Genome-Wide Analysis Reveals Copy Number Variant Gene TGFBR3 Regulates Pig Back Fat Deposition
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Pig Samples and Sequencing Data
2.2. CNV Calling
2.3. Comparison between LW and MZ Pigs
2.4. Screening of CNVR-Related Genes Associated with Fat Deposition by Multi-Omics Association Analysis
2.5. QTL Analyses
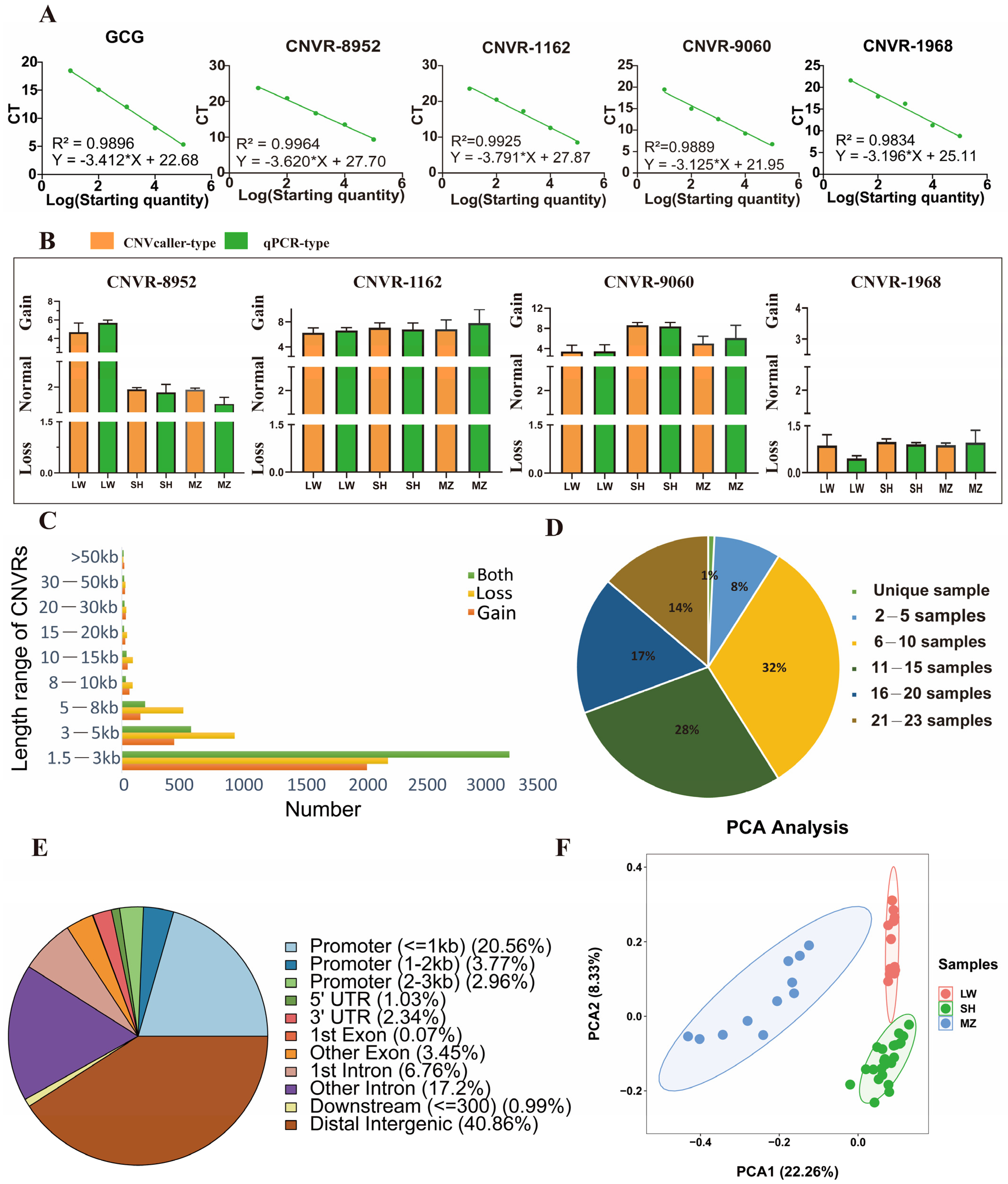
2.6. Validation of CNV Type
2.7. Cell Culture, Transfection, and Differentiation
2.8. RNA Isolation, cDNA Library Preparation, and qPCR
2.9. Oil Red O Staining and Triglyceride Assay
2.10. EdU Assay
2.11. Statistical Analysis
3. Results
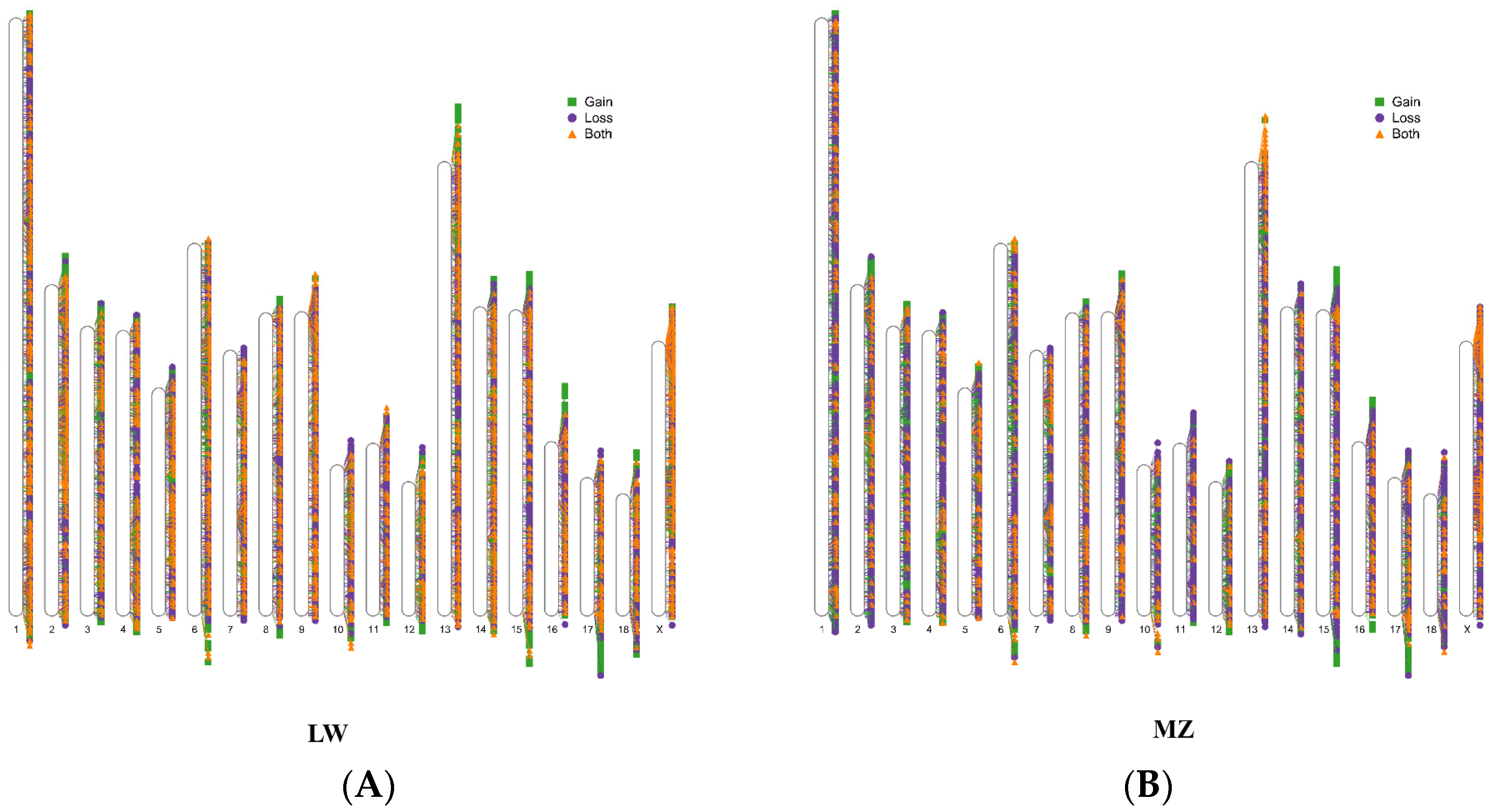
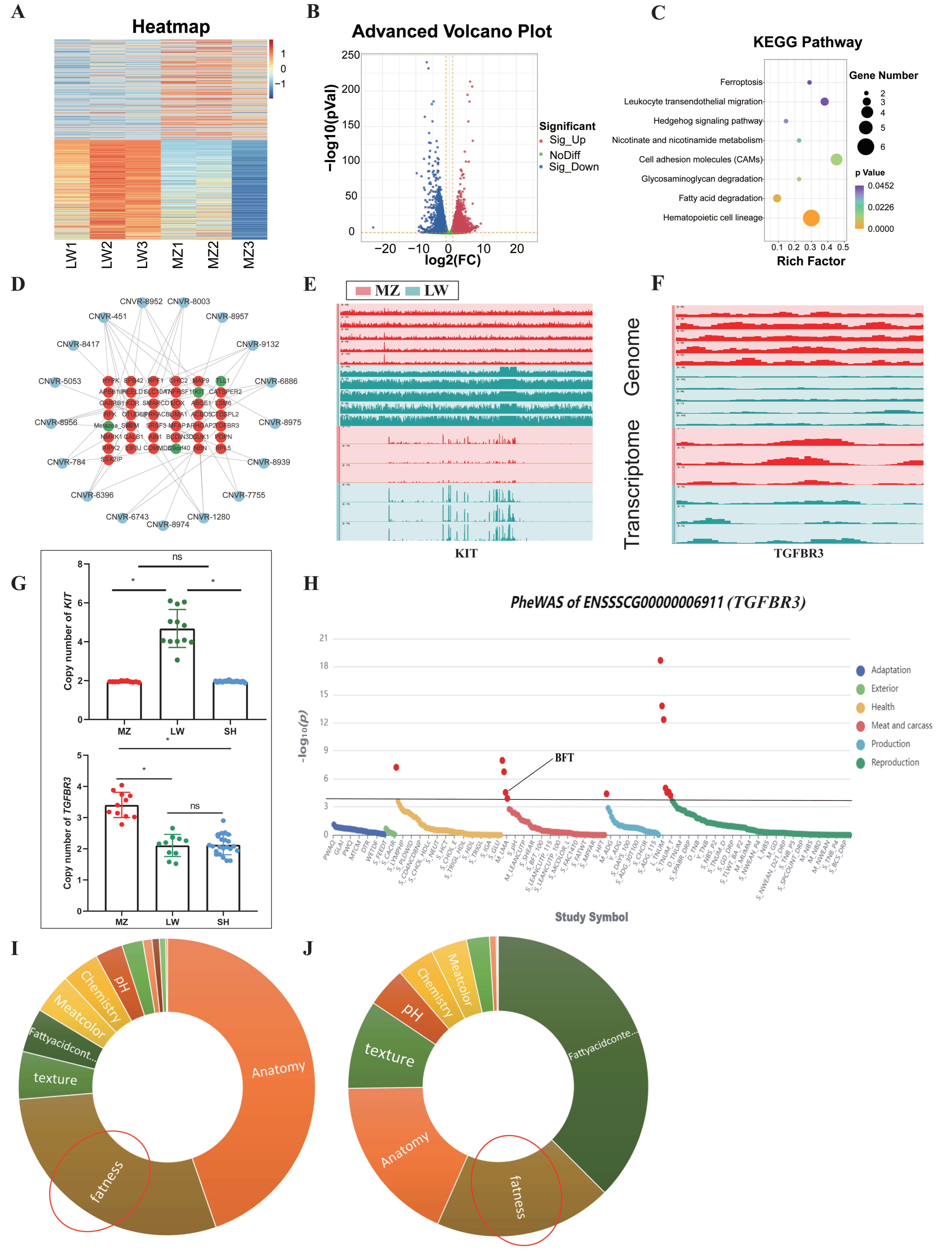
3.1. CNVR Detection in LW and MZ Pigs
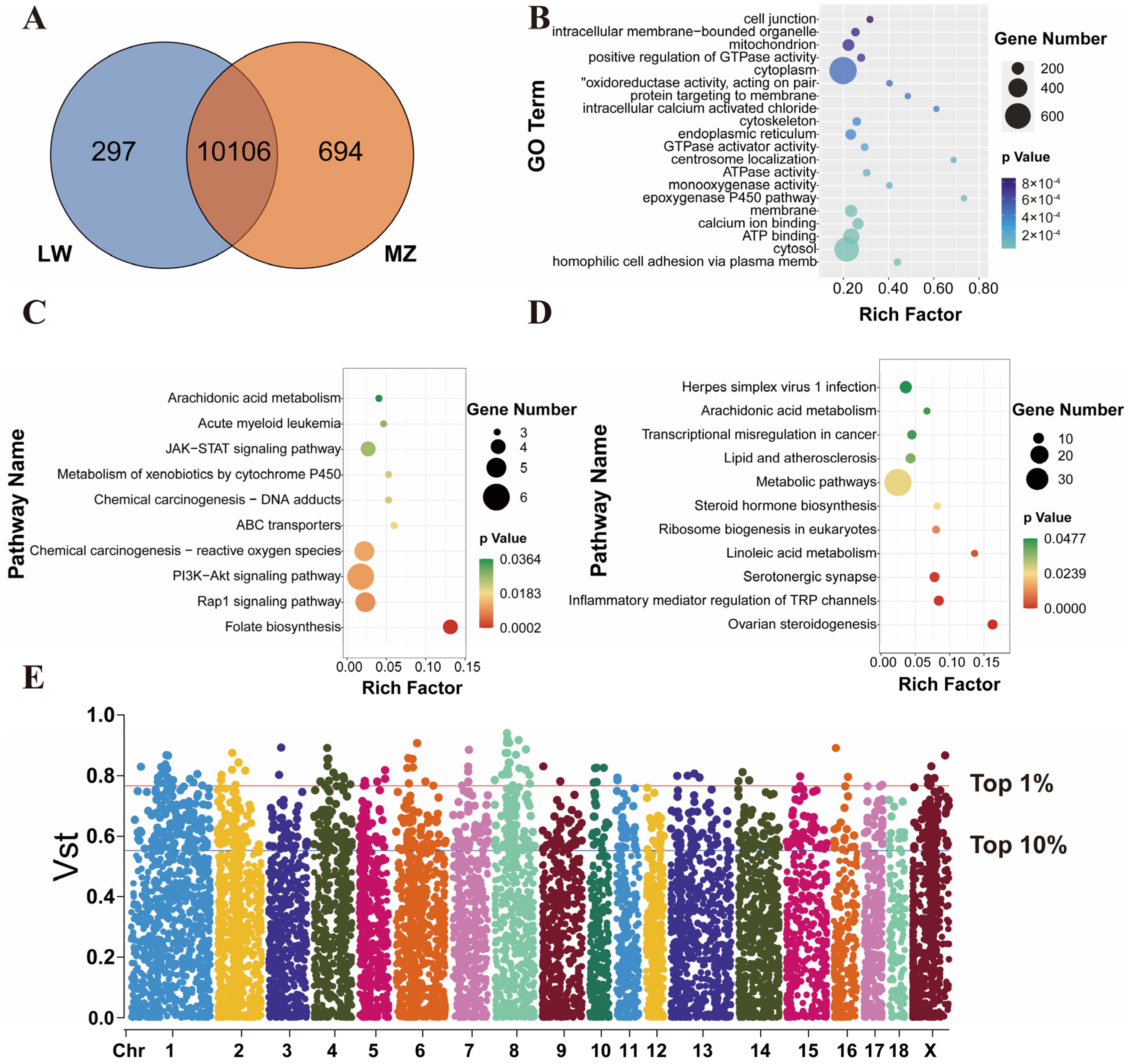
3.2. Shared and Breed-Specific CNVRs
3.3. Multi-Omics Integration Analysis
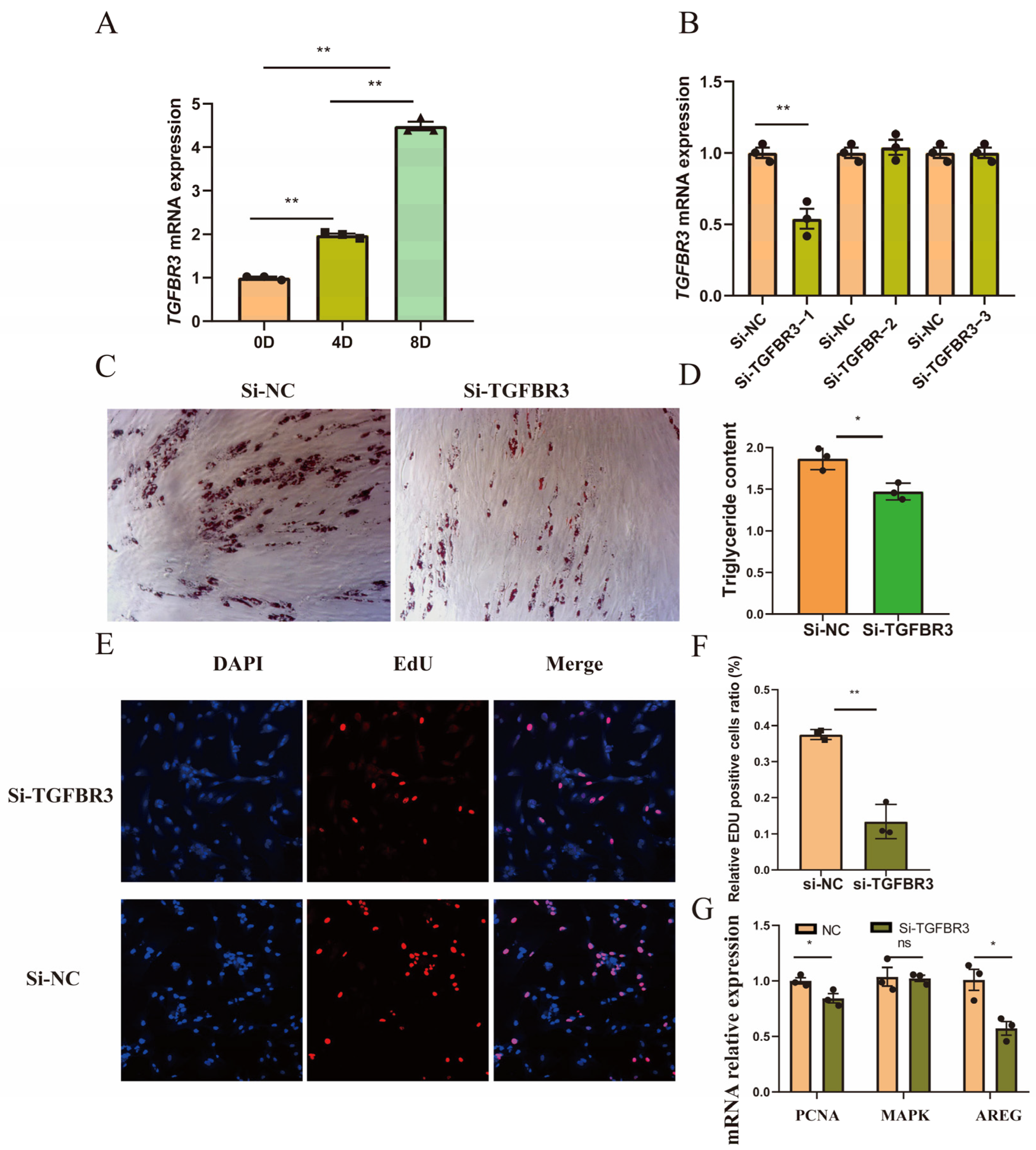
3.4. Decreased Expression of TGFBR3 Inhibited Preadipocyte Proliferation and Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mas, G.; Llavall, M.; Coll, D.; Roca, R.; Diaz, I.; Oliver, M.A.; Gispert, M.; Realini, C.E. Effect of an elevated monounsaturated fat diet on pork carcass and meat quality traits and tissue fatty acid composition from York-crossed barrows and gilts. Meat Sci. 2011, 89, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Lassaletta, L.; Estelles, F.; Beusen, A.H.W.; Bouwman, L.; Calvet, S.; van Grinsven, H.J.M.; Doelman, J.C.; Stehfest, E.; Uwizeye, A.; Westhoek, H. Future global pig production systems according to the Shared Socioeconomic Pathways. Sci. Total Environ. 2019, 665, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, Y.; Sun, B.F.; Zhao, Y.L.; Yang, Y.G. FTO and obesity: Mechanisms of association. Curr. Diab Rep. 2014, 14, 486. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, Z.; Sun, Y.; Wang, H.; Wang, C.; Yu, S.; Liu, J.; Zhang, Y.; Fan, B.; Li, K.; et al. Analysis of genome-wide copy number variations in Chinese indigenous and western pig breeds by 60 K SNP genotyping arrays. PLoS ONE 2014, 9, e106780. