The Use of Genomic Screening for the Detection of Chromosomal Abnormalities in the Domestic Horse: Five New Cases of 65,XXY Syndrome in the Pura Raza Español Breed
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Enrolment, Parentage Testing, and Cytogenetic Screening
2.2. Resampling of CNA Candidates
2.3. STR-Based Chromosomal Diagnosis
2.4. SNP Genotyping and CNA Genomic Analysis
2.5. Cell Culture and Fluorescent In Situ Hybridization (FISH) Chromosomal Analysis
2.6. Genetic Analysis and Inheritance Estimation
3. Results
3.1. Detection of 65,XXY CNA by Molecular Screening
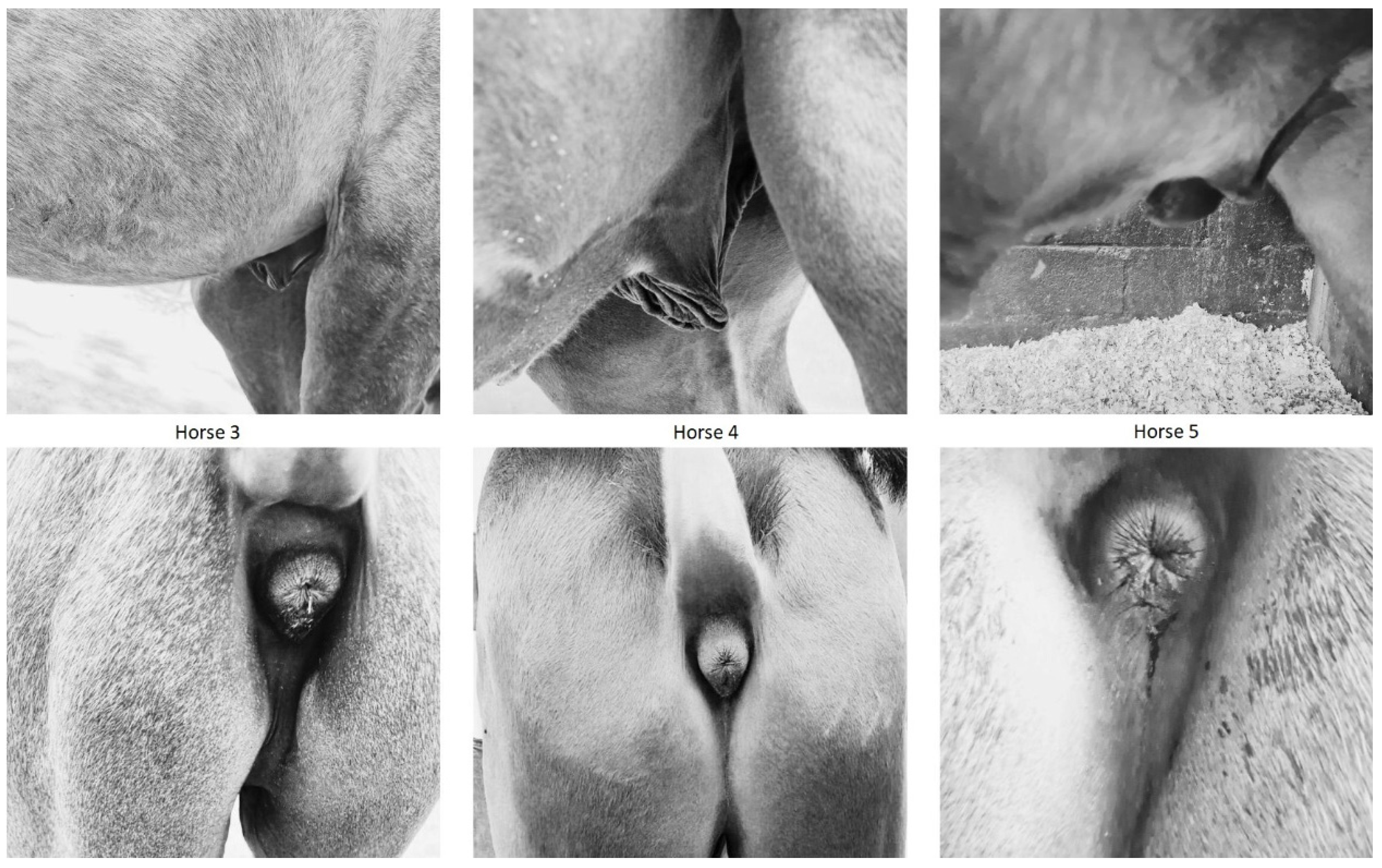
3.2. Animal Phenotyping
3.3. Sex-Linked STR Chromosomal Analysis
3.4. SNP-Based Chromosomal Analysis
3.5. In Situ Hybridization Analysis
3.6. Genetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Payne, H.W.; Ellsworth, K.; DeGroot, A. Aneuploidy in an infertile mare. J. Am. Vet. Med. Assoc. 1968, 153, 1293–1299. [Google Scholar]
- Basrur, P.K.; Kanagawa, H.; Gilman, J.P. An equine intersex with unilateral gonadal agenesis. Can. J. Comp. Med. 1969, 33, 297–306. [Google Scholar] [PubMed]
- Power, M.M. Chromosomes of the horse. In Domestic Animal Cytogenetics; Advances in Veterinary Science and Comparative Medicine; Academic Press, Inc.: San Diego, CA, USA, 1990; Volume 34, pp. 131–167. [Google Scholar]
- Bugno-Poniewierska, M.; Raudsepp, T. Horse Clinical Cytogenetics: Recurrent Themes and Novel Findings. Animals 2021, 11, 831. [Google Scholar] [CrossRef] [PubMed]
- Lear, T.L.; McGee, R.B. Disorders of sexual development in the domestic horse, Equus caballus. Sex. Dev. 2012, 6, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Albarella, S.; De Lorenzi, L.; Catone, G.; Magi, G.E.; Petrucci, L.; Vullo, C.; D’Anza, E.; Parma, P.; Raudsepp, T.; Ciotola, F.; et al. Diagnosis of XX/XY Blood Cell Chimerism at a Low Percentage in Horses. J. Equine Vet. Sci. 2018, 69, 129–135. [Google Scholar] [CrossRef]
- Anaya, G.; Moreno-Millan, M.; Bugno-Poniewierska, M.; Pawlina, K.; Membrillo, A.; Molina, A.; Demyda-Peyras, S. Sex reversal syndrome in the horse: Four new cases of feminization in individuals carrying a 64,XY SRY negative chromosomal complement. Anim. Reprod. Sci. 2014, 151, 22–27. [Google Scholar] [CrossRef]
