Preliminary Functional Analysis of the Gut Microbiome in Colic Horses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sampling Location
2.3. Criteria Inclusion
2.4. Microbiome Study
2.5. Statistical Analyses
2.6. Data Availability
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jorgenson, J.K.; Bennett, J.A.; Burk, S.V. A Comparative Study of Equine Gut Microbiomes Using 16S and 18S rRNA Sequencing. FASEB J. 2019, 33, 484.8. [Google Scholar] [CrossRef]
- Al Jassim, R.A.M.; Andrews, F.M. The Bacterial Community of the Horse Gastrointestinal Tract and Its Relation to Fermentative Acidosis, Laminitis, Colic, and Stomach Ulcers. Vet. Clin. N. Am. Equine Pract. 2009, 25, 199–215. [Google Scholar] [CrossRef]
- Julliand, V.; de Vaux, A.; Millet, L.; Fonty, G. Identification of Ruminococcus flavefaciens as the Predominant Cellulolytic Bacterial Species of the Equine Cecum. Appl. Environ. Microbiol. 1999, 65, 3738–3741. [Google Scholar] [CrossRef]
- Cerqueira, F.M.; Photenhauer, A.L.; Pollet, R.M.; Brown, H.A.; Koropatkin, N.M. Starch Digestion by Gut Bacteria: Crowdsourcing for Carbs. Trends Microbiol. 2020, 28, 95–108. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Żak-Bochenek, A.; Bajzert, J.; Sambor, D.; Siwińska, N.; Szponar, B.; Łaczmański, Ł.; Żebrowska, P.; Czajkowska, A.; Karczewski, M.; Chełmońska-Soyta, A. Homeostasis of the Intestinal Mucosa in Healthy Horses-Correlation between the Fecal Microbiome, Secretory Immunoglobulin A and Fecal Egg Count. Animals 2022, 12, 3094. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Theelen, M.J.P.; Luiken, R.E.C.; Wagenaar, J.A.; Sloet van Oldruitenborgh-Oosterbaan, M.M.; Rossen, J.W.A.; Zomer, A.L. The Equine Faecal Microbiota of Healthy Horses and Ponies in The Netherlands: Impact of Host and Environmental Factors. Animals 2021, 11, 1762. [Google Scholar] [CrossRef]
- Daly, K.; Proudman, C.J.; Duncan, S.H.; Flint, H.J.; Dyer, J.; Shirazi-Beechey, S.P. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease. Br. J. Nutr. 2012, 107, 989–995. [Google Scholar] [CrossRef]
- Ganda, E.; Chakrabarti, A.; Sardi, M.I.; Tench, M.; Kozlowicz, B.K.; Norton, S.A.; Warren, L.K.; Khafipour, E. Saccharomyces cerevisiae fermentation product improves robustness of equine gut microbiome upon stress. Front. Vet. Sci. 2023, 10, 1134092. [Google Scholar] [CrossRef]