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, N.; Li, J.; Zhang, W.; Hei, W.; Ji, M.; Yang, Y.; Cao, G.; Guo, X.; Li, B. MiR-23b Promotes Porcine Preadipocyte Differentiation via SESN3 and ACSL4. Cells 2022, 11, 2339. [Google Scholar] [CrossRef]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef]
- Klopocki, E.; Mundlos, S. Copy-number variations, noncoding sequences, and human phenotypes. Annu. Rev. Genom. Hum. Genet. 2011, 12, 53–72. [Google Scholar] [CrossRef]
- Fadista, J.; Nygaard, M.; Holm, L.E.; Thomsen, B.; Bendixen, C. A snapshot of CNVs in the pig genome. PLoS ONE 2008, 3, e3916. [Google Scholar] [CrossRef]
- Zheng, X.; Zhao, P.; Yang, K.; Ning, C.; Wang, H.; Zhou, L.; Liu, J. CNV analysis of Meishan pig by next-generation sequencing and effects of AHR gene CNV on pig reproductive traits. J. Anim. Sci. Biotechnol. 2020, 11, 42. [Google Scholar] [CrossRef]
- Wang, H.; Wang, C.; Yang, K.; Liu, J.; Zhang, Y.; Wang, Y.; Xu, X.; Michal, J.J.; Jiang, Z.; Liu, B. Genome Wide Distributions and Functional Characterization of Copy Number Variations between Chinese and Western Pigs. PLoS ONE 2015, 10, e0131522. [Google Scholar] [CrossRef]
- Liu, M.; Li, B.; Shi, T.; Huang, Y.; Liu, G.E.; Lan, X.; Lei, C.; Chen, H. Copy number variation of bovine SHH gene is associated with body conformation traits in Chinese beef cattle. J. Appl. Genet. 2019, 60, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, X.; Cheng, J.; Jiang, R.; Huang, R.; Wang, D.; Huang, Y.; Pi, L.; Hu, L.; Chen, H. Copy Number Variation of the PIGY Gene in Sheep and Its Association Analysis with Growth Traits. Animals 2020, 10, 688. [Google Scholar] [CrossRef]
- Khatri, B.; Kang, S.; Shouse, S.; Anthony, N.; Kuenzel, W.; Kong, B.C. Copy number variation study in Japanese quail associated with stress related traits using whole genome re-sequencing data. PLoS ONE 2019, 14, e0214543. [Google Scholar] [CrossRef]
