Mitochondrial Genome and Phylogenetic Analysis of the Narrownose Smooth-Hound Shark Mustelus schmitti Springer, 1939
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
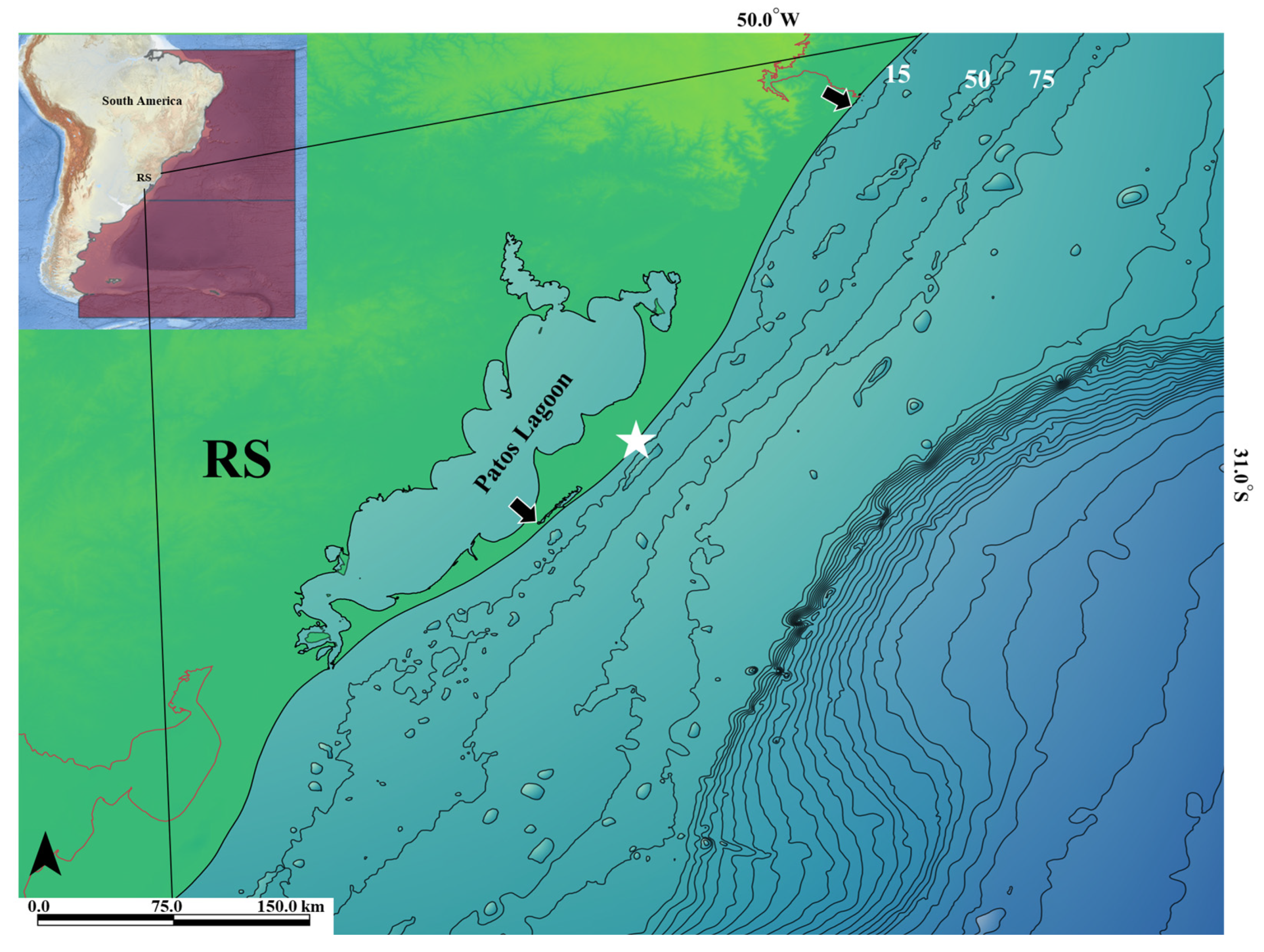
2.1. Specimen Collection
2.2. DNA Extraction, Sequencing and mtDNA Assembly
2.3. Annotation and Sequence Analysis
2.4. Phylogenetic Analyses
3. Results
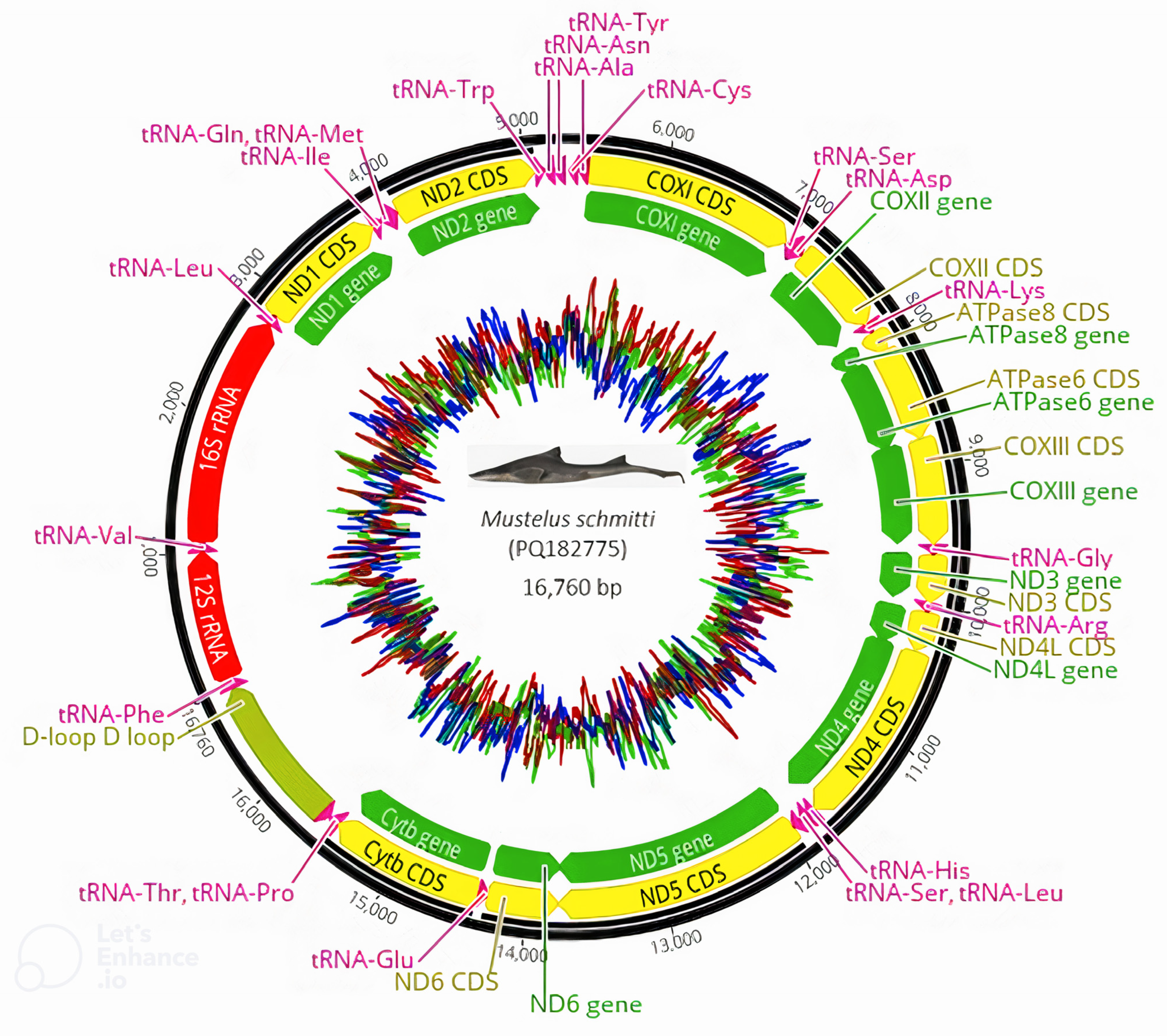
3.1. Genome Structure, Composition and Asymmetry
3.2. Protein Coding Regions, Transfer RNA and Ribosomal RNA
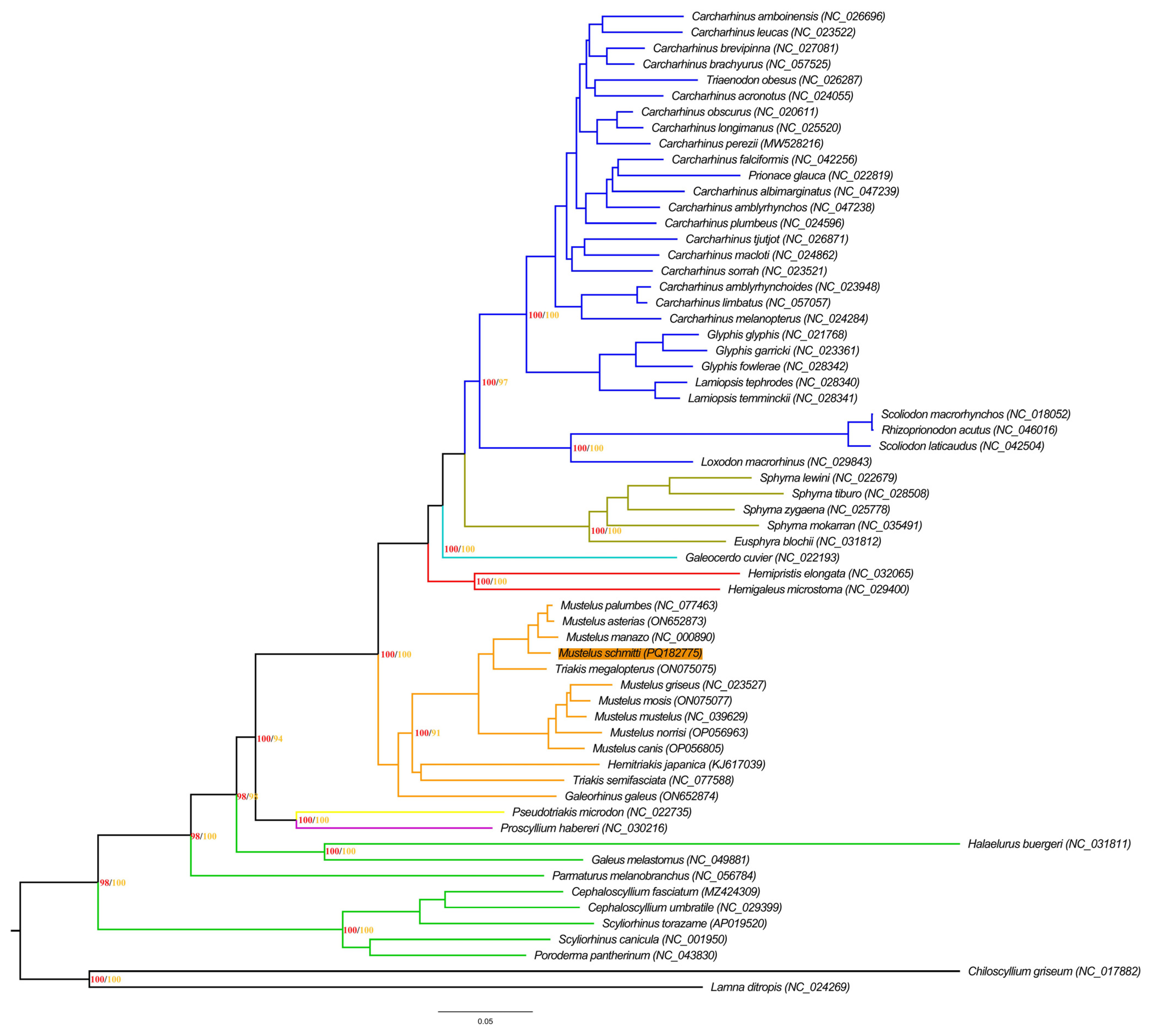
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Compagno, L.J. Phyletic relationships of living sharks and rays. Am. Zool. 1977, 17, 303–322. [Google Scholar] [CrossRef]
- Stein, R.W.; Mull, C.G.; Kuhn, T.S.; Aschliman, N.C.; Davidson, L.N.; Joy, J.B.; Smith, G.J.; Dulvy, N.K.; Mooers, A.O. Global priorities for conserving the evolutionary history of sharks, rays and chimaeras. Nat. Ecol. Evol. 2018, 2, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Ebert, D.A.; Dando, M.; Fowler, S. Sharks of the World: A Complete Guide; Princeton University Press: Princeton, NJ, USA, 2021; Volume 22. [Google Scholar]
