a standardized weight loss plan in obese cats. BMC Vet. Res. 2020, 16, 112. [Google Scholar] [CrossRef] [PubMed]
- Laflamme, D.P. Development and validation of a body condition score system for cats: Clinical tool. Feline Pract. 1997, 25, 13–18. [Google Scholar]
- National Research Council. Nutrient Requirements of Dogs and Cats; National Academies Press: Washington, DC, USA, 2006. [Google Scholar]
- Baldwin, K.; Bartges, J.; Buffington, T.; Freeman, L.M.; Grabow, M.; Legred, J.; Ostwald, D., Jr. AAHA nutritional assessment guidelines for dogs and cats. J. Am. Anim. Hosp. Assoc. 2010, 4, 285–296. [Google Scholar] [CrossRef]
- Nelson, R.W.; Himsel, C.A.; Feldman, E.C.; Bottoms, G.D. Glucose tolerance and insulin response in normal-weight and obese cats. Am. J. Vet. Res. 1990, 51, 1357–1362. [Google Scholar] [CrossRef]
- Verbrugghe, A.; Hesta, M.; Daminet, S.; Polis, I.; Holst, J.J.; Buyse, J.; Wuyts, B.; Janssens, G.P. Propionate absorbed from the colon acts as gluconeogenic substrate in a strict carnivore, the domestic cat (Felis catus). J. Anim. Physiol. Anim. Nutr. 2012, 96, 1054–1064. [Google Scholar] [CrossRef]
- Xia, Y.; Sun, J. Bioinformatic and Statistical Analysis of Microbiome Data: From Raw Sequences to Advanced Modeling with QIIME 2 and R; Springer Nature: Berlin, Germany, 2023. [Google Scholar]
- Gong, H.; Gao, H.; Ren, Q.; He, J. The abundance of bifidobacterium in relation to visceral obesity and serum uric acid. Sci. Rep. 2022, 12, 13073. [Google Scholar] [CrossRef] [PubMed]