- Locke, M.E.; Milojevic, M.; Eitutis, S.T.; Patel, N.; Wishart, A.E.; Daley, M.; Hill, K.A. Genomic copy number variation in Mus musculus. BMC Genom. 2015, 16, 497. [Google Scholar] [CrossRef] [PubMed]
- Schiavo, G.; Dolezal, M.A.; Scotti, E.; Bertolini, F.; Calo, D.G.; Galimberti, G.; Russo, V.; Fontanesi, L. Copy number variants in Italian Large White pigs detected using high-density single nucleotide polymorphisms and their association with back fat thickness. Anim. Genet. 2014, 45, 745–749. [Google Scholar] [CrossRef]
- Panda, S.; Kumar, A.; Gaur, G.K.; Ahmad, S.F.; Chauhan, A.; Mehrotra, A.; Dutt, T. Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs. Anim. Biotechnol. 2022, 34, 1891–1899. [Google Scholar] [CrossRef]
- Ellis, M.; Lympany, C.; Haley, C.S.; Brown, I.; Warkup, C.C. The eating quality of pork from Meishan and Large White pigs and their reciprocal crosses. Anim. Sci. 2010, 60, 125–131. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Fang, L.; Li, M.; Tian, J.; Wang, L.; Zhao, F. Integrating genome-wide association studies and population genomics analysis reveals the genetic architecture of growth and backfat traits in pigs. Front. Genet. 2022, 13, 1078696. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, Z.; Cai, Y.; Chen, T.; Li, C.; Fu, W.; Jiang, Y. CNVcaller: Highly efficient and widely applicable software for detecting copy number variations in large populations. Gigascience 2017, 6, gix115. [Google Scholar] [CrossRef]
- Hao, Z.; Lv, D.; Ge, Y.; Shi, J.; Weijers, D.; Yu, G.; Chen, J. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 2020, 6, e251. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Murphy, D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e79. [Google Scholar] [CrossRef]
- Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ballester, M.; Castelló, A.; Ibáñez, E.; Sánchez, A.; Folch, J.M. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. BioTechniques 2004, 37, 610–613. [Google Scholar] [CrossRef]
