Analysis of the Genetic Diversity and Genetic Structure of Jiangshan Black Pigs Using Single Nucleotide Polymorphism (SNP) Chips
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. DNA Extraction and Quality Control
2.3. Chip Detection
2.4. Genetic Diversity Analysis
2.5. Genetic Relationships and Population Structure Analysis
2.6. Inbreeding Coefficient Analysis
3. Results
3.1. Genotype Quality Control
3.2. Genetic Diversity
3.3. Analysis of Kinship
3.4. Inbreeding Coefficient
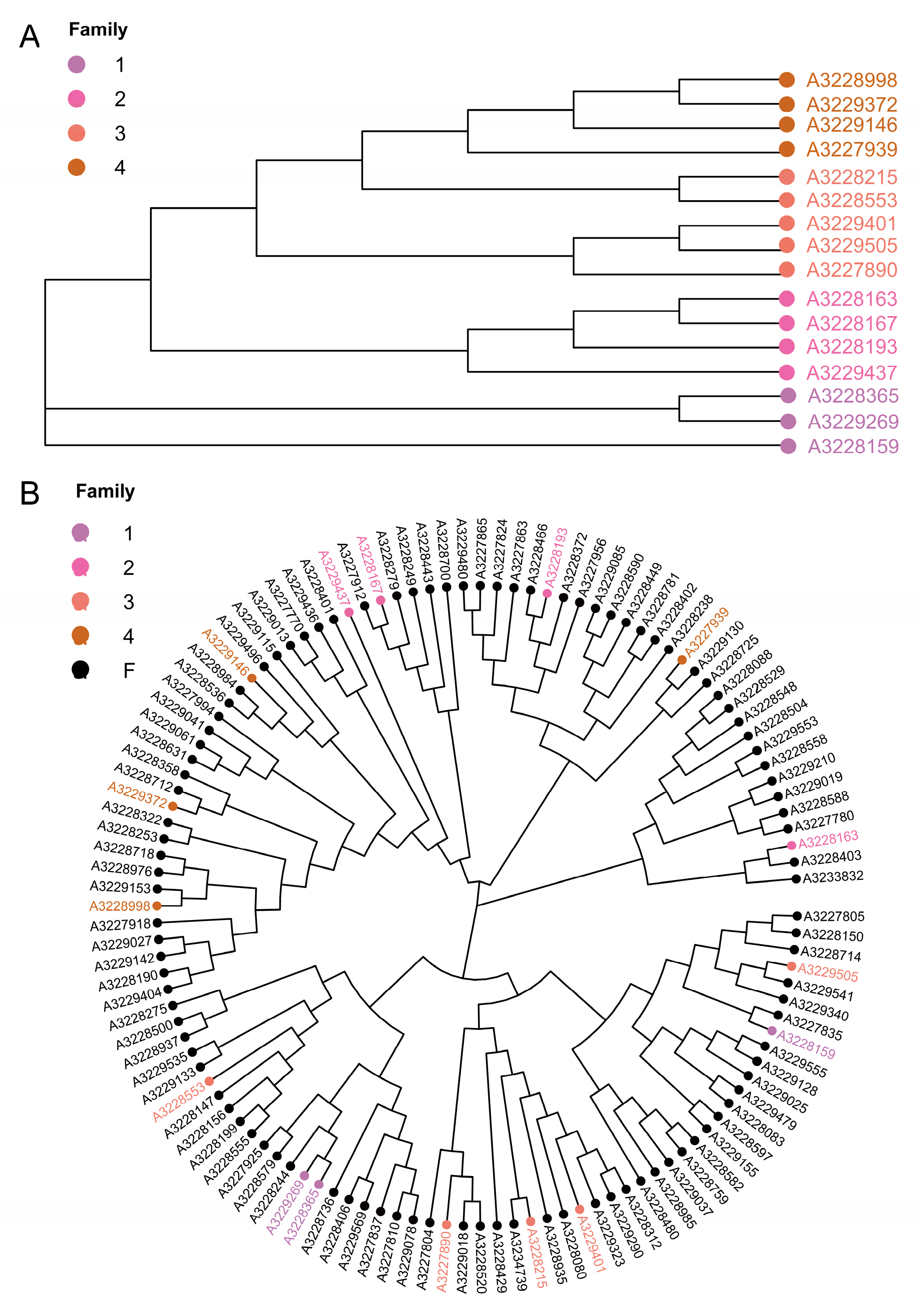
3.5. Cluster Analysis and Family Construction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, M.; Wang, L.; Lou, P.; Mao, Y.; Liu, J.; Wang, W.; Wang, G.; Zheng, R.; Zuo, B.; Wang, Y. Study on Germplasm Characteristics of Jiangshan Black Pig. Chin. J. Anim. Sci. 2023, 59, 112–117. [Google Scholar]
- Zhang, H.; Dai, X.; Lu, J.; Tao, Y. Jiangshan black pig. In Annals of Livestock and Poultry Genetic Resources in Zhejiang Province; Zhejiang Science and Technology Press: Hangzhou, China, 2016; Volume 1, pp. 45–49. [Google Scholar]