- An, H.M.; Park, S.Y.; Lee, D.K.; Kim, J.R.; Cha, M.K.; Lee, S.W.; Lim, H.T.; Kim, K.J.; Ha, N.J. Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids Health Dis. 2011, 10, 116. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.N.; Yu, Q.F.; Fu, N.; Liu, X.W.; Lu, F.G. Effects of four bifidobacteria on obesity in high-fat diet induced rats. World J. Gastroenterol. 2010, 16, 3394–3401. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wu, Z. Gut probiotics and health of dogs and cats: Benefits, applications, and underlying mechanisms. Microorganisms 2023, 11, 2452. [Google Scholar] [CrossRef]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef]
- Hooda, S.; Vester Boler, B.M.; Kerr, K.R.; Dowd, S.E.; Swanson, K.S. The gut microbiome of kittens is affected by dietary protein: Carbohydrate ratio and associated with blood metabolite and hormone concentrations. Br. J. Nutr. 2013, 109, 1637–1646. [Google Scholar] [CrossRef]
- Bell, E.T.; Suchodolski, J.S.; Isaiah, A.; Fleeman, L.M.; Cook, A.K.; Steiner, J.M.; Mansfield, C.S. Faecal microbiota of cats with insulin-treated diabetes mellitus. PLoS ONE 2014, 9, e108729. [Google Scholar] [CrossRef]
- Suchodolski, J.S.; Foster, M.L.; Sohail, M.U.; Leutenegger, C.; Queen, E.V.; Steiner, J.M.; Marks, S.L. The fecal microbiome in cats with diarrhea. PLoS ONE 2015, 10, e0127378. [Google Scholar] [CrossRef]
- Fischer, M.M.; Kessler, A.M.; Kieffer, D.A.; Knotts, T.A.; Kim, K.; Wei, A.; Ramsey, J.J.; Fascetti, A.J. Effects of obesity, energy restriction and neutering on the faecal microbiota of cats. Br. J. Nutr. 2017, 118, 513–524. [Google Scholar] [CrossRef]