- Murray, J.-A.M.D.; Brown, S.; O’Shaughnessy, P.; Monteiro, A.; Warren, H.; Hastie, P.M. Effect of Live Yeast Culture Supplementation on Fibrolytic and Saccharolytic Bacterial Populations in the Feces of Horses Fed a High-Fiber or High-Starch Diet. J. Equine Vet. Sci. 2017, 51, 41–45. [Google Scholar] [CrossRef]
- Edwards, J.E.; Shetty, S.A.; van den Berg, P.; Burden, F.; van Doorn, D.A.; Pellikaan, W.F.; Dijkstra, J.; Smidt, H. Multi-kingdom characterization of the core equine fecal microbiota based on multiple equine (sub)species. Anim. Microbiome 2020, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, M.L.; Swecker, W.S., Jr.; Jensen, R.V.; Ponder, M.A. Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V4 gene amplicons. FEMS Microbiol. Lett. 2012, 326, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Dougal, K.; Harris, P.A.; Girdwood, S.E.; Creevey, C.J.; Curtis, G.C.; Barfoot, C.F.; Argo, C.M.; Newbold, C.J. Changes in the Total Fecal Bacterial Population in Individual Horses Maintained on a Restricted Diet Over 6 Weeks. Front. Microbiol. 2017, 8, 1502. [Google Scholar] [CrossRef]
- Costa, M.C.; Arroyo, L.G.; Allen-Vercoe, E.; Stämpfli, H.R.; Kim, P.T.; Sturgeon, A.; Weese, J.S. Comparison of the Fecal Microbiota of Healthy Horses and Horses with Colitis by High Throughput Sequencing of the V3-V5 Region of the 16S rRNA Gene. PLoS ONE 2012, 7, e41484. [Google Scholar] [CrossRef]
- Massacci, F.R.; Clark, A.; Ruet, A.; Lansade, L.; Costa, M.; Mach, N. Inter-breed diversity and temporal dynamics of the faecal microbiota in healthy horses. J. Anim. Breed. Genet. 2020, 137, 103–120. [Google Scholar] [CrossRef]
- Lee, J.; Kang, Y.-J.; Kim, Y.-K.; Choi, J.-Y.; Shin, S.-M.; Shin, M.-C. Exploring the Influence of Growth-Associated Host Genetics on the Initial Gut Microbiota in Horses. Genes 2023, 14, 1354. [Google Scholar] [CrossRef]