- Groth, K.A.; Skakkebæk, A.; Høst, C.; Gravholt, C.H.; Bojesen, A. Klinefelter syndrome—A clinical update. J. Clin. Endocrinol. Metab. 2013, 98, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Bugno-Poniewierska, M.; Jankowska, M.; Raudsepp, T.; Kowalska, K.; Pawlina-Tyszko, K.; Szmatola, T. Molecular cytogenetic screening of sex chromosome abnormalities in young horse populations. Equine Vet. J. 2024, 56, 786–795. [Google Scholar] [CrossRef]
- Makinen, A.; Katila, T.; Andersson, M.; Gustavsson, I. Two sterile stallions with XXY-syndrome. Equine Vet. J. 2000, 32, 358–360. [Google Scholar] [CrossRef]
- Iannuzzi, L.; Di Meo, G.P.; Perucatti, A.; Spadetta, M.; Incarnato, D.; Parma, P.; Iannuzzi, A.; Ciotola, F.; Peretti, V.; Perrotta, G.; et al. Clinical, cytogenetic and molecular studies on sterile stallion and mare affected by XXY and sex reversal syndromes, respectively. Caryologia 2004, 57, 400–404. [Google Scholar]
- Kakoi, H.; Hirota, K.; Gawahara, H.; Kurosawa, M.; Kuwajima, M. Genetic diagnosis of sex chromosome aberrations in horses based on parentage test by microsatellite DNA and analysis of X- and Y-linked markers. Equine Vet. J. 2005, 37, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Laseca, N.; Anaya, G.; Pena, Z.; Pirosanto, Y.; Molina, A.; Demyda Peyras, S. Impaired Reproductive Function in Equines: From Genetics to Genomics. Animals 2021, 11, 393. [Google Scholar] [CrossRef]
- Anaya, G.; Fernandez, M.E.; Valera, M.; Molina, A.; Azcona, F.; Azor, P.; Sole, M.; Moreno-Millan, M.; Demyda-Peyras, S. Prevalence of twin foaling and blood chimaerism in purebred Spanish horses. Vet. J. 2018, 234, 142–144. [Google Scholar] [CrossRef]
- Demyda-Peyras, S.; Membrillo, A.; Bugno-Poniewierska, M.; Pawlina, K.; Anaya, G.; Moreno-Millan, M. The use of molecular and cytogenetic methods as a valuable tool in the detection of chromosomal abnormalities in horses: A case of sex chromosome chimerism in a Spanish purebred colt. Cytogenet. Genome Res. 2013, 141, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Demyda-Peyras, S.; Anaya, G.; Bugno-Poniewierska, M.; Pawlina, K.; Membrillo, A.; Valera, M.; Moreno-Millan, M. The use of a novel combination of diagnostic molecular and cytogenetic approaches in horses with sexual karyotype abnormalities: A rare case with an abnormal cellular chimerism. Theriogenology 2014, 81, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Pirosanto, Y.; Laseca, N.; Valera, M.; Molina, A.; Moreno-Millan, M.; Bugno-Poniewierska, M.; Ross, P.; Azor, P.; Demyda-Peyras, S. Screening and detection of chromosomal copy number alterations in the domestic horse using SNP-array genotyping data. Anim. Genet. 2021, 52, 431–439. [Google Scholar] [CrossRef]