- Yang, H. Livestock development in China: Animal production, consumption and genetic resources. J. Anim. Breed. Genet. 2013, 130, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Chai, Z. Consideration on Protection, Exploitation and Utilization of Livestock and Poultry Germplasm Resources in Jiangshan City. Zhejiang J. Anim. Sci. Vet. Med. 2021, 46, 19–20+23. [Google Scholar]
- Yuan, J.; Zhou, X.; Xu, G.; Xu, S.; Liu, B. Genetic diversity and population structure of Tongcheng pigs in China using whole-genome SNP chip. Front. Genet. 2022, 13, 910521. [Google Scholar] [CrossRef]
- Gebreyesus, G.; Lund, M.S.; Sahana, G.; Su, G. Reliabilities of Genomic Prediction for Young Stock Survival Traits Using 54K SNP Chip Augmented With Additional Single-Nucleotide Polymorphisms Selected From Imputed Whole-Genome Sequencing Data. Front. Genet. 2021, 12, 667300. [Google Scholar] [CrossRef]
- Zhao, H.H.; Ma, Z.; Ying, Z.X.; Niu, F.N.; Luo, M.T.; Wang, Z.; Cheng, X.; Zhang, Q.Q.; Niu, Q. Clinical manifestations and acid alpha-glucosidase mutation characterisation of a cohort of patients with late-onset Pompe disease in eastern China. Ann. Transl. Med. 2021, 9, 1803. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Jing, W.; Samuels, D.C.; Sheng, Q.; Shyr, Y.; Guo, Y. Strategies for processing and quality control of Illumina genotyping arrays. Brief. Bioinform. 2018, 19, 765–775. [Google Scholar] [CrossRef]