- Xu, Z.; Jiang, W.; Huang, W.; Lin, Y.; Chan, F.K.; Ng, S.C. Gut microbiota in patients with obesity and metabolic disorders—A systematic review. Genes Nutr. 2022, 17, 2. [Google Scholar] [CrossRef]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Loviselli, A.; Manzin, A.; et al. Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 2021, 11, 5532. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.S.; Chen, L.; Yang, J.; Wu, Z.H.; Wang, X.Y.; Zhang, Q.; Liu, W.J.; Liu, H.X. Altered gut microbial profile accompanied by abnormal fatty acid metabolism activity exacerbates endometrial cancer progression. Microbiol. Spectr. 2022, 10, e0261222. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; van den Munckhof, I.C.; Chen, L.; Bonder, M.J.; Schraa, K.; Rutten, J.H.; Riksen, N.P.; de Graaf, J.; Oosting, M.; Sanna, S.; et al. Gut microbial associations to plasma metabolites linked to cardiovascular phenotypes and risk. Circ. Res. 2019, 124, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.N.; Yao, Y.; Ju, S.Y. Short chain fatty acids and fecal microbiota abundance in humans with obesity: A systematic review and meta-analysis. Nutrients 2019, 11, 2512. [Google Scholar] [CrossRef]
- Tirosh, A.; Calay, E.S.; Tuncman, G.; Claiborn, K.C.; Inouye, K.E.; Eguchi, K.; Alcala, M.; Rathaus, M.; Hollander, K.; Ron, I.; et al. The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci. Transl. Med. 2019, 11, eaav0120. [Google Scholar] [CrossRef]
- Choo, J.; Leong, L.; Rogers, G. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015, 5, 16350. [Google Scholar] [CrossRef]