- Weigmann, S. Annotated checklist of the living sharks, batoids and chimaeras (Chondrichthyes) of the world, with a focus on biogeographical diversity. J. Fish Biol. 2016, 88, 837–1037. [Google Scholar] [CrossRef]
- Compagno, L. Pt. 2: Carcharhiniformes. In Sharks of the World. An Annotated and Illustrated Catalogue of Shark Species Known to Date; FAO Species Catalogue; FAO: Rome, Italy, 1984. [Google Scholar]
- Rosa, M.R.; Gadig, O.B.F. Taxonomic comments and an identification key to species for the smooth-hound sharks genus Mustelus Link, 1790 (Chondrichthyes: Triakidae) from the western South Atlantic. Pan-Am. J. Aquat. Sci. 2010, 5, 401–413. [Google Scholar]
- BRASIL. Portaria MMA Nº 148, de 07 de junho de 2022. Lista Nacional Oficial de Espécies da Fauna Ameaçadas de Extinção. Anexo I e II. Portaria MMA Nº 148, de 07 de Junho de 2022 2022a, Portaria MMA Nº 148/2022, 116. Available online: https://www.icmbio.gov.br/cepsul/images/stories/legislacao/Portaria/2020/P_mma_148_2022_altera_anexos_P_mma_443_444_445_2014_atualiza_especies_ameacadas_extincao.pdf (accessed on 21 May 2024).