- Sun, H.; Wang, Z.; Zhang, Z.; Xiao, Q.; Zhong, X.U.; Zhang, X.Z.; Yang, H.J.; Zhu, M.X.; Xue, M.; Liu, X.H.; et al. Exploring the Current Situation of Conservation of Meishan Pigs Based on Genome Sequencing Data. J. Shanghai Jiaotong Univ. Agric. Sci. 2017, 4, 69–74. [Google Scholar]
- Barbato, M.; Orozco-Terwengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef]
- Sved, J.A. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 1971, 2, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Herrero-Medrano, J.M.; Megens, H.J.; Groenen, M.A.; Ramis, G.; Bosse, M.; Pérez-Enciso, M.; Crooijmans, R.P. Conservation genomic analysis of domestic and wild pig populations from the Iberian Peninsula. BMC Genet. 2013, 14, 106. [Google Scholar] [CrossRef] [PubMed]
- Uimari, P.; Tapio, M. Extent of linkage disequilibrium and effective population size in Finnish Landrace and Finnish Yorkshire pig breeds. J. Anim. Sci. 2011, 89, 609–614. [Google Scholar] [CrossRef]
- Vanraden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Lee, S.H.; Seo, D.W.; Cho, E.S.; Choi, B.H.; Kim, Y.M.; Hong, J.K.; Han, H.D.; Jung, Y.B.; Kim, D.J.; Choi, T.J.; et al. Genetic Diversity and Ancestral Study for Korean Native Pigs Using 60K SNP Chip. Animals 2020, 10, 760. [Google Scholar] [CrossRef]