- Shilton, C.A.; Kahler, A.; Davis, B.W.; Crabtree, J.R.; Crowhurst, J.; McGladdery, A.J.; Wathes, D.C.; Raudsepp, T.; de Mestre, A.M. Whole genome analysis reveals aneuploidies in early pregnancy loss in the horse. Sci. Rep. 2020, 10, 13314. [Google Scholar] [CrossRef]
- Nogueira, P.P.O.; Amorim, G.; Oliveira, O.M.; Demyda-Peyras, S.; Santos, B.M.; Mota, L. Sex Reversal Syndrome in an Egyptian Arabian Horse Detected Using Genomic Data—A case report. J. Equine Vet. Sci. 2021, 104, 103692. [Google Scholar] [CrossRef]
- Raudsepp, T.; Das, P.J.; Avila, F.; Chowdhary, B.P. The Pseudoautosomal Region and Sex Chromosome Aneuploidies in Domestic Species. Sex. Dev. 2012, 6, 72–83. [Google Scholar] [CrossRef]
- Raudsepp, T.; Chowdhary, B.P. The horse pseudoautosomal region (PAR): Characterization and comparison with the human, chimp and mouse PARs. Cytogenet. Genome Res. 2008, 121, 102–109. [Google Scholar] [CrossRef]
- Janecka, J.E.; Davis, B.W.; Ghosh, S.; Paria, N.; Das, P.J.; Orlando, L.; Schubert, M.; Nielsen, M.K.; Stout, T.A.E.; Brashear, W.; et al. Horse Y chromosome assembly displays unique evolutionary features and putative stallion fertility genes. Nat. Commun. 2018, 9, 2945. [Google Scholar] [CrossRef] [PubMed]
- Solé, M.; Valera, M.; Fernández, J. Genetic structure and connectivity analysis in a large domestic livestock meta-population: The case of the Pura Raza Español horses. J. Anim. Breed. Gen. 2018, 135, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Demyda-Peyras, S.; Laseca, N.; Anaya, G.; Kij-Mitka, B.; Molina, A.; Karlau, A.; Valera, M. Prevalence of Sex-Related Chromosomal Abnormalities in a Large Cohort of Spanish Purebred Horses. Animals 2023, 13, 539. [Google Scholar] [CrossRef]
- ANCCE. Pura Raza Español Horse Breed Studbook. Available online: https://www.lgancce.com/web/ (accessed on 28 August 2024).
- ISAG. Equine Genetics and Thoroughbred Parentage Testing Guideline. Available online: https://www.isag.us/committee_members.asp?comm=IS-EGTP (accessed on 1 June 2024).