- Plancade, S.; Clark, A.; Philippe, C.; Helbling, J.-C.; Moisan, M.-P.; Esquerré, D.; Le Moyec, L.; Robert, C.; Barrey, E.; Mach, N. Unraveling the effects of the gut microbiota composition and function on horse endurance physiology. Sci. Rep. 2019, 9, 9620. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Guo, R.; Ni, W.; Liu, K.; Liu, Z.; Dai, J.; Xu, Y.; Abduriyim, S.; Wu, Z.; et al. Expanded catalogue of metagenome-assembled genomes reveals resistome characteristics and athletic performance-associated microbes in horse. Microbiome 2023, 11, 7. [Google Scholar] [CrossRef]
- Blackmore, T.M.; Dugdale, A.; Argo, C.M.; Curtis, G.; Pinloche, E.; Harris, P.A.; Worgan, H.J.; Girdwood, S.E.; Dougal, K.; Newbold, C.J.; et al. Strong Stability and Host Specific Bacterial Community in Faeces of Ponies. PLoS ONE 2013, 8, e75079. [Google Scholar] [CrossRef]
- Venable, E.B.; Bland, S.D.; McPherson, J.L.; Francis, J. Role of the gut microbiota in equine health and disease. Anim. Front. 2016, 6, 43–49. [Google Scholar] [CrossRef]
- Salem, S.E.; Maddox, T.W.; Berg, A.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period. Sci. Rep. 2018, 8, 8510. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Holcombe, S.J.; Embertson, R.M.; Kurtz, K.A.; Roessner, H.A.; Jalali, M.; Wismer, S.E. Changes in the faecal microbiota of mares precede the development of post partum colic. Equine Vet. J. 2015, 47, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Elzinga, S.E.; Weese, J.S.; Adams, A.A. Comparison of the Fecal Microbiota in Horses with Equine Metabolic Syndrome and Metabolically Normal Controls Fed a Similar All-Forage Diet. J. Equine Vet. Sci. 2016, 44, 9–16. [Google Scholar] [CrossRef]
- Lara, F.; Castro, R.; Thomson, P. Changes in the gut microbiome and colic in horses: Are they causes or consequences? Open Vet. J. 2022, 12, 242–249. [Google Scholar] [CrossRef]
- Durham, A.E. The Role of Nutrition in Colic. Vet. Clin. N. Am. Equine Pract. 2009, 25, 67–78. [Google Scholar] [CrossRef]
- Costa, M.C.; Weese, J.S. Understanding the Intestinal Microbiome in Health and Disease. Vet. Clin. N. Am. Equine Pract. 2018, 34, 1–12. [Google Scholar] [CrossRef]
- Erwin, S.J.; Blikslager, A.T.; Ziegler, A.L. Age-Dependent Intestinal Repair: Implications for Foals with Severe Colic. Animals 2021, 11, 3337. [Google Scholar] [CrossRef]