- Rubin, C.J.; Megens, H.J.; Martinez Barrio, A.; Maqbool, K.; Sayyab, S.; Schwochow, D.; Wang, C.; Carlborg, O.; Jern, P.; Jorgensen, C.B.; et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef]
- Jeong, H.; Song, K.D.; Seo, M.; Caetano-Anolles, K.; Kim, J.; Kwak, W.; Oh, J.D.; Kim, E.; Jeong, D.K.; Cho, S.; et al. Exploring evidence of positive selection reveals genetic basis of meat quality traits in Berkshire pigs through whole genome sequencing. BMC Genet. 2015, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Gao, Y.; Yin, H.; Bai, Z.; Liu, S.; Zeng, H.; Bai, L.; Cai, Z.; Zhao, B.; Li, X.; et al. A compendium of genetic regulatory effects across pig tissues. Nat. Genet. 2024, 56, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef] [PubMed]
- Bussotti, G.; Gouzelou, E.; Cortes Boite, M.; Kherachi, I.; Harrat, Z.; Eddaikra, N.; Mottram, J.C.; Antoniou, M.; Christodoulou, V.; Bali, A.; et al. Leishmania Genome Dynamics during Environmental Adaptation Reveal Strain-Specific Differences in Gene Copy Number Variation, Karyotype Instability, and Telomeric Amplification. mBio 2018, 9, e01399-18. [Google Scholar] [CrossRef]
- Zhao, Q.B.; Sun, H.; Zhang, Z.; Xu, Z.; Olasege, B.S.; Ma, P.P.; Zhang, X.Z.; Wang, Q.S.; Pan, Y.C. Exploring the Structure of Haplotype Blocks and Genetic Diversity in Chinese Indigenous Pig Populations for Conservation Purpose. Evol. Bioinform. Online 2019, 15, 1176934318825082. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, Y.; Wang, Y.; Zhang, Y.; Shen, Y.; Su, Q.; Gao, G.; Xu, H.; Zhou, X.; Liu, B. Whole-genome sequencing reveals breed-differential CNVs between Tongcheng and Large White pigs. Anim. Genet. 2020, 51, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Velagaleti, G.V.; Bien-Willner, G.A.; Northup, J.K.; Lockhart, L.H.; Hawkins, J.C.; Jalal, S.M.; Withers, M.; Lupski, J.R.; Stankiewicz, P. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am. J. Hum. Genet. 2005, 76, 652–662. [Google Scholar] [CrossRef]
