Pleiotropic Gene HMGA2 Regulates Myoblast Proliferation and Affects Body Size of Sheep
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals and Isolation of Sheep Myoblasts
2.3. DNA and RNA Extraction and cDNA Synthesis
2.4. Overexpression and Interference of HMGA2 Gene
2.5. Detection of Sheep Myoblast Proliferation
2.6. Determination of HMGA2 Promoter Activity
2.7. Identification and Genotyping of SNP in the HMGA2 Core Promoter
2.8. Statistical Analysis
3. Results
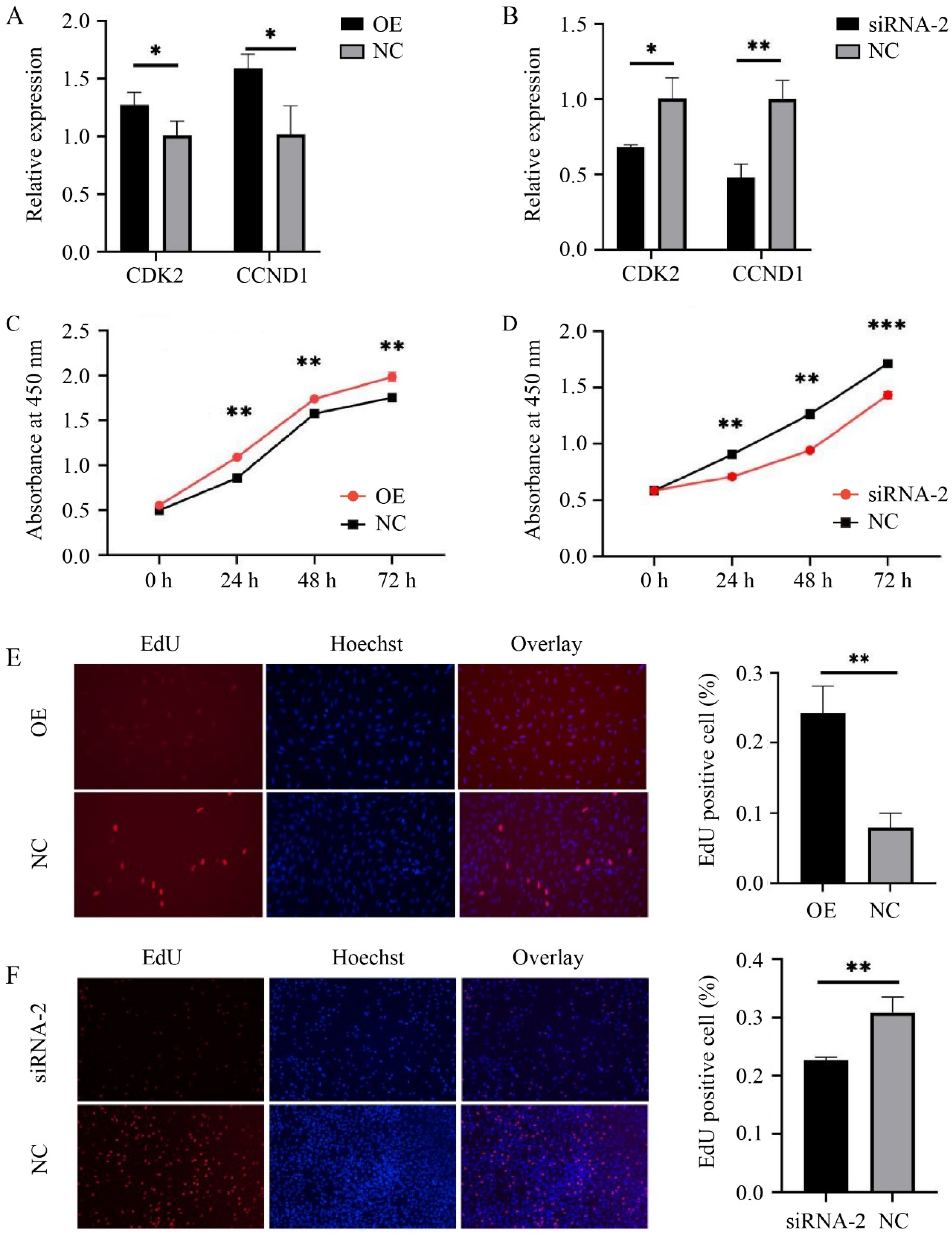
3.1. Efficiencies of Overexpression and Interference
3.2. HMGA2 Promotes Proliferation of Sheep Myoblasts
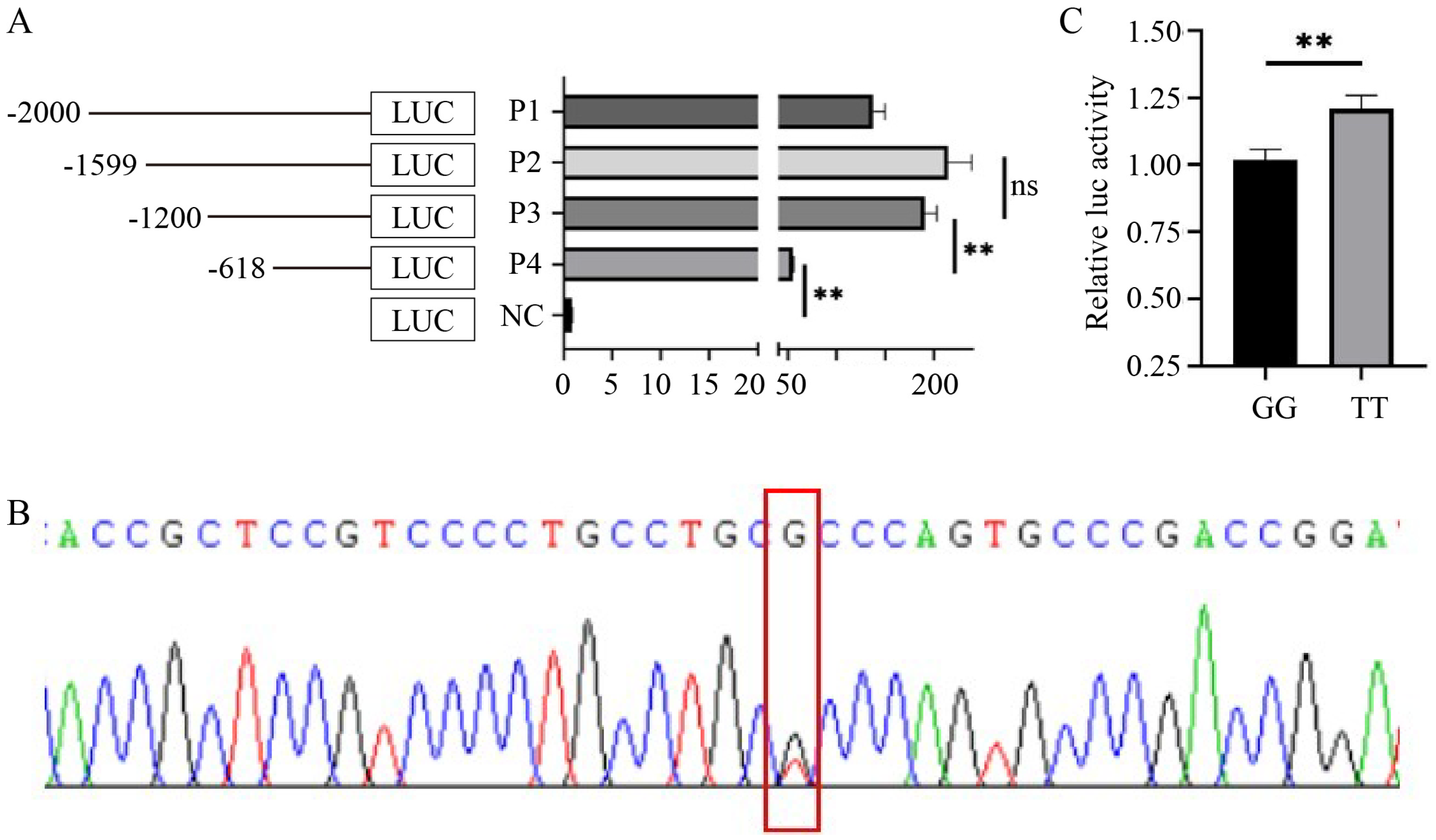
3.3. SNP rs428001129 within HMGA2 Promoter Affects Promoter Activity
3.4. Efficiencies of Overexpression and Interference
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yue, G. Reproductive characteristics of Chinese Hu sheep. Anim. Reprod. Sci. 1996, 44, 223–230. [Google Scholar] [CrossRef]
- Matarneh, S.K.; Silva, S.L.; Gerrard, D.E. New insights in muscle biology that alter meat quality. Annu. Rev. Anim. Biosci. 2021, 9, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. CSH Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Millay, D.P.; O’Rourke, J.R.; Sutherland, L.B.; Bezprozvannaya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 2013, 499, 301–305. [Google Scholar] [CrossRef]
- Quinn, M.E.; Goh, Q.; Kurosaka, M.; Gamage, D.G.; Petrany, M.J.; Prasad, V.; Millay, D.P. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat. Commun. 2017, 8, 15665. [Google Scholar] [CrossRef]
- Chen, B.; You, W.; Wang, Y.; Shan, T. The regulatory role of Myomaker and Myomixer–Myomerger–Minion in muscle development and regeneration. Cell. Mol. Life Sci. 2020, 77, 1551–1569. [Google Scholar] [CrossRef]
- Xiang, X.; Benson, K.F.; Chada, K. Mini-mouse: Disruption of the pygmy locus in a transgenic insertional mutant. Science 1990, 247, 967–969. [Google Scholar] [CrossRef]
- Benson, K.F.; Chada, K. Mini-mouse: Phenotypic characterization of a transgenic insertional mutant allelic to pygmy. Genet. Res. 1994, 64, 27–33. [Google Scholar] [CrossRef][Green Version]