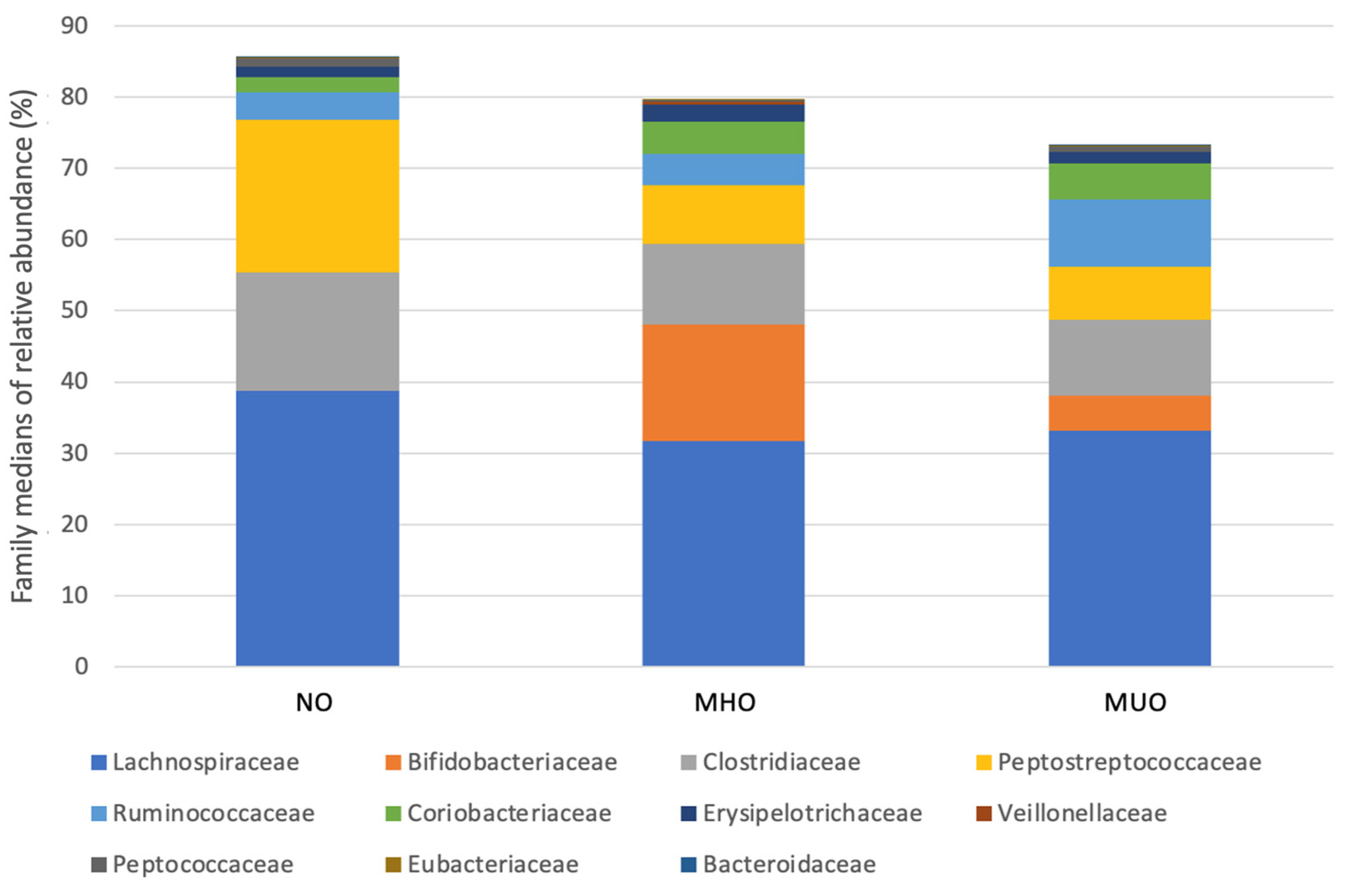
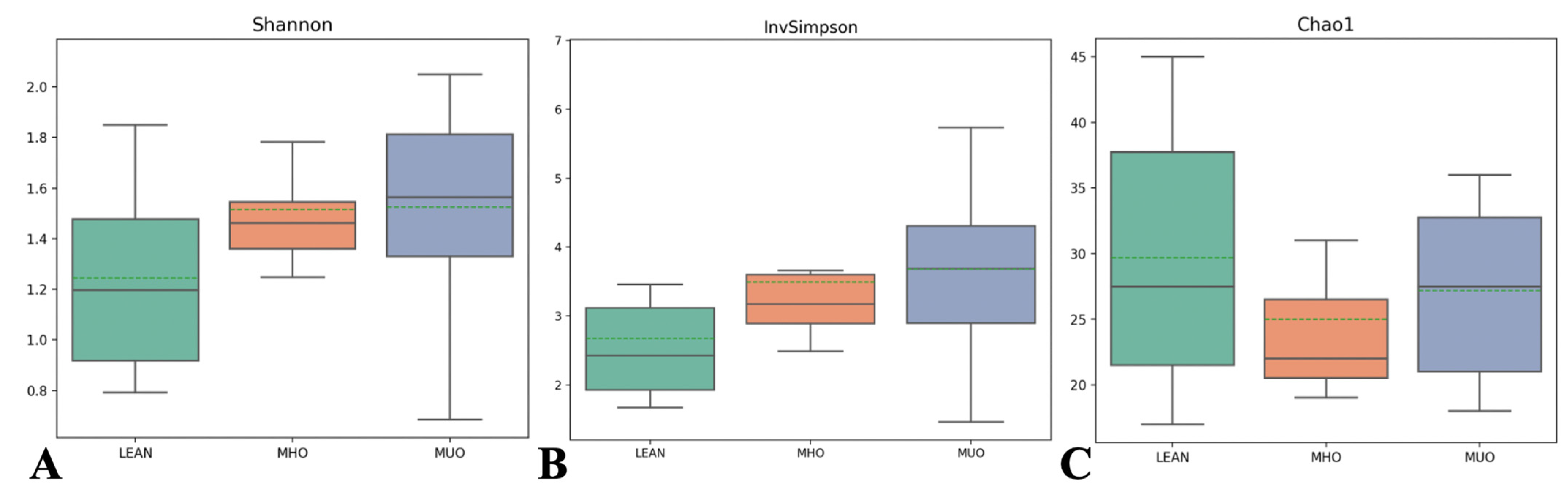
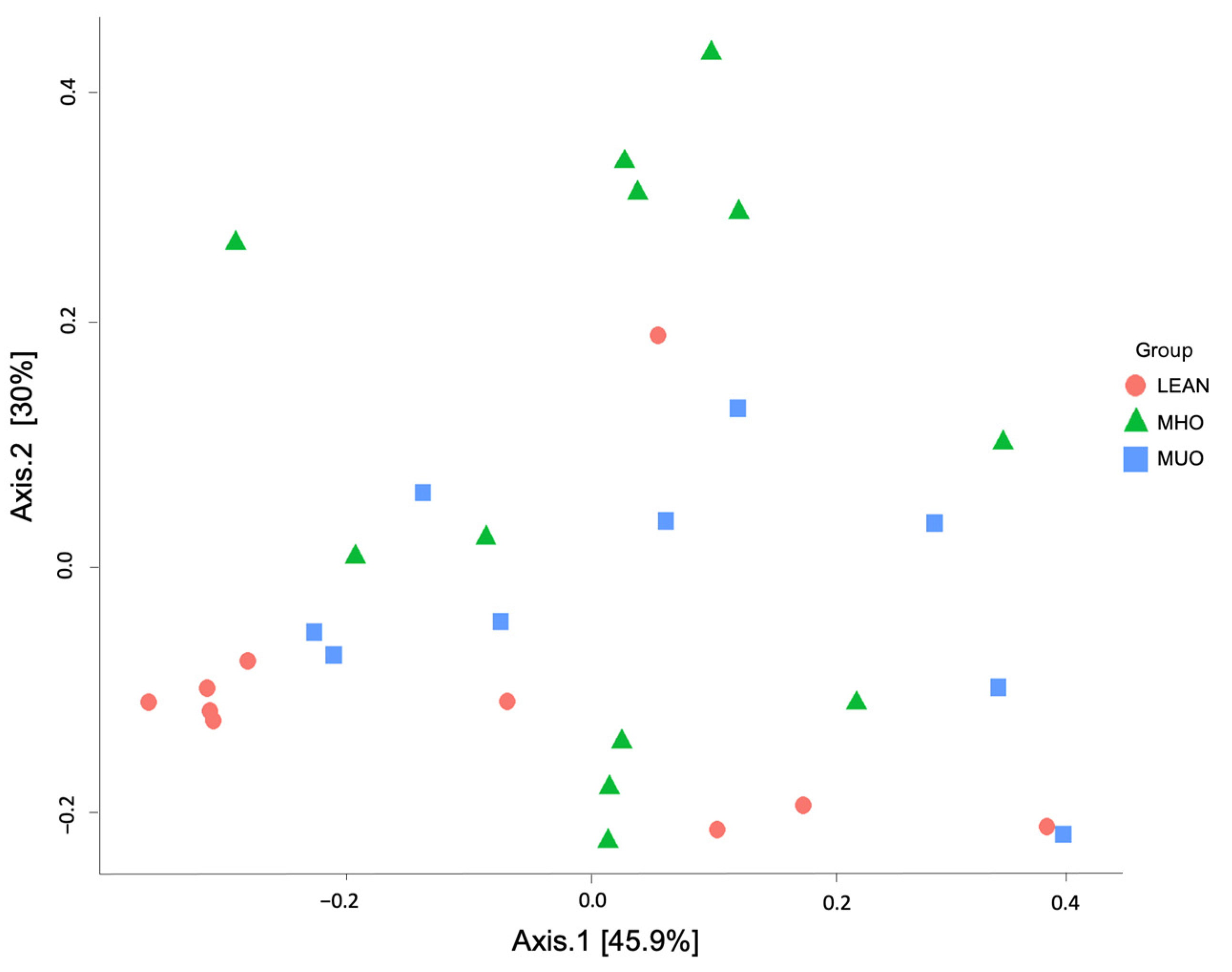
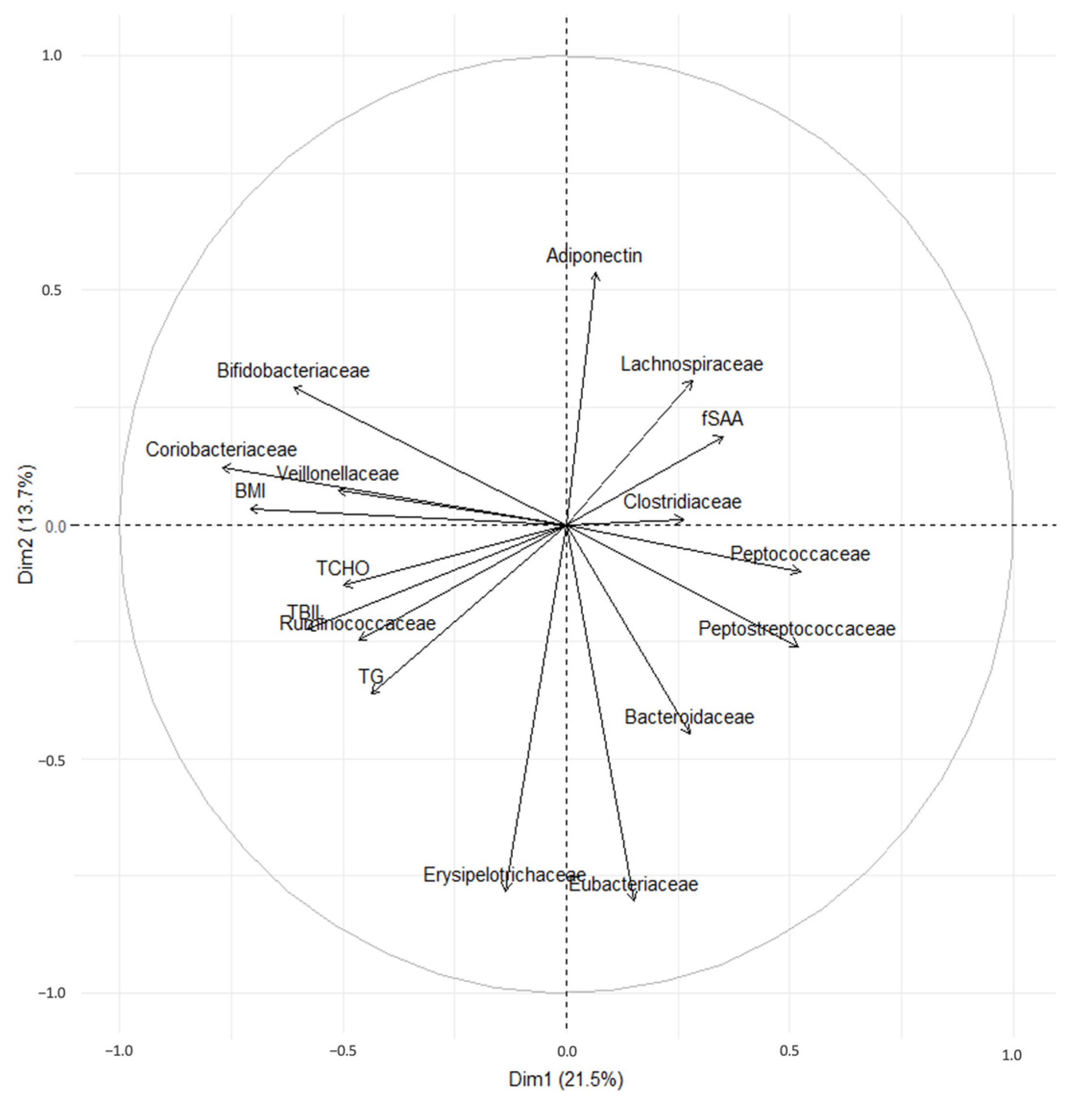
| NO | MHO | MUO | |
|---|---|---|---|
| N | 10 | 12 | 9 |
| Sex | |||
| Castrated male | 7 | 10 | 5 |
| Spayed female | 3 | 2 | 4 |
| Age | 4 (2–9) | 5 (2–9) | 5 (3–9) |
| BW (kg) | 5.0 ± 0.76 b | 8.4 ± 1.40 a | 7.3 ± 0.96 a |
| BMI (kg/m2) | 19.5 ± 3.93 b | 37.3 ± 4.68 a | 39.1 ± 4.79 a |
| Girth (cm) | 37.7 ± 1.95 b | 49.0 ± 3.52 a | 51.2 ± 4.89 a |
| BCS (1–9/9) | 4 (4–5) | 9 (8–9) | 9 (8–9) |
| NO | MHO | MUO | |
|---|---|---|---|
| TG (mg/dL) | 93 (56–188) a,b | 107.4 (60–272) a | 279.7 (169–375) b |
| Adiponectin (μg/mL) | 1.5 ± 0.3 | 1.5 ± 0.8 | 0.9 ± 0.5 |
| fSAA (ng/mL) | 226.8 (7.1–805.4) | 128.2 (1.6–465.2) | 111.57 (0.0–712.6) |
| NO vs. MHO | NO vs. MUO | MHO vs. MUO | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | SE | p | FDR | E | SE | p | FDR | E | SE | p | FDR | |
| Phylum (4/7) | −1.320 | 3.639 | 0.002 | 0.005 | −0.997 | 2.025 | 0.006 | 0.022 | 0.750 | 3.349 | 0.256 | 0.341 |
| Actinobacteria | 1.200 | 3.565 | 0.002 | 0.005 | 0.778 | 2.005 | 0.027 | 0.055 | −0.753 | 3.321 | 0.227 | 0.341 |
| Firmicutes | 0.482 | 0.358 | 0.598 | 0.797 | 0.374 | 0.419 | 0.935 | 0.935 | −0.235 | 0.104 | 0.256 | 0.341 |
| Bacteroidetes | 0.375 | 0.187 | 0.920 | 0.920 | 0.455 | 0.211 | 0.901 | 0.935 | 0.310 | 0.060 | 0.914 | 0.914 |
| Proteobacteria | ||||||||||||