- Liu, B.; Shen, L.; Guo, Z.; Gan, M.; Chen, Y.; Yang, R.; Niu, L.; Jiang, D.; Zhong, Z.; Li, X.; et al. Single nucleotide polymorphism-based analysis of the genetic structure of Liangshan pig population. Anim. Biosci. 2021, 34, 1105–1115. [Google Scholar] [CrossRef]
- Suwannasing, R.; Duangjinda, M.; Boonkum, W.; Taharnklaew, R.; Tuangsithtanon, K. The identification of novel regions for reproduction trait in Landrace and Large White pigs using a single step genome-wide association study. Asian-Australas. J. Anim. Sci. 2018, 31, 1852–1862. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Kumar, A.; Gaur, G.K.; Ahmad, S.F.; Chauhan, A.; Mehrotra, A.; Dutt, T. Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs. Anim. Biotechnol. 2023, 34, 1891–1899. [Google Scholar] [CrossRef]
- Leng, D.; Ge, L.; Sun, J. Characterization analysis of Rongchang pig population based on the Zhongxin-1 Porcine Breeding Array PLUS. Anim. Biosci. 2023, 36, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dong, R.; Li, X.; Cui, C.; Yu, G. Analysis of the Genetic Diversity and Family Structure of the Licha Black Pig Population on Jiaodong Peninsula, Shandong Province, China. Animals 2022, 12, 1045. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Zhang, Z.; Sun, H.; Wang, Q.; Pan, Y. Pudong White pig: A unique genetic resource disclosed by sequencing data. Anim. Int. J. Anim. Biosci. 2017, 11, 1117–1124. [Google Scholar] [CrossRef]
- Wang, Z.; Zhong, Z.; Xie, X.; Wang, F.; Pan, D.; Wang, Q.; Pan, Y.; Xiao, Q.; Tan, Z. Detection of Runs of Homozygosity and Identification of Candidate Genes in the Whole Genome of Tunchang Pigs. Animals 2024, 14, 201. [Google Scholar] [CrossRef]
- Long, X.; Chen, L.; Wu, P.-X.; Zhang, T.-H.; Pan, H.-M.; Zhang, L.; Wang, J.-Y.; Guo, Z.-Y.; Chai, J. Evaluation of the Genetic Structure and Selection Signatures in Hechuan Black Pigs Conserved Population. Acta Vet. Et Zootech. Sin. 2023, 54, 1854–1867. [Google Scholar]
- Valluzzi, C.; Rando, A.; Macciotta, N.P.P.; Gaspa, G.; Di Gregorio, P. The Nero Lucano Pig Breed: Recovery and Variability. Animals 2021, 11, 1331. [Google Scholar] [CrossRef]