- Zhou, X.; Benson, K.F.; Ashar, H.R.; Chada, K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376, 771–774. [Google Scholar] [CrossRef]
- Chung, J.; Zhang, X.; Collins, B.; Sper, R.B.; Gleason, K.; Simpson, S.; Koh, S.; Sommer, J.; Flowers, W.L.; Petters, R.M. High mobility group A2 (HMGA2) deficiency in pigs leads to dwarfism, abnormal fetal resource allocation, and cryptorchidism. Proc. Natl. Acad. Sci. USA 2018, 115, 5420–5425. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.P.; de Groof, A.; van der Poel, J.J.; Herbergs, J.; Masabanda, J.; Fries, R.; Groenen, M. The HMGI-C gene is a likely candidate for the autosomal dwarf locus in the chicken. J. Hered. 1998, 89, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Ruyter-Spira, C.; Herbergs, J.; Limpens, E.; Marsh, J.; van Der Poel, J.; Ayoubi, T.; Groenen, M. Nucleotide sequence of the chicken HMGI-C cDNA and expression of the HMGI-C and IGF1 genes in autosomal dwarf chicken embryos. BBA-Gene Struct. Expr. 1998, 1399, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.; Hu, D.; Archer, J.; Feng, C.; Afonso, S.; Chen, C.; Blanco-Aguiar, J.A.; Garreau, H.; Boucher, S.; Ferreira, P.G. Dwarfism and altered craniofacial development in rabbits is caused by a 12.1 kb deletion at the HMGA2 locus. Genetics 2017, 205, 955–965. [Google Scholar] [CrossRef]
- Hammond, S.M.; Sharpless, N.E. HMGA2, microRNAs, and stem cell aging. Cell 2008, 135, 1013–1016. [Google Scholar] [CrossRef]
- Smeti, I.; Watabe, I.; Savary, E.; Fontbonne, A.; Zine, A. HMGA2, the architectural transcription factor high mobility group, is expressed in the developing and mature mouse cochlea. PLoS ONE 2014, 9, e88757. [Google Scholar] [CrossRef]
- Reeves, R.; Beckerbauer, L. HMGI/Y proteins: Flexible regulators of transcription and chromatin structure. BBA-Gene Struct. Expr. 2001, 1519, 13–29. [Google Scholar] [CrossRef]
- Cui, T.; Leng, F. Specific recognition of AT-rich DNA sequences by the mammalian high mobility group protein AT-hook 2: A SELEX study. Biochemistry 2007, 46, 13059–13066. [Google Scholar] [CrossRef]