| Class (7/19) | −1.168 | 2.831 | 0.002 | 0.013 | −0.660 | 1.458 | 0.003 | 0.023 | 0.811 | 2.702 | 0.227 | 0.501 |
| Actinomycetes | −0.442 | 1.188 | 0.198 | 0.278 | −0.697 | 0.540 | 0.236 | 0.413 | 0.144 | 1.301 | 0.915 | 0.915 |
| Bacilli | 0.484 | 0.355 | 0.468 | 0.468 | 0.373 | 0.416 | 1.000 | 1.000 | −0.242 | 0.104 | 0.227 | 0.501 |
| Bacteroidia | 1.295 | 4.310 | 0.004 | 0.013 | 1.061 | 2.652 | 0.027 | 0.079 | −0.624 | 4.018 | 0.286 | 0.501 |
| Clostridia | −1.125 | 1.168 | 0.008 | 0.019 | −1.057 | 0.948 | 0.034 | 0.079 | 0.260 | 1.252 | 0.722 | 0.915 |
| Coriobacteriia | −0.491 | 0.673 | 0.468 | 0.468 | −0.495 | 1.394 | 0.683 | 0.956 | −0.231 | 1.351 | 0.887 | 0.915 |
| Erysipelotrichia | −0.615 | 0.282 | 0.070 | 0.123 | 0.191 | 0.047 | 0.825 | 0.963 | 0.633 | 0.295 | 0.036 | 0.250 |
| Negativicutes | ||||||||||||
| Order (7/38) | 0.484 | 0.355 | 0.468 | 0.468 | 0.373 | 0.416 | 1.000 | 1.000 | −0.242 | 0.104 | 0.227 | 0.501 |
| Bacteroidales | −1.169 | 2.829 | 0.002 | 0.011 | −0.661 | 1.458 | 0.003 | 0.023 | 0.811 | 2.700 | 0.227 | 0.501 |
| Bifidobacteriales | −1.125 | 1.168 | 0.008 | 0.019 | −1.057 | 0.948 | 0.034 | 0.079 | 0.260 | 1.252 | 0.722 | 0.915 |
| Coriobacteriales | −0.491 | 0.673 | 0.468 | 0.468 | −0.495 | 1.394 | 0.683 | 0.836 | −0.231 | 1.351 | 0.887 | 0.915 |
| Erysipelotrichales | 1.295 | 4.310 | 0.004 | 0.013 | 1.061 | 2.652 | 0.027 | 0.079 | −0.624 | 4.018 | 0.286 | 0.501 |
| Eubacteriales | −0.443 | 1.188 | 0.175 | 0.245 | −0.698 | 0.540 | 0.204 | 0.356 | 0.144 | 1.301 | 0.915 | 0.915 |
| Lactobacillales | −0.640 | 0.283 | 0.044 | 0.078 | 0.053 | 0.043 | 0.716 | 0.836 | 0.635 | 0.295 | 0.023 | 0.164 |