- Stewart, H.L.; Southwood, L.L.; Indugu, N.; Vecchiarelli, B.; Engiles, J.B.; Pitta, D. Differences in the equine faecal microbiota between horses presenting to a tertiary referral hospital for colic compared with an elective surgical procedure. Equine Vet. J. 2019, 51, 336–342. [Google Scholar] [CrossRef]
- Faubladier, C.; Chaucheyras-Durand, F.; da Veiga, L.; Julliand, V. Effect of transportation on fecal bacterial communities and fermentative activities in horses: Impact of Saccharomyces cerevisiae CNCM I-1077 supplementation. J. Anim. Sci. 2013, 91, 1736–1744. [Google Scholar] [CrossRef]
- Dougal, K.; de la Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Geor, R.J.; Nielsen, B.D.; Schott, H.C., II; Elzinga, S.; Newbold, C.J. Characterisation of the Faecal Bacterial Community in Adult and Elderly Horses Fed a High Fibre, High Oil or High Starch Diet Using 454 Pyrosequencing. PLoS ONE 2014, 9, e87424. [Google Scholar] [CrossRef] [PubMed]
- Proudman, C.J.; Hunter, J.O.; Darby, A.C.; Escalona, E.E.; Batty, C.; Turner, C. Characterisation of the faecal metabolome and microbiome of Thoroughbred racehorses. Equine Vet. J. 2015, 47, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Schoster, A.; Mosing, M.; Jalali, M.; Staempfli, H.R.; Weese, J.S. Effects of transport, fasting and anaesthesia on the faecal microbiota of healthy adult horses. Equine Vet. J. 2016, 48, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Park, T.; Cheong, H.; Yoon, J.; Kim, A.; Yun, Y.; Unno, T. Comparison of the Fecal Microbiota of Horses with Intestinal Disease and Their Healthy Counterparts. Vet. Sci. 2021, 8, 113. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Salem, S.E.; Maddox, T.W.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Acute changes in the colonic microbiota are associated with large intestinal forms of surgical colic. BMC Vet. Res. 2019, 15, 468. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Pitta, D.; Indugu, N.; Hennessy, M.; Vecchiarelli, B.; Stewart, H.; Willette, J.; Dobbie, T.; Engiles, J.; Southwood, L. 358 Understanding the role of the fecal bacterial microbiota in equine colic. J. Anim. Sci. 2020, 98, 94. [Google Scholar] [CrossRef]
- Barton, M.H.; Hallowell, G.D. Current Topics in Medical Colic. Vet. Clin. N. Am. Equine Pract. 2023, 39, 229–248. [Google Scholar] [CrossRef]