- Vegesna, R.; Tomaszkiewicz, M.; Medvedev, P.; Makova, K.D. Dosage regulation, and variation in gene expression and copy number of human Y chromosome ampliconic genes. PLoS Genet. 2019, 15, e1008369. [Google Scholar] [CrossRef]
- Ran, X.Q.; Pan, H.; Huang, S.H.; Liu, C.; Niu, X.; Li, S.; Wang, J.F. Copy number variations of MTHFSD gene across pig breeds and its association with litter size traits in Chinese indigenous Xiang pig. J. Anim. Physiol. Anim. Nutr. 2018, 102, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, C.; Xiong, X.; Fang, S.; Yang, H.; Zhang, Z.; Ren, J.; Guo, Y.; Huang, L. Copy number variation in the MSRB3 gene enlarges porcine ear size through a mechanism involving miR-584-5p. Genet. Sel. Evol. 2018, 50, 72. [Google Scholar] [CrossRef]
- Abbott, B.D.; Schmid, J.E.; Pitt, J.A.; Buckalew, A.R.; Wood, C.R.; Held, G.A.; Diliberto, J.J. Adverse reproductive outcomes in the transgenic Ah receptor-deficient mouse. Toxicol. Appl. Pharmacol. 1999, 155, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Xin, K.; Pan, T.; Gao, S.; Yan, S. A Transcription Factor Regulates Gene Expression in Chloroplasts. Int. J. Mol. Sci. 2021, 22, 6769. [Google Scholar] [CrossRef]
- Jiao, Y.; Zan, L.S.; Liu, Y.F.; Wang, H.B. Molecular characterization, polymorphism of the ACOX1 gene and association with ultrasound traits in Bos taurus. Genet. Mol. Res. 2011, 10, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Fu, S.; Chen, Y.; Jin, W.; Zhai, B.; Li, Y.; Sun, G.; Han, R.; Wang, Y.; Tian, Y.; et al. MicroRNA-15a Regulates the Differentiation of Intramuscular Preadipocytes by Targeting ACAA1, ACOX1 and SCP2 in Chickens. Int. J. Mol. Sci. 2019, 20, 4063. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Zhang, Y.T.; Tseng, Y.J.; Zhang, J. miR-222 targets ACOX1, promotes triglyceride accumulation in hepatocytes. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Fernandez, M.E.; Giles, D.A.; Stankiewicz, T.E.; Sheridan, R.; Karns, R.; Cappelletti, M.; Lampe, K.; Mukherjee, R.; Sina, C.; Sallese, A.; et al. Peroxisomal beta-oxidation regulates whole body metabolism, inflammatory vigor, and pathogenesis of nonalcoholic fatty liver disease. JCI Insight 2018, 3, e93626. [Google Scholar] [CrossRef]