| Veillonellales | ||||||||||||
| Family (11/72) | 0.743 | 0.118 | 0.323 | 0.435 | −0.049 | −0.215 | 0.806 | 0.806 | −0.589 | 0.166 | 0.831 | 0.887 |
| Eubacteriaceae | −0.640 | 0.283 | 0.044 | 0.128 | 0.053 | 0.043 | 0.716 | 0.806 | 0.635 | 0.295 | 0.023 | 0.216 |
| Veillonellaceae | −0.491 | 0.673 | 0.468 | 0.515 | −0.495 | 1.394 | 0.683 | 0.806 | −0.231 | 1.351 | 0.887 | 0.887 |
| Erysipelotrichaceae | 0.055 | 1.932 | 0.356 | 0.435 | 0.376 | 1.559 | 0.288 | 0.453 | 0.204 | 2.168 | 0.887 | 0.887 |
| Clostridiaceae | 0.574 | 4.509 | 0.187 | 0.385 | 0.309 | 4.925 | 0.253 | 0.453 | −0.250 | 4.437 | 0.722 | 0.887 |
| Lachnospiraceae | −0.424 | 1.108 | 0.692 | 0.692 | −1.824 | 1.062 | 0.002 | 0.018 | −0.758 | 1.224 | 0.039 | 0.216 |
| Ruminococcaceae | 0.827 | 2.968 | 0.210 | 0.385 | 0.470 | 3.639 | 0.288 | 0.453 | −0.299 | 2.443 | 0.887 | 0.887 |
| Peptostreptococcaceae | 0.505 | 0.263 | 0.355 | 0.435 | 0.344 | 0.312 | 0.806 | 0.806 | −0.341 | 0.092 | 0.135 | 0.497 |
| Bacteroidaceae | −1.125 | 1.168 | 0.008 | 0.046 | −1.057 | 0.948 | 0.034 | 0.124 | 0.260 | 1.252 | 0.722 | 0.887 |
| Coriobacteriaceae | 0.941 | 0.157 | 0.047 | 0.128 | 0.598 | 0.170 | 0.161 | 0.444 | −0.331 | 0.132 | 0.609 | 0.887 |
| Peptococcaceae | −1.169 | 2.829 | 0.002 | 0.017 | −0.661 | 1.458 | 0.003 | 0.018 | 0.811 | 2.700 | 0.227 | 0.624 |
| Bifidobacteriaceae | ||||||||||||
| Genus (16/88) | 0.479 | 0.314 | 0.320 | 0.565 | 0.296 | 0.381 | 0.870 | 0.928 | −0.308 | 0.142 | 0.239 | 0.893 |