- Zha, L.; Wang, Z.; Tang, W.; Zhang, N.; Liao, G.; Huang, Z. Genome-wide analysis of HMGA2 transcription factor binding sites by ChIP on chip in gastric carcinoma cells. Mol. Cell. Biochem. 2012, 364, 243–251. [Google Scholar] [CrossRef]
- Vignali, R.; Marracci, S. HMGA genes and proteins in development and evolution. Int. J. Mol. Sci. 2020, 21, 654. [Google Scholar] [CrossRef]
- Li, Z.; Gilbert, J.A.; Zhang, Y.; Zhang, M.; Qiu, Q.; Ramanujan, K.; Shavlakadze, T.; Eash, J.K.; Scaramozza, A.; Goddeeris, M.M. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev. Cell 2012, 23, 1176–1188. [Google Scholar] [CrossRef]
- Makvandi-Nejad, S.; Hoffman, G.E.; Allen, J.J.; Chu, E.; Gu, E.; Chandler, A.M.; Loredo, A.I.; Bellone, R.R.; Mezey, J.G.; Brooks, S.A. Four loci explain 83% of size variation in the horse. PLoS ONE 2012, 7, e39929. [Google Scholar] [CrossRef] [PubMed]
- Hayward, J.J.; Castelhano, M.G.; Oliveira, K.C.; Corey, E.; Balkman, C.; Baxter, T.L.; Casal, M.L.; Center, S.A.; Fang, M.; Garrison, S.J. Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 2016, 7, 10460. [Google Scholar] [CrossRef] [PubMed]
- Cesar, A.S.; Regitano, L.C.; Reecy, J.M.; Poleti, M.D.; Oliveira, P.S.; de Oliveira, G.B.; Moreira, G.C.; Mudadu, M.A.; Tizioto, P.C.; Koltes, J.E. Identification of putative regulatory regions and transcription factors associated with intramuscular fat content traits. BMC Genom. 2018, 19, 499. [Google Scholar] [CrossRef]
- Bolormaa, S.; Pryce, J.E.; Reverter, A.; Zhang, Y.; Barendse, W.; Kemper, K.; Tier, B.; Savin, K.; Hayes, B.J.; Goddard, M.E. A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 2014, 10, e1004198. [Google Scholar] [CrossRef]
- Selionova, M.; Aibazov, M.; Sermyagin, A.; Belous, A.; Deniskova, T.; Mamontova, T.; Zharkova, E.; Zinovieva, N. Genome-Wide Association and Pathway Analysis of Carcass and Meat Quality Traits in Karachai Young Goats. Animals 2023, 13, 3237. [Google Scholar] [CrossRef]
- Saatchi, M.; Schnabel, R.D.; Taylor, J.F.; Garrick, D.J. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genom. 2014, 15, 442. [Google Scholar] [CrossRef]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef]