- Morita, A.; Enokizono, T.; Ohto, T.; Tanaka, M.; Watanabe, S.; Takada, Y.; Iwama, K.; Mizuguchi, T.; Matsumoto, N.; Morita, M.; et al. Novel ACOX1 mutations in two siblings with peroxisomal acyl-CoA oxidase deficiency. Brain Dev. 2021, 43, 475–481. [Google Scholar] [CrossRef]
- Morais, S.; Knoll-Gellida, A.; Andre, M.; Barthe, C.; Babin, P.J. Conserved expression of alternative splicing variants of peroxisomal acyl-CoA oxidase 1 in vertebrates and developmental and nutritional regulation in fish. Physiol. Genom. 2007, 28, 239–252. [Google Scholar] [CrossRef]
- Zeng, J.; Deng, S.; Wang, Y.; Li, P.; Tang, L.; Pang, Y. Specific Inhibition of Acyl-CoA Oxidase-1 by an Acetylenic Acid Improves Hepatic Lipid and Reactive Oxygen Species (ROS) Metabolism in Rats Fed a High Fat Diet. J. Biol. Chem. 2017, 292, 3800–3809. [Google Scholar] [CrossRef]
- Zhang, F.; Xiong, Q.; Tao, H.; Liu, Y.; Zhang, N.; Li, X.F.; Suo, X.J.; Yang, Q.P.; Chen, M.X. ACOX1, regulated by C/EBPalpha and miR-25-3p, promotes bovine preadipocyte adipogenesis. J. Mol. Endocrinol. 2021, 66, 195–205. [Google Scholar] [CrossRef]
- Zuo, B.; Yang, H.; Wang, J.; Lei, M.G.; Xiong, Y.Z. Molecular characterization, sequence variation and association with fat deposition traits of ACOX1 gene in pigs. J. Anim. Feed. Sci. 2007, 16, 433–444. [Google Scholar] [CrossRef]
- Kuwata, H.; Hara, S. Role of acyl-CoA synthetase ACSL4 in arachidonic acid metabolism. Prostaglandins Other Lipid Mediat. 2019, 144, 106363. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Zhang, Y.; Xin, L.; Kong, S.; Chen, Y.; Yang, S.; Li, K. Global Transcriptomic Profiling of Cardiac Hypertrophy and Fatty Heart Induced by Long-Term High-Energy Diet in Bama Miniature Pigs. PLoS ONE 2015, 10, e0132420. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Zhang, B.; Zhong, H.; Lu, Y.; Zhang, H. Candidate gene screening for lipid deposition using combined transcriptomic and proteomic data from Nanyang black pigs. BMC Genom. 2021, 22, 441. [Google Scholar] [CrossRef]