- Dimsoski, P. Development of a 17-plex microsatellite polymerase chain reaction kit for genotyping horses. Croat. Med. J. 2003, 44, 332–335. [Google Scholar]
- Hasegawa, T.; Sato, F.; Ishida, N.; Fukushima, Y.; Mukoyama, H. Sex determination by simultaneous amplification of equine SRY and amelogenin genes. J. Vet. Med. Sci. 2000, 62, 1109–1110. [Google Scholar] [CrossRef] [PubMed]
- Anaya, G.; Molina, A.; Valera, M.; Moreno-Millan, M.; Azor, P.; Peral-Garcia, P.; Demyda-Peyras, S. Sex chromosomal abnormalities associated with equine infertility: Validation of a simple molecular screening tool in the Purebred Spanish Horse. Anim. Genet. 2017, 48, 412–419. [Google Scholar] [CrossRef]
- R-Core-Team. A Language and Environment for Statistical Computing; V4.4.1 (Race for Your life); Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Hartigan, J.A.; Hartigan, P.M. The Dip Test of Unimodality. Ann. Stat. 1985, 13, 70–84. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Sourc. Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Dowle, M.; Srinivasan, A. Data.Table: Extension of Data.Frame, R Package Version 1.12. 8. Manual. 2019. Available online: https://www.rdocumentation.org/packages/data.table/versions/1.12.8 (accessed on 1 August 2024).
- Maechler, M. Diptest: Hartigan’s Dip Test Statistic for Unimodality R Package, v0.77. 2024. Available online: https://cran.r-project.org/web/packages/diptest/index.html (accessed on 1 August 2024).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: New York, NY, USA, 2016. [Google Scholar]
- Pieńkowska-Schelling, A.; Bugno, M.; Owczarek-Lipska, M.; Schelling, C.; Słota, E. Probe generated by Y chromosome microdissection is useful for analysing the sex chromosomes of the domestic horse. J. Anim. Feed Sci. 2006, 15, 173–178. [Google Scholar] [CrossRef]
- Bugno, M.; Slota, E.; Pienkowska-Schelling, A.; Schelling, C. Identification of chromosome abnormalities in the horse using a panel of chromosome-specific painting probes generated by microdissection. Acta Vet. Hung. 2009, 57, 369–381. [Google Scholar] [CrossRef]
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution in Mendelian Populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Luo, Z. Computing Inbreeding Coefficients in Large Populations. Genet. Sel. Evol. 1992, 24, 305–313. [Google Scholar] [CrossRef]
- Falconer, D.; MCKay, T. Introduction to Quantitative Genetics; Harlow: Essex, UK, 1996. [Google Scholar]
- Gutierrez, J.P.; Goyache, F. A note on ENDOG: A computer program for analysing pedigree information. J. Anim. Breed. Genet. 2005, 122, 172–176. [Google Scholar] [CrossRef]
- Poyato-Bonilla, J.; Laseca, N.; Demyda-Peyras, S.; Molina, A.; Valera, M. 500 years of breeding in the Carthusian Strain of Pura Raza Espanol horse: An evolutional analysis using genealogical and genomic data. J. Anim. Breed. Genet. 2022, 139, 84–99. [Google Scholar] [CrossRef] [PubMed]
- NHI. How Many People Are Affected by or at Risk for Klinefelter Syndrome (KS)? Available online: https://www.nichd.nih.gov/health/topics/klinefelter/conditioninfo/risk (accessed on 28 August 2024).