- Danoviz, M.E.; Yablonka-Reuveni, Z. Skeletal muscle satellite cells: Background and methods for isolation and analysis in a primary culture system. Myogenesis Methods Protoc. 2012, 798, 21–52. [Google Scholar]
- Di Pietro, F.; Ortenzi, F.; Tilio, M.; Concetti, F.; Napolioni, V. Genomic DNA extraction from whole blood stored from 15-to 30-years at− 20 C by rapid phenol–chloroform protocol: A useful tool for genetic epidemiology studies. Mol. Cell. Probes 2011, 25, 44–48. [Google Scholar] [CrossRef]
- Simms, D.; Cizdziel, P.E.; Chomczynski, P. TRIzol: A new reagent for optimal single-step isolation of RNA. Focus 1993, 15, 532–535. [Google Scholar]
- Wang, S.; Cao, X.; Ge, L.; Gu, Y.; Lv, X.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Sun, W. MiR-22-3p inhibits proliferation and promotes differentiation of skeletal muscle cells by targeting IGFBP3 in Hu sheep. Animals 2022, 12, 114. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.; He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15, 97. [Google Scholar]
- Ashar, H.R.; Chouinard, R.A., Jr.; Dokur, M.; Chada, K. In vivo modulation of HMGA2 expression. BBA-Gene Regul. Mech. 2010, 1799, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Ditzel, H.J.; Duijf, P.H.; Khaze, V.; Gjerstorff, M.F.; Baradaran, B. HMGA2 as a critical regulator in cancer development. Genes 2021, 12, 269. [Google Scholar] [CrossRef]
- Zhu, H.; Shah, S.; Shyh-Chang, N.; Shinoda, G.; Einhorn, W.S.; Viswanathan, S.R.; Takeuchi, A.; Grasemann, C.; Rinn, J.L.; Lopez, M.F. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 2010, 42, 626–630. [Google Scholar] [CrossRef]
- Copley, M.R.; Babovic, S.; Benz, C.; Knapp, D.J.; Beer, P.A.; Kent, D.G.; Wohrer, S.; Treloar, D.Q.; Day, C.; Rowe, K. The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 2013, 15, 916–925. [Google Scholar] [CrossRef]
- Powers, J.T.; Tsanov, K.M.; Pearson, D.S.; Roels, F.; Spina, C.S.; Ebright, R.; Seligson, M.; De Soysa, Y.; Cahan, P.; Theißen, J. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 2016, 535, 246–251. [Google Scholar] [CrossRef]