| Bacteroides | −1.164 | 3.191 | 0.001 | 0.020 | −0.668 | 1.619 | 0.003 | 0.053 | 0.811 | 3.040 | 0.227 | 0.893 |
| Bifidobacterium | 0.165 | 3.626 | 0.598 | 0.638 | −0.182 | 4.604 | 0.683 | 0.850 | −0.336 | 4.247 | 0.670 | 0.893 |
| Blautia | 0.949 | 4.690 | 0.065 | 0.296 | 0.749 | 5.454 | 0.086 | 0.410 | −0.123 | 3.729 | 0.831 | 0.943 |
| Clostridium | −0.818 | 0.740 | 0.129 | 0.345 | −0.455 | 0.480 | 0.514 | 0.822 | 0.492 | 0.768 | 0.394 | 0.893 |
| Collinsella | 0.012 | 0.020 | 0.947 | 0.947 | 0.528 | 0.018 | 0.413 | 0.735 | 0.461 | 0.019 | 0.477 | 0.893 |
| Coprococcus | 0.268 | 0.399 | 0.210 | 0.481 | 0.433 | 0.371 | 0.624 | 0.850 | 0.145 | 0.285 | 0.434 | 0.893 |
| Dorea | −0.239 | 0.033 | 0.260 | 0.519 | 0.201 | 0.019 | 0.307 | 0.614 | 0.338 | 0.035 | 0.617 | 0.893 |
| Erysipelatoclostridium | −0.414 | 0.627 | 0.510 | 0.582 | −0.509 | 0.688 | 0.935 | 0.935 | −0.083 | 0.770 | 0.943 | 0.943 |
| Eubacterium | −0.607 | 0.029 | 0.077 | 0.296 | −0.533 | 0.035 | 0.167 | 0.446 | 0.002 | 0.040 | 0.886 | 0.943 |
| Faecalibacterium | −0.835 | 0.082 | 0.092 | 0.296 | −1.024 | 0.054 | 0.048 | 0.381 | 0.192 | 0.090 | 0.940 | 0.943 |
| Gemmiger | 0.196 | 0.013 | 0.466 | 0.582 | −0.535 | 0.213 | 0.744 | 0.850 | −0.580 | 0.193 | 0.118 | 0.893 |
| Lachnoclostridium | −0.738 | 0.257 | 0.353 | 0.565 | −0.986 | 0.180 | 0.131 | 0.418 | 0.108 | 0.289 | 0.668 | 0.893 |
| Peptoclostridium | 1.034 | 0.050 | 0.078 | 0.296 | 0.887 | 0.057 | 0.305 | 0.614 | −0.236 | 0.014 | 0.534 | 0.893 |
| Robinsoniella | 0.027 | 1.624 | 0.510 | 0.582 | −0.747 | 1.938 | 0.102 | 0.410 | −0.918 | 1.606 | 0.055 | 0.880 |