- Garber, A.; Hastie, P.; Murray, J.-A. Factors Influencing Equine Gut Microbiota: Current Knowledge. J. Equine Vet. Sci. 2020, 88, 102943. [Google Scholar] [CrossRef]
- Boucher, L.; Leduc, L.; Leclère, M.; Costa, M.C. Current Understanding of Equine Gut Dysbiosis and Microbiota Manipulation Techniques: Comparison with Current Knowledge in Other Species. Animals 2024, 14, 758. [Google Scholar] [CrossRef] [PubMed]
- Alicia, F.; Emmanuel, J.; Véronique, J. Whole-Genome Sequencing and Annotation of Fibrobacter succinogenes HC4, Isolated from the Horse Cecum. Microbiol. Resour. Announc. 2022, 11, e00440-22. [Google Scholar] [CrossRef]
- Koike, S.; Shingu, Y.; Inaba, H.; Kawai, M.; Kobayashi, Y.; Hata, H.; Tanaka, K.; Okubo, M. Fecal Bacteria in Hokkaido Native Horses as Characterized by Microscopic Enumeration and Competitive Polymerase Chain Reaction Assays. J. Equine Sci. 2000, 11, 45–50. [Google Scholar] [CrossRef][Green Version]
- Milinovich, G.J.; Klieve, A.V.; Pollitt, C.C.; Trott, D.J. Microbial Events in the Hindgut During Carbohydrate-induced Equine Laminitis. Vet. Clin. N. Am. Equine Pract. 2010, 26, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Jokisalo, J.; Bryan, J.; Legget, B.; Abbott, Y.; Katz, L.M. Multiple-drug resistant Acinetobacter baumannii bronchopneumonia in a colt following intensive care treatment. Equine Vet. Educ. 2010, 22, 281–286. [Google Scholar] [CrossRef]
- van der Kolk, J.H.; Endimiani, A.; Graubner, C.; Gerber, V.; Perreten, V. Acinetobacter in veterinary medicine, with an emphasis on Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2019, 16, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Walther, B.; Klein, K.-S.; Barton, A.-K.; Semmler, T.; Huber, C.; Wolf, S.A.; Tedin, K.; Merle, R.; Mitrach, F.; Guenther, S.; et al. Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Acinetobacter baumannii among horses entering a veterinary teaching hospital: The contemporary “Trojan Horse”. PLoS ONE 2018, 13, e0191873. [Google Scholar] [CrossRef]
- Dicks, L.M.T.; Botha, M.; Dicks, E.; Botes, M. The equine gastro-intestinal tract: An overview of the microbiota, disease and treatment. Livest. Sci. 2014, 160, 69–81. [Google Scholar] [CrossRef]