- Song, K.; Yan, Z.; Wang, P.; Cheng, W.; Li, J.; Bai, Y.; Sun, G.; Gun, S. Analysis on Genetic Diversity and Genetic Structure Based on SNP Chips of Huixian Qingni Black Pig. Acta Vet. Et Zootech. Sin. 2024, 55, 995–1006. [Google Scholar]
- Zhao, Q.B.; Sun, H.; Zhang, Z.; Xu, Z.; Olasege, B.S.; Ma, P.P.; Zhang, X.Z.; Wang, Q.S.; Pan, Y.C. Exploring the Structure of Haplotype Blocks and Genetic Diversity in Chinese Indigenous Pig Populations for Conservation Purpose. Evol. Bioinform. Online 2019, 15, 1176934318825082. [Google Scholar] [CrossRef]
- Gibson, J.; Morton, N.E.; Collins, A. Extended tracts of homozygosity in outbred human populations. Hum. Mol. Genet. 2006, 15, 789–795. [Google Scholar] [CrossRef]
- Ghoreishifar, S.M.; Moradi-Shahrbabak, H.; Parna, N.; Davoudi, P.; Khansefid, M. Linkage disequilibrium and within-breed genetic diversity in Iranian Zandi sheep. Arch. Anim. Breed. 2019, 62, 143–151. [Google Scholar] [CrossRef]
- Tao, L.; He, X.Y.; Wang, F.Y.; Pan, L.X.; Wang, X.Y.; Gan, S.Q.; Di, R.; Chu, M.X. Identification of genes associated with litter size combining genomic approaches in Luzhong mutton sheep. Anim. Genet. 2021, 52, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Machmoum, M.; Boujenane, I.; Azelhak, R.; Badaoui, B.; Petit, D.; Piro, M. Genetic Diversity and Population Structure of Arabian Horse Populations Using Microsatellite Markers. J. Equine Vet. Sci. 2020, 93, 103200. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.-H.; Zhang, Z.; Xiao, Q.; Wang, Q.-S.; Chen, G.-H.; Pan, Y.-C. Genetic Diversity and Population Structure Analysis of Shanghai White Pig (Sus scrofa). J. Agric. Biotechnol. 2016, 24, 1293–1301. [Google Scholar]
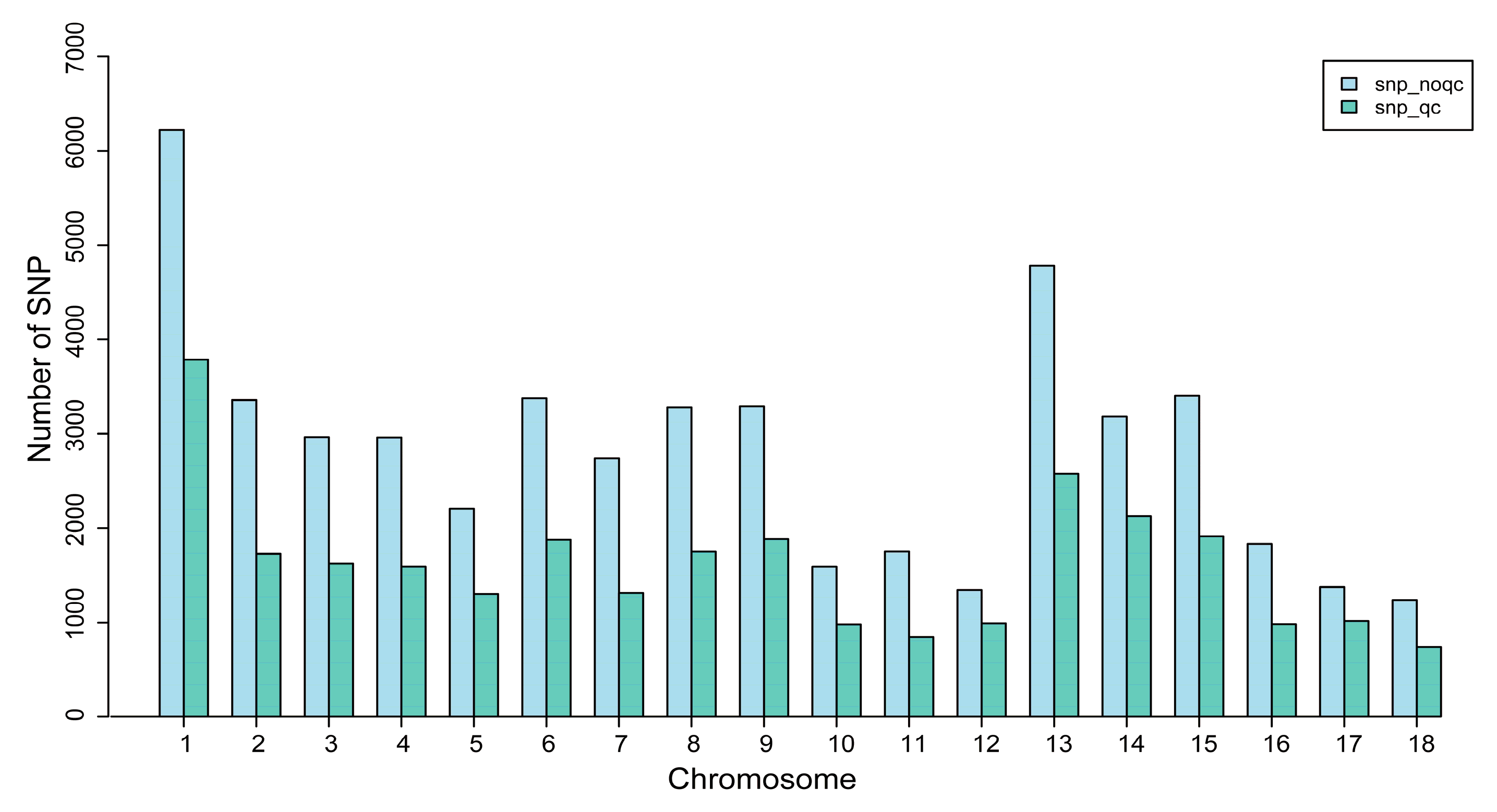
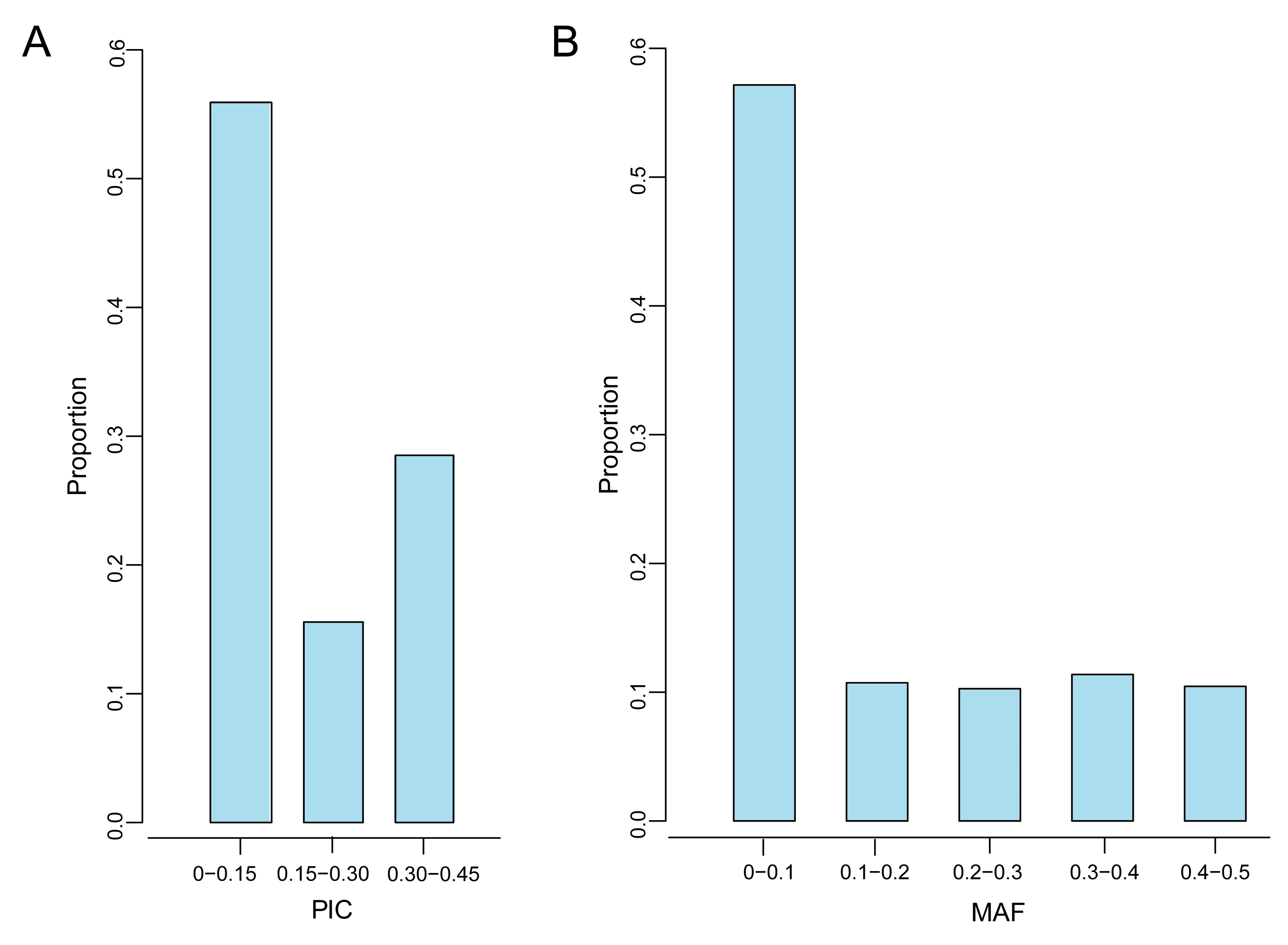
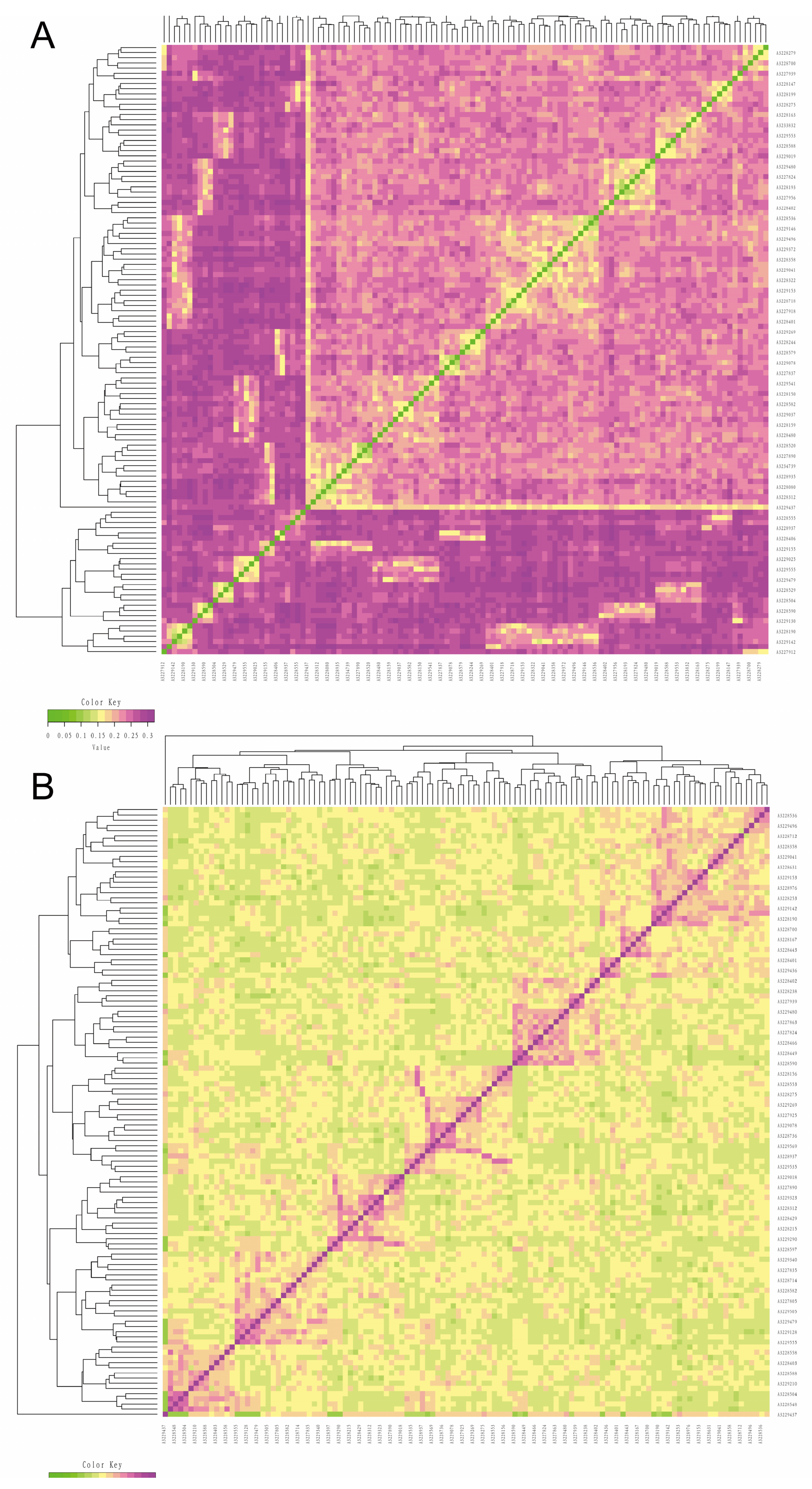
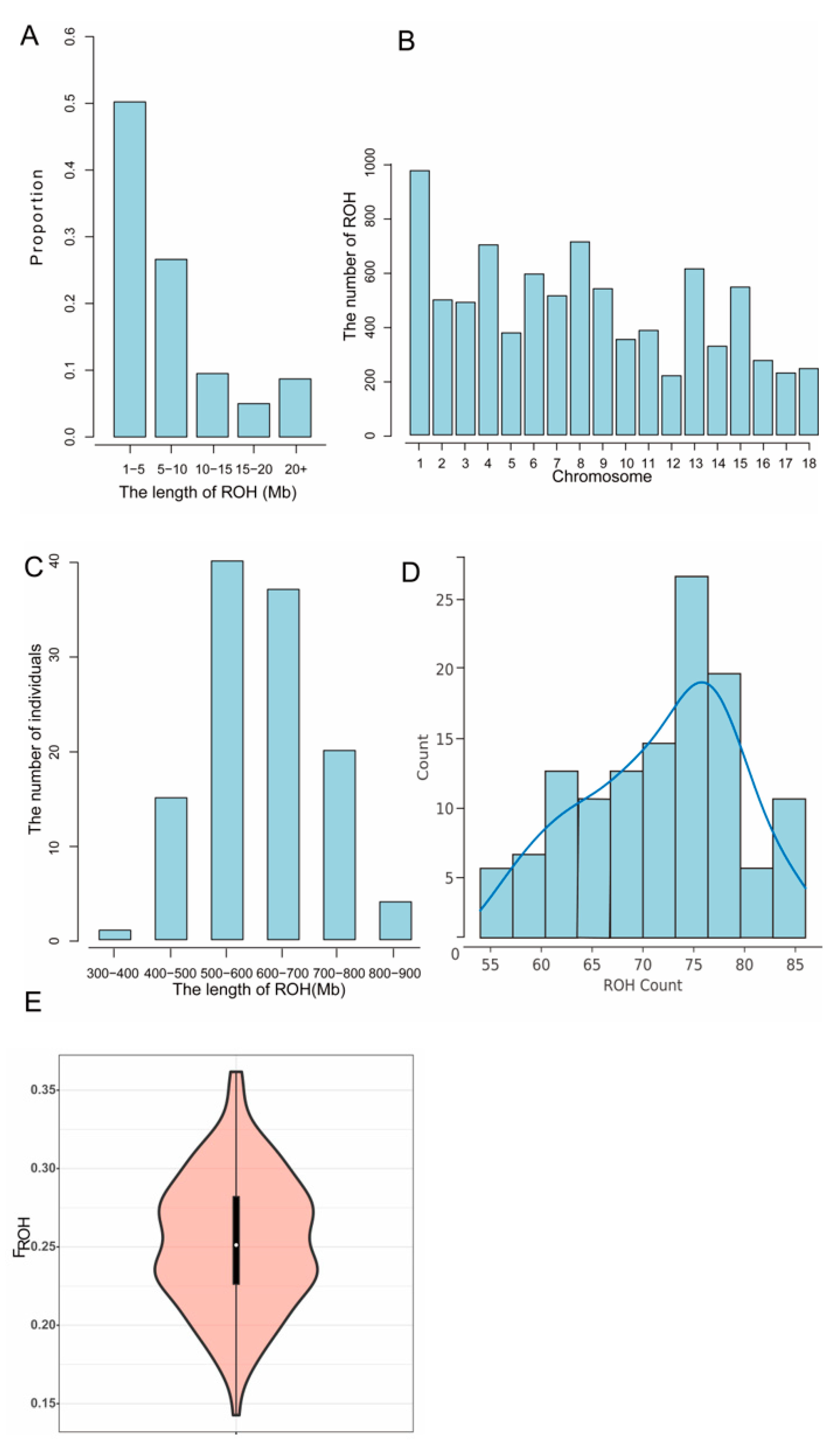
| Quality Control Standards | Number of SNPs Tags |
|---|---|
| Total number of SNPs | 57,466 |
| SNP with MAF < 0.01 | 20,888 |
| SNP noting Hardy–Weinberg equilibrium p < 10−6 | 147 |
| SNP with call rate < 0.90 | 830 |
| SNPs on chromosome X | 4252 |
| SNPs on chromosome 0 | 2310 |
| Insertion/deletion | 6 |
| SNPs used after quality control | 29,033 |
| Genetic Diversity Parameters | Parameter |
|---|---|
| Ne | 4.9 |
| PN | 0.507 |
| HE | 0.315 |
| HO | 0.345 |
| PIC | 0.147 |
| Ae | 1.528 |
| MAF | 0.137 |
| Family | Sex | Amount |
|---|---|---|
| 1 | boar | 5 |
| sow | 64 | |
| 2 | boar | 13 |
| sow | 99 | |
| 3 | boar | 9 |
| sow | 84 | |
| 4 | boar | 7 |