- Dai, N.; Zhao, L.; Wrighting, D.; Krämer, D.; Majithia, A.; Wang, Y.; Cracan, V.; Borges-Rivera, D.; Mootha, V.K.; Nahrendorf, M. IGF2BP2/IMP2-deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins. Cell Metab. 2015, 21, 609–621. [Google Scholar] [CrossRef]
- Abi Habib, W.; Brioude, F.; Edouard, T.; Bennett, J.T.; Lienhardt-Roussie, A.; Tixier, F.; Salem, J.; Yuen, T.; Azzi, S.; Le Bouc, Y. Genetic disruption of the oncogenic HMGA2–PLAG1–IGF2 pathway causes fetal growth restriction. Genet. Med. 2018, 20, 250–258. [Google Scholar] [CrossRef]
- Preissl, S.; Gaulton, K.J.; Ren, B. Characterizing cis-regulatory elements using single-cell epigenomics. Nat. Rev. Genet. 2023, 24, 21–43. [Google Scholar] [CrossRef]
- Shirai, K.; Nagae, G.; Seki, M.; Kudo, Y.; Kamio, A.; Hayashi, A.; Okabe, A.; Ota, S.; Tsutsumi, S.; Fujita, T. TET1 upregulation drives cancer cell growth through aberrant enhancer hydroxymethylation of HMGA2 in hepatocellular carcinoma. Cancer Sci. 2021, 112, 2855–2869. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Cao, X.; Wang, X.; Wang, J.; Yue, B.; Sun, W.; Huang, Y.; Lan, X.; Ren, G.; Lei, C. Dynamic chromatin architectures provide insights into the genetics of cattle myogenesis. J. Anim. Sci. Biotechnol. 2023, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Zhang, Y.; Li, L.; Xie, X.; Huang, J.; Zhang, M.; Ni, X.; Li, X. A dual-luciferase reporter system for characterization of small RNA target genes in both mammalian and insect cells. Insect Sci. 2022, 29, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Z.; Kanagaratham, C.; Jancik, S.; Radzioch, D. Promoter deletion analysis using a dual-luciferase reporter system. Gene Regul. Methods Protoc. 2013, 977, 79–93. [Google Scholar]
- Aljubouri, T.R.S.; Al-Shuhaib, M.B.S.; Javadmanesh, A. HMGA2 gene polymorphisms and their effects on main growth traits indices in Awassi and Karakul sheep. Agric. Nat. Resour. 2020, 54, 587–594. [Google Scholar]

| ID | Sequence (5′-3′) | Product |
|---|---|---|
| siRNA-1 | CCGGUGAGCCCUCUCCUAATT | \ |
| UAGGAGAGGGCUCACCGGTT | ||
| siRNA-2 | GAGACAUCCUCACAAGAGUTT | \ |
| ACUCUUGUGAGGAUGUCUCTT | ||
| siRNA-3 | GCCAGCAUUCAAUUUCUACTT | \ |
| GUAGAAAUUGAAUGCUGGCTT | ||
| siRNA-NC | UUCUCCGAACGUGUCACGUTT | \ |