- Sharkey, L.C.; DeWitt, S.; Stockman, C. Neurologic signs and hyperammonemia in a horse with colic. Vet. Clin. Pathol. 2006, 35, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.S.; Ijssennagger, N.; Kies, A.K.; van Mil, S.W.C. Protein fermentation in the gut; implications for intestinal dysfunction in humans, pigs, and poultry. Am. J. Physiol. Liver Physiol. 2018, 315, G159–G170. [Google Scholar] [CrossRef]
- Diether, N.E.; Willing, B.P. Microbial fermentation of dietary protein: An important factor in diet–microbe–host interaction. Microorganisms 2019, 7, 19. [Google Scholar] [CrossRef]


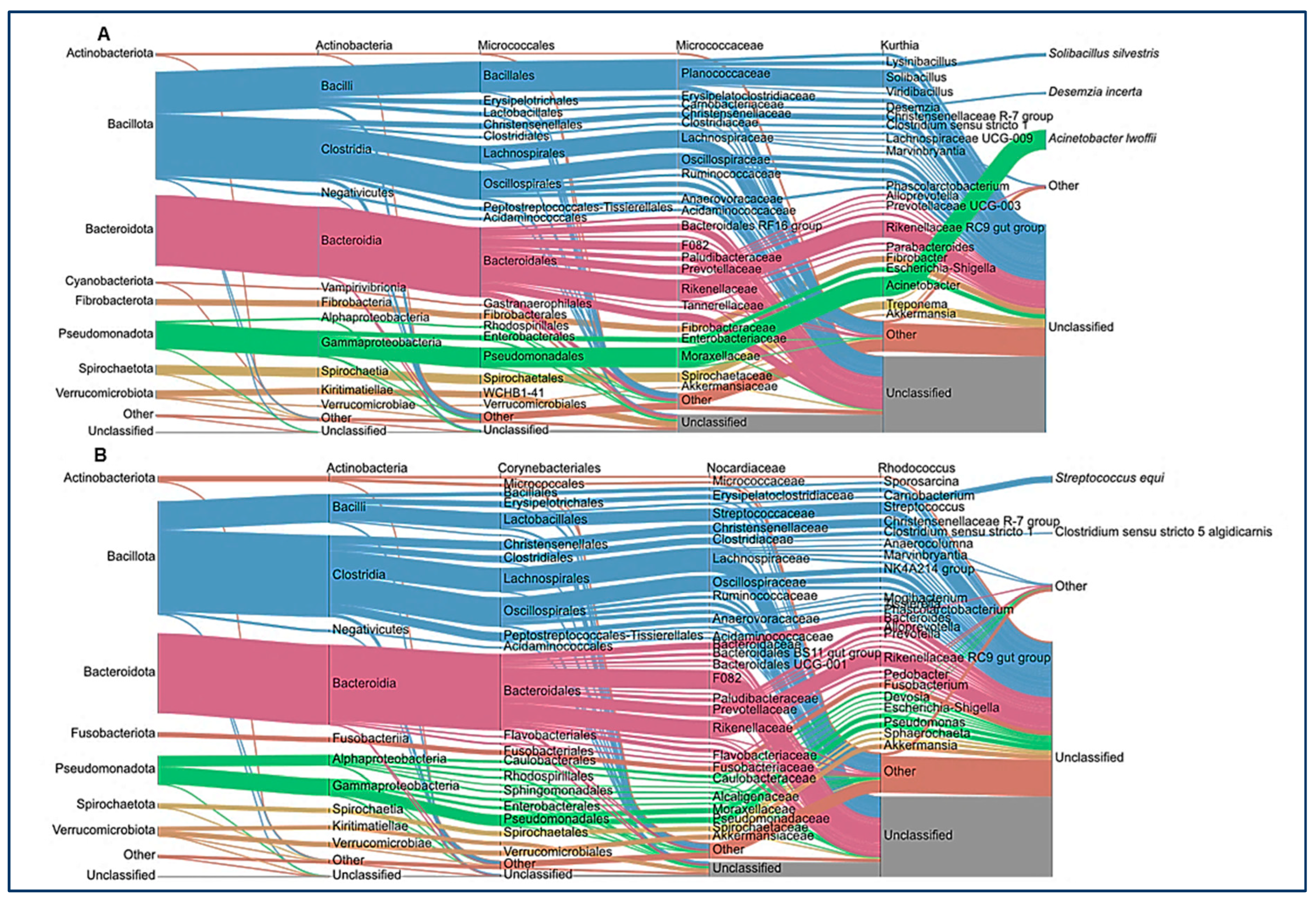

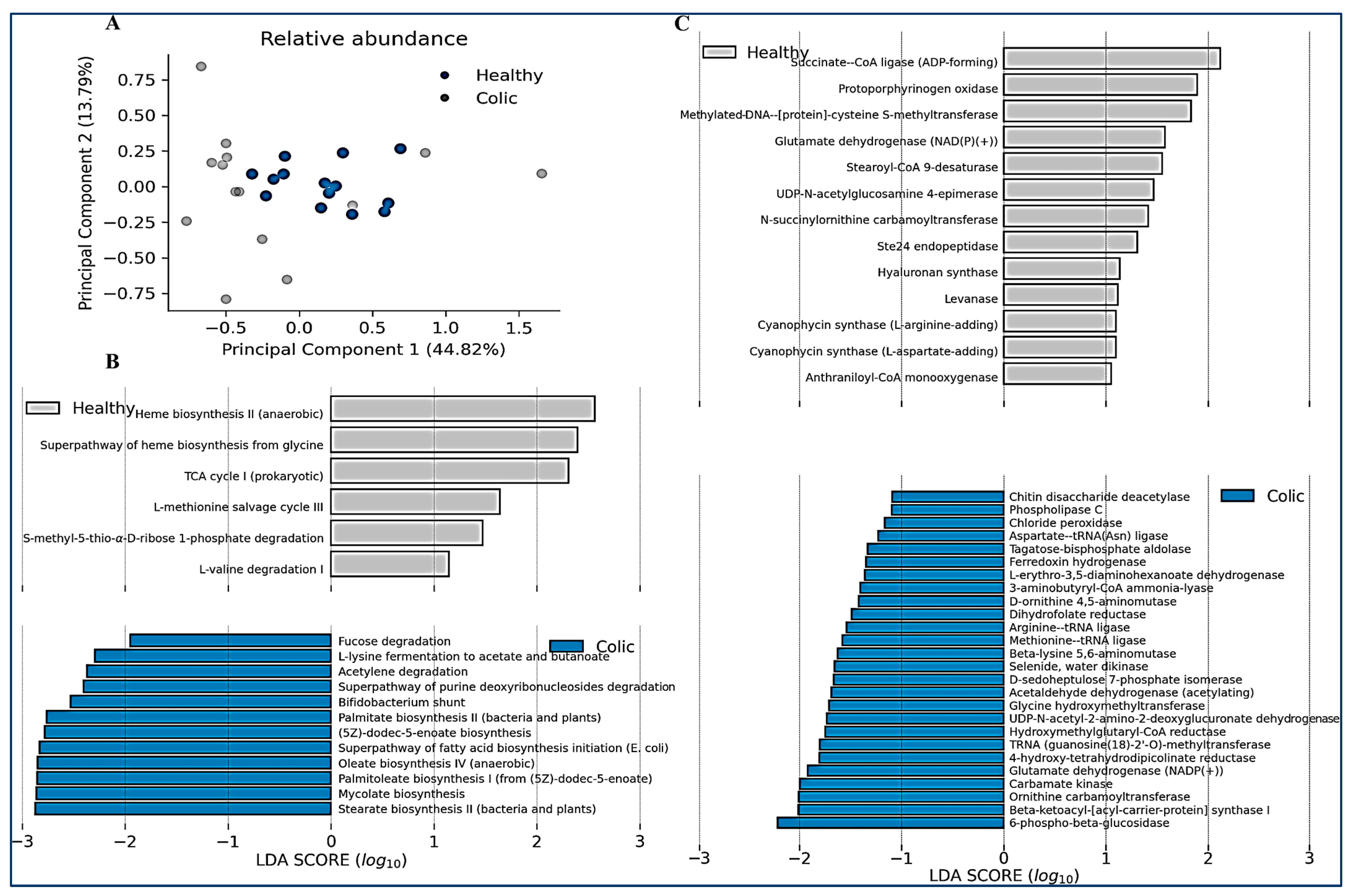
| Category | Variable | Control (N°/%) | Colic (N°/%) |
|---|---|---|---|
| Age | Young (3–4 years) | 3/21% | 1/7.1% |
| Mature (5–15 years) | 10/71% | 12/90% | |
| Geriatric (>15 years) | 1/7% | 1/7.1% | |
| Breed | Chilean | 9/64.3% | 8/57.2% |
| Warmblood | 1/7.1% | 4/28.5% | |
| Thoroughbred | 3/21% | 1/7.1% | |
| Quarter horse | 1/7.1% | 1/7.1% | |
| Sex | Gelding | 7/50% | 7/50% |
| Female | 6/43% | 4/30% | |
| Stallion | 1/7% | 3/20% |
| Diagnosis | Treatment Category | Number |
|---|---|---|
| Large colon impaction | Surgical | 7 |
| Large colon displacement | Surgical | 2 |
| Large intestinal rupture | Surgical | 1 |
| Large colon torsion | Surgical | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomson, P.; Garrido, D.; Santibáñez, R.; Lara, F. Preliminary Functional Analysis of the Gut Microbiome in Colic Horses. Animals 2024, 14, 3222. https://doi.org/10.3390/ani14223222
Thomson P, Garrido D, Santibáñez R, Lara F. Preliminary Functional Analysis of the Gut Microbiome in Colic Horses. Animals. 2024; 14(22):3222. https://doi.org/10.3390/ani14223222
Chicago/Turabian StyleThomson, Pamela, Daniel Garrido, Rodrigo Santibáñez, and Felipe Lara. 2024. "Preliminary Functional Analysis of the Gut Microbiome in Colic Horses" Animals 14, no. 22: 3222. https://doi.org/10.3390/ani14223222
APA StyleThomson, P., Garrido, D., Santibáñez, R., & Lara, F. (2024). Preliminary Functional Analysis of the Gut Microbiome in Colic Horses. Animals, 14(22), 3222. https://doi.org/10.3390/ani14223222