- Bugno, M.; Slota, E.; Koscielny, M. Karyotype evaluation among young horse populations in Poland. Schweiz. Arch. Tierheilkd. 2007, 149, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Ducos, A.; Revay, T.; Kovacs, A.; Hidas, A.; Pinton, A.; Bonnet-Garnier, A.; Molteni, L.; Slota, E.; Switonski, M.; Arruga, M.V.; et al. Cytogenetic screening of livestock populations in Europe: An overview. Cytogenet. Genome Res. 2008, 120, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Pienkowska-Schelling, A.; Kaul, A.; Schelling, C. X chromosome aneuploidy and micronuclei in fertile mares. Theriogenology 2020, 147, 34–38. [Google Scholar] [CrossRef]
- Szczerbal, I.; Nowacka-Woszuk, J.; Kopp-Kuhlman, C.; Mackowski, M.; Switonski, M. Application of droplet digital PCR in diagnosing of X monosomy in mares. Equine Vet. J. 2020, 52, 627–631. [Google Scholar] [CrossRef]
- Encina, A.; Valera, M.; Laseca, N.; Rodrigues, A.; Demyda Peyrás, S.; Perdomo-González, D.; Azor-Ortiz, P.J.; Gil, A.; Ripolles, M.; Anaya, G.; et al. Design of a Medium-Density chip for the genetic management of the Pura Raza Español and related breeds. In Proceedings of the International Committee for Animal Recording Conferece, Bled, Eslovenia, 19–24 May 2024. [Google Scholar]
- Aminou, O.; Badaoui, B.; Machmoum, M.; Piro, M. Evaluation of the effectiveness of single nucleotide polymorphisms compared to microsatellite markers for parentage verification in Moroccan horses. Anim. Genet. 2024, 55, 404–409. [Google Scholar] [CrossRef]
- Ishige, T.; Kikuchi, M.; Kakoi, H.; Hirota, K.I.; Ohnuma, A.; Tozaki, T.; Hirosawa, Y.; Tanaka, S.; Nagata, S.I. Evaluation of parentage testing using single nucleotide polymorphism markers for draft horses in Japan. Anim. Sci. J. 2023, 94, e13854. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, S.M.; Oyungerel, B.; Cho, G.J. Single nucleotide polymorphisms for parentage testing of horse breeds in Korea. Anim. Biosci. 2024, 37, 600–608. [Google Scholar] [CrossRef]
- Nolte, W.; Alkhoder, H.; Wobbe, M.; Stock, K.F.; Kalm, E.; Vosgerau, S.; Krattenmacher, N.; Thaller, G.; Tetens, J.; Kühn, C. Replacement of microsatellite markers by imputed medium-density SNP arrays for parentage control in German warmblood horses. J. Appl. Genet. 2022, 63, 783–792. [Google Scholar] [CrossRef]
- Villagómez, D.A.F.; Parma, P.; Radi, O.; Di Meo, G.; Pinton, A.; Iannuzzi, L.; King, W.A. Classical and molecular cytogenetics of disorders of sex development in domestic animals. Cytogenet. Genome Res. 2009, 126, 110–131. [Google Scholar] [CrossRef] [PubMed]
- Kubień, E.M.; Pozor, M.A.; Tischner, M. Clinical, cytogenetic and endocrine evaluation of a horse with a 65,XXY karyotype. Equine Vet. J. 1993, 25, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Tüttelmann, F.; Gromoll, J. Novel genetic aspects of Klinefelter’s syndrome. Mol. Hum. Reprod. 2010, 16, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Perdomo-Gonzalez, D.I.; Laseca, N.; Demyda-Peyras, S.; Valera, M.; Cervantes, I.; Molina, A. Fine-tuning genomic and pedigree inbreeding rates in equine population with a deep and reliable stud book: The case of the Pura Raza Espanola horse. J. Anim. Sci. Biotechnol. 2022, 13, 127. [Google Scholar] [CrossRef]
- Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Gen. 2012, 13, 493–504. [Google Scholar] [CrossRef]
- Bojesen, A.; Juul, S.; Gravholt, C.H. Prenatal and postnatal prevalence of Klinefelter syndrome: A national registry study. J. Clin. Endocrinol. Metab. 2003, 88, 622–626. [Google Scholar] [CrossRef]