| Ruminococcus | −0.502 | 0.331 | 0.468 | 0.582 | 0.145 | 0.192 | 0.744 | 0.850 | 0.637 | 0.316 | 0.433 | 0.893 |
| Subdoligranulum | −1.320 | 3.639 | 0.002 | 0.005 | −0.997 | 2.025 | 0.006 | 0.022 | 0.750 | 3.349 | 0.256 | 0.341 |
| Group | Dissimilarity | ANOSIM(R) | p-Value |
|---|---|---|---|
| NO VS. MHO | Bray–Curtis | 0.061411 | 0.155 |
| weighted UniFrac | 0.10961 | 0.068 | |
| unweighted UniFrac | 0.068168 | 0.177 | |
| NO VS. MUO | Bray–Curtis | 0.034568 | 0.24 |
| weighted UniFrac | 0.05679 | 0.168 | |
| unweighted UniFrac | 0.010425 | 0.393 | |
| NO VS. MUO | Bray–Curtis | −0.068446 | 0.826 |
| weighted UniFrac | −0.046296 | 0.713 | |
| unweighted UniFrac | −0.055192 | 0.781 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeon, K.-D.; Kim, S.-M.; Kim, J.-H. Association between Gut Microbiota and Metabolic Health and Obesity Status in Cats. Animals 2024, 14, 2524. https://doi.org/10.3390/ani14172524
Yeon K-D, Kim S-M, Kim J-H. Association between Gut Microbiota and Metabolic Health and Obesity Status in Cats. Animals. 2024; 14(17):2524. https://doi.org/10.3390/ani14172524
Chicago/Turabian StyleYeon, Kyu-Duk, Sun-Myung Kim, and Jung-Hyun Kim. 2024. "Association between Gut Microbiota and Metabolic Health and Obesity Status in Cats" Animals 14, no. 17: 2524. https://doi.org/10.3390/ani14172524
APA StyleYeon, K.-D., Kim, S.-M., & Kim, J.-H. (2024). Association between Gut Microbiota and Metabolic Health and Obesity Status in Cats. Animals, 14(17), 2524. https://doi.org/10.3390/ani14172524