| ACGUGACACGUUCGGAGAATT | ||
| CDK2-F | TGGGCCAGGCAGGATTTTAG | 166 bp |
| CDK2-R | GTCGAAGGTGAGGTACTGGC | |
| CCND1-F | GCTTCCTCTCCTATCACCGC | 149 bp |
| CCND1-R | GGCTTTGGGGTCCAAGTTCT | |
| GAPDH-F | GTCGGAGTGAACGGATTTGG | 196 bp |
| GAPDH-R | CATTGATGACGAGATTCCCG | |
| HMGA2-F | AGACCCAAAGGCAGCAAAAAC | 100 bp |
| HMGA2-R | GCCATTTCCTAGGTCTGCCTC | |
| P1 | cgagctcttacgcgtgctagcAAAAGTTTTTATTTTGGAATTG | 1942 bp |
| P2 | cgagctcttacgcgtgctagcTCAGTGGAGGCTGGTGCG | 1541 bp |
| P3 | cgagctcttacgcgtgctagcCAGGTAAAGGCCAAGCCCC | 1142 bp |
| P4 | cgagctcttacgcgtgctagcGGATCCCCGCAGAATCTCC | 560 bp |
| R | acttagatcgcagatctcgagTCGCCTCTGTCGCCCTGA | \ |
| SNP-F | GCCTCCCTCCTCCTCATACT | 492 bp |
| SNP-R | CGGCTTGGAAAGGGAAGAGA |
| Age | Traits | Genotype | p Value | ||
|---|---|---|---|---|---|
| GG | GT | TT | |||
| birth | body weight | 3.67 ± 0.81 | 3.73 ± 0.84 | 3.95 ± 0.94 | 0.250 |
| withers height | 37.63 ± 3.13 | 37.90 ± 2.88 | 38.85 ± 2.47 | 0.141 | |
| body length | 30.46 ± 2.78 | 30.88 ± 3.01 | 31.17 ± 2.99 | 0.264 | |
| chest girth | 35.62 ± 2.89 | 35.82 ± 3.03 | 37.17 ± 4.54 | 0.055 | |
| chest depth | 15.60 ± 2.16 | 15.58 ± 1.52 | 15.90 ± 1.85 | 0.704 | |
| chest width | 10.16 ± 1.48 b | 10.44 ± 1.50 b | 11.06 ± 1.65 a | 0.009 | |
| cannon circumference | 5.83 ± 0.68 | 5.93 ± 0.70 | 6.06 ± 0.62 | 0.183 | |
| 1 mth | body weight | 10.22 ± 2.43 | 9.82 ± 2.24 | 10.30 ± 2.32 | 0.545 |
| withers height | 45.74 ± 3.06 | 44.93 ± 3.11 | 43.85 ± 3.48 | 0.095 | |
| body length | 43.43 ± 3.83 | 42.71 ± 3.48 | 42.70 ± 2.91 | 0.455 | |
| chest girth | 50.06 ± 4.34 | 49.66 ± 3.65 | 50.30 ± 3.56 | 0.781 | |
| chest depth | 22.23 ± 1.86 | 21.95 ± 1.71 | 21.85 ± 1.62 | 0.568 | |
| chest width | 14.43 ± 1.51 | 14.24 ± 1.47 | 14.30 ± 1.67 | 0.725 | |
| cannon circumference | 6.50 ± 0.49 | 6.38 ± 0.45 | 6.45 ± 0.37 | 0.292 | |
| 2 mth | body weight | 17.85 ± 3.81 | 18.28 ± 3.89 | 18.64 ± 4.49 | 0.411 |
| withers height | 49.75 ± 3.41 | 50.28 ± 3.58 | 50.68 ± 3.95 | 0.216 | |
| body length | 51.93 ± 4.54 | 52.53 ± 4.94 | 52.75 ± 4.81 | 0.409 | |
| chest girth | 59.85 ± 5.08 | 60.66 ± 4.82 | 60.96 ± 5.66 | 0.232 | |