| Horse 1 | Horse 2 | Horse 3 | Horse 4 | Horse 5 | ||
|---|---|---|---|---|---|---|
| ECAX markers | TKY38 | 105/129 | 129 | 111/129 | 107/129 | 105/129 |
| UCDEQ502 | 164 | 164/174 | 164 | 116/174 | 162/164 | |
| LEX003 | 194/210 | 214/216 | 206/210 | 208/210 | 212/214 | |
| LEX026 | 312/314 | 316 | 308 | 314 | 300/314 | |
| TKY270 | 168 | 168/170 | 168/172 | 170/172 | 170/172 | |
| AMEX | + | + | + | + | + | |
| ECAY markers | AMEY | + | + | + | + | + |
| EcaYH12 | + | + | + | + | + | |
| SRY | + | + | + | + | + | |
| ECA | 5 | 15 | 25 | PAR | NO-PAR | Y | |
|---|---|---|---|---|---|---|---|
| Horse 1 | HET (%) | 31.61 | 31.78 | 29.59 | 40.00 | 32.83 | 0.00 |
| LRR | 0.04 | 0.08 | 0.04 | 0.14 | 0.03 | −0.05 | |
| BAF dip test | 0.95 | 0.99 | 0.99 | 0.25 | 0.99 | 1 | |
| Horse 2 | HET (%) | 24.95 | 30.87 | 26.92 | 63.89 | 26.47 | 4.13 |
| LRR | 0.03 | 0.06 | −0.01 | 0.10 | 0.07 | −0.22 | |
| BAF dip test | 1 | 1 | 0.89 | 0.04 | 0.93 | 0.84 | |
| Horse 3 | HET (%) | 22.87 | 25.19 | 27.55 | 50.00 | 29.19 | 3.25 |
| LRR | 0.02 | 0.02 | −0.03 | 0.13 | 0.04 | −0.16 | |
| BAF dip test | 0.97 | 0.99 | 0.97 | 0.11 | 1 | 0.92 | |
| Horse 4 | HET (%) | 31.87 | 32.95 | 29.99 | 58.33 | 28.63 | 2.44 |
| LRR | 0.04 | 0.06 | 0.03 | 0.12 | 0.03 | −0.38 | |
| BAF dip test | 0.96 | 0.98 | 1 | 0 | 1 | 1 | |
| Horse 5 | HET (%) | 30.87 | 29.87 | 23.04 | 52.78 | 32.56 | 2.44 |
| LRR | 0.02 | 0.04 | 0.01 | 0.12 | 0.03 | −0.39 | |
| BAF dip test | 0.98 | 0.94 | 0.79 | 0.09 | 0.99 | 1 |
| Id | Born | F | F of the Mare | Foals per Mare | Age of the Mare | Average fij |
|---|---|---|---|---|---|---|
| Horse 1 | 27 April 2021 | 5.13% | 18.89% | 2 | 8 | 5.19% |
| Horse 2 | 3 March 2022 | 5.86% | 6.37% | 3 | 9 | 5.91% |
| Horse 3 | 12 December 2022 | 5.58% | 5.03% | 6 | 10 | 5.75% |
| Horse 4 | 28 March 2023 | 7.59% | 4.56% | 8 | 16 | 5.06% |
| Horse 5 | 15 July 2023 | 4.54% | 5.92% | 1 | 9 | 5.09% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valera, M.; Karlau, A.; Anaya, G.; Bugno-Poniewierska, M.; Molina, A.; Encina, A.; Azor, P.J.; Demyda-Peyrás, S. The Use of Genomic Screening for the Detection of Chromosomal Abnormalities in the Domestic Horse: Five New Cases of 65,XXY Syndrome in the Pura Raza Español Breed. Animals 2024, 14, 2560. https://doi.org/10.3390/ani14172560
Valera M, Karlau A, Anaya G, Bugno-Poniewierska M, Molina A, Encina A, Azor PJ, Demyda-Peyrás S. The Use of Genomic Screening for the Detection of Chromosomal Abnormalities in the Domestic Horse: Five New Cases of 65,XXY Syndrome in the Pura Raza Español Breed. Animals. 2024; 14(17):2560. https://doi.org/10.3390/ani14172560
Chicago/Turabian StyleValera, Mercedes, Ayelén Karlau, Gabriel Anaya, Monika Bugno-Poniewierska, Antonio Molina, Ana Encina, Pedro J. Azor, and Sebastián Demyda-Peyrás. 2024. "The Use of Genomic Screening for the Detection of Chromosomal Abnormalities in the Domestic Horse: Five New Cases of 65,XXY Syndrome in the Pura Raza Español Breed" Animals 14, no. 17: 2560. https://doi.org/10.3390/ani14172560
APA StyleValera, M., Karlau, A., Anaya, G., Bugno-Poniewierska, M., Molina, A., Encina, A., Azor, P. J., & Demyda-Peyrás, S. (2024). The Use of Genomic Screening for the Detection of Chromosomal Abnormalities in the Domestic Horse: Five New Cases of 65,XXY Syndrome in the Pura Raza Español Breed. Animals, 14(17), 2560. https://doi.org/10.3390/ani14172560