- BRASIL. Portaria MMA Nº 300, de 13 de dezembro de 2022. Lista Nacional Oficial de Espécies da Fauna Ameaçadas de Extinção. Anexo I e II. Portaria MMA Nº 300, de 13 de Dezembro de 2022 2022b, Portaria MMA Nº 300/2022, 90. Available online: https://www.icmbio.gov.br/cepsul/images/stories/legislacao/Portaria/2022/P_gm_mma_300_2022_reconhece_lista_nacional_spp_ameacadas_extincao.pdf (accessed on 21 May 2024).
- Nisa-Castro-Neto, W. Análise de Pesca de Carcharias Taurus Rafinesque, 1810 (Chondrichthyes, Odontaspididae) e Seu Declínio Nas Regiões Sul e Sudeste do Brasil; Pontifícia Universidade Católica do Rio Grande do Sul (PUCRS): Porto Alegre, Brazil, 2001. [Google Scholar]
- Santos, P.R.S.; Balanin, S.; Gadig, O.B.F.; Garrone-Neto, D. The historical and contemporary knowledge on the elasmobranchs of Cananeia and adjacent waters, a coastal marine hotspot of southeastern Brazil. Reg. Stud. Mar. Sci. 2022, 51, 102224. [Google Scholar] [CrossRef]