| chest depth | 23.54 ± 2.22 b | 23.88 ± 2.19 ab | 24.61 ± 2.31 a | 0.040 | |
| chest width | 16.34 ± 2.23 b | 16.94 ± 2.40 a | 17.79 ± 2.94 a | 0.002 | |
| cannon circumference | 6.92 ± 0.58 | 7.00 ± 0.62 | 7.20 ± 0.79 | 0.062 | |
| 3 mth | body weight | 24.41 ± 3.95 | 24.44 ± 4.08 | 24.70 ± 3.90 | 0.981 |
| withers height | 54.70 ± 3.02 | 54.87 ± 3.30 | 55.19 ± 3.27 | 0.912 | |
| body length | 56.64 ± 3.06 | 56.98 ± 3.24 | 58.06 ± 3.12 | 0.490 | |
| chest girth | 67.32 ± 4.27 | 67.13 ± 4.70 | 67.38 ± 3.42 | 0.973 | |
| chest depth | 26.83 ± 1.69 | 26.90 ± 1.70 | 26.81 ± 1.28 | 0.975 | |
| chest width | 17.71 ± 1.64 | 17.95 ± 1.53 | 18.13 ± 1.30 | 0.659 | |
| cannon circumference | 7.44 ± 0.44 | 7.36 ± 0.54 | 7.50 ± 0.38 | 0.632 | |
| 6 mth | body weight | 37.33 ± 7.39 | 37.50 ± 7.43 | 37.90 ± 8.65 | 0.848 |
| withers height | 61.44 ± 4.25 | 61.83 ± 3.81 | 62.43 ± 4.26 | 0.292 | |
| body length | 66.18 ± 4.68 | 65.90 ± 4.62 | 67.33 ± 5.07 | 0.313 | |
| chest girth | 77.90 ± 6.00 | 77.70 ± 5.95 | 78.00 ± 7.40 | 0.969 | |
| chest depth | 28.66 ± 2.13 b | 28.82 ± 2.15 b | 29.72 ± 2.93 a | 0.047 | |
| chest width | 19.33 ± 2.21 | 19.47 ± 2.32 | 20.02 ± 2.30 | 0.269 | |
| cannon circumference | 8.05 ± 0.84 | 8.02 ± 0.81 | 8.38 ± 1.07 | 0.107 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Ling, C.; Liu, Y.; Gu, Y.; Huang, J.; Sun, W. Pleiotropic Gene HMGA2 Regulates Myoblast Proliferation and Affects Body Size of Sheep. Animals 2024, 14, 2721. https://doi.org/10.3390/ani14182721
Cao X, Ling C, Liu Y, Gu Y, Huang J, Sun W. Pleiotropic Gene HMGA2 Regulates Myoblast Proliferation and Affects Body Size of Sheep. Animals. 2024; 14(18):2721. https://doi.org/10.3390/ani14182721
Chicago/Turabian StyleCao, Xiukai, Chen Ling, Yongqi Liu, Yifei Gu, Jinlin Huang, and Wei Sun. 2024. "Pleiotropic Gene HMGA2 Regulates Myoblast Proliferation and Affects Body Size of Sheep" Animals 14, no. 18: 2721. https://doi.org/10.3390/ani14182721
APA StyleCao, X., Ling, C., Liu, Y., Gu, Y., Huang, J., & Sun, W. (2024). Pleiotropic Gene HMGA2 Regulates Myoblast Proliferation and Affects Body Size of Sheep. Animals, 14(18), 2721. https://doi.org/10.3390/ani14182721