| sow | 78 | |
| other | sow | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, M.; Wang, L.; Ma, Z.; Mao, Y.; Wang, G.; Zheng, R.; Zuo, B.; Wang, Y. Analysis of the Genetic Diversity and Genetic Structure of Jiangshan Black Pigs Using Single Nucleotide Polymorphism (SNP) Chips. Animals 2024, 14, 2660. https://doi.org/10.3390/ani14182660
Zhu M, Wang L, Ma Z, Mao Y, Wang G, Zheng R, Zuo B, Wang Y. Analysis of the Genetic Diversity and Genetic Structure of Jiangshan Black Pigs Using Single Nucleotide Polymorphism (SNP) Chips. Animals. 2024; 14(18):2660. https://doi.org/10.3390/ani14182660
Chicago/Turabian StyleZhu, Mingfei, Litong Wang, Zhibo Ma, Yangcang Mao, Guoshui Wang, Rong Zheng, Bo Zuo, and Yizhen Wang. 2024. "Analysis of the Genetic Diversity and Genetic Structure of Jiangshan Black Pigs Using Single Nucleotide Polymorphism (SNP) Chips" Animals 14, no. 18: 2660. https://doi.org/10.3390/ani14182660
APA StyleZhu, M., Wang, L., Ma, Z., Mao, Y., Wang, G., Zheng, R., Zuo, B., & Wang, Y. (2024). Analysis of the Genetic Diversity and Genetic Structure of Jiangshan Black Pigs Using Single Nucleotide Polymorphism (SNP) Chips. Animals, 14(18), 2660. https://doi.org/10.3390/ani14182660