- Fang, C.; Guo, F.; Zhao, X.; Zhang, Z.; Lu, J.; Pan, H.; Xu, T.; Li, W.; Yang, M.; Huang, Y.; et al. Biological mechanisms of growth performance and meat quality in porcine muscle tissue. Anim. Biotechnol. 2021, 33, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Zhang, H.; Hua, Z.; Zhu, Z.; Tao, J.; Xiao, H.; Zhang, L.; Bi, Y.; Wang, H. ACSL4 Directs Intramuscular Adipogenesis and Fatty Acid Composition in Pigs. Animals 2022, 12, 119. [Google Scholar] [CrossRef]
- Liu, M.; Tao, G.; Cao, Y.; Hu, Y.; Zhang, Z. Silencing of IGF2BP1 restrains ox-LDL-induced lipid accumulation and inflammation by reducing RUNX1 expression and promoting autophagy in macrophages. J. Biochem. Mol. Toxicol. 2022, 36, e22994. [Google Scholar] [CrossRef]
- Ma, J.; Gilbert, H.; Iannuccelli, N.; Duan, Y.; Guo, B.; Huang, W.; Ma, H.; Riquet, J.; Bidanel, J.P.; Huang, L.; et al. Fine mapping of fatness QTL on porcine chromosome X and analyses of three positional candidate genes. BMC Genet. 2013, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; Jiang, Y.Z.; Cen, W.M.; Xing, S.H.; Zhu, L.; Tang, G.Q.; Li, M.Z.; Jiang, A.A.; Lou, P.E.; Wen, A.X.; et al. Distribution of H-FABP and ACSL4 gene polymorphisms and their associations with intramuscular fat content and backfat thickness in different pig populations. Genet. Mol. Res. 2014, 13, 6759–6772. [Google Scholar] [CrossRef]
- Daily, J.W.; Yang, H.J.; Liu, M.; Kim, M.J.; Park, S. Subcutaneous fat mass is associated with genetic risk scores related to proinflammatory cytokine signaling and interact with physical activity in middle-aged obese adults. Nutr. Metab. 2019, 16, 75. [Google Scholar] [CrossRef]
- Shishido, E.; Aleksic, B.; Ozaki, N. Copy-number variation in the pathogenesis of autism spectrum disorder. Psychiatry Clin. Neurosci. 2014, 68, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; Riaz, S.; Morales-Cruz, A.; Amrine, K.C.; McGuire, B.; Gubler, W.D.; Walker, M.A.; Cantu, D. Adaptive genomic structural variation in the grape powdery mildew pathogen, Erysiphe necator. BMC Genom. 2014, 15, 1081. [Google Scholar] [CrossRef] [PubMed]
| Breed | No. Sample | No. CNVR | No. Gains | No. Losses | No. Both | Length (bp) | Coverage, % |
|---|---|---|---|---|---|---|---|
| LW | 12 | 10,403 | 3400 | 4454 | 2549 | 40,296,797 | 1.68 |
| MZ | 11 | 10,800 | 3827 | 5541 | 1432 | 41,886,800 | 1.75 |
| Merge | 23 | 11,097 | 2885 | 4019 | 4193 | 44,147,303 | 1.85 |
| Group | Total Number | Exterior | Health | Meat | Production | Reproduction | Fatness (Contained in Meat) | Fatness Proportion of Total QTLs (%) | Fatness Proportion of the Meat (%) |
|---|---|---|---|---|---|---|---|---|---|
| cis-eQTL_CNVR | 1414 | 51 | 153 | 1007 | 141 | 62 | 292 | 20.65 | 28.99 |
| Total QTLs | 32,475 | 2492 | 6716 | 17,191 | 2911 | 3165 | 3289 | 10.13 | 19.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Yang, H.; Xu, Q.; Liu, M.; Chao, X.; Chen, J.; Zhou, B. Genome-Wide Analysis Reveals Copy Number Variant Gene TGFBR3 Regulates Pig Back Fat Deposition. Animals 2024, 14, 2657. https://doi.org/10.3390/ani14182657
Zhang C, Yang H, Xu Q, Liu M, Chao X, Chen J, Zhou B. Genome-Wide Analysis Reveals Copy Number Variant Gene TGFBR3 Regulates Pig Back Fat Deposition. Animals. 2024; 14(18):2657. https://doi.org/10.3390/ani14182657
Chicago/Turabian StyleZhang, Chunlei, Huan Yang, Qinglei Xu, Mingzheng Liu, Xiaohuan Chao, Jiahao Chen, and Bo Zhou. 2024. "Genome-Wide Analysis Reveals Copy Number Variant Gene TGFBR3 Regulates Pig Back Fat Deposition" Animals 14, no. 18: 2657. https://doi.org/10.3390/ani14182657
APA StyleZhang, C., Yang, H., Xu, Q., Liu, M., Chao, X., Chen, J., & Zhou, B. (2024). Genome-Wide Analysis Reveals Copy Number Variant Gene TGFBR3 Regulates Pig Back Fat Deposition. Animals, 14(18), 2657. https://doi.org/10.3390/ani14182657