- Dulvy, N.K.; Pacoureau, N.; Rigby, C.L.; Pollom, R.A.; Jabado, R.W.; Ebert, D.A.; Finucci, B.; Pollock, C.M.; Cheok, J.; Derrick, D.H. Overfishing drives over one-third of all sharks and rays toward a global extinction crisis. Curr. Biol. 2021, 31, 4773–4787. [Google Scholar] [CrossRef]
- Instituto Chico Mendes de Conservação da Biodiversidade. Livro Vermelho da Fauna Brasileira Ameaçada de Extinção; Instituto Chico Mendes de Conservação da Biodiversidade: Rio de Janeiro, Brazil, 2018; Volume VI-Peixes. [Google Scholar]
- Pollom, R.; Barreto, R.; Charvet, P.; Chiaramonte, G.E.; Cuevas, J.M.; Herman, K.; Montealegre-Quijano, S.; Motta, F.; Paesch, L.; Rincon, G. Mustelus fasciatus. In The IUCN Red List of Threatened Species 2020; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2020; e.T44581A2995765. [Google Scholar]
- Pollom, R.; Barreto, R.; Charvet, P.; Chiaramonte, G.E.; Cuevas, J.M.; Herman, K.; Montealegre-Quijano, S.; Motta, F.; Paesch, L.; Rincon, G. Mustelus schmitti. In The IUCN Red List of Threatened Species 2020; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2020; e.T60203A3092243. [Google Scholar]
- Essington, T.E.; Moriarty, P.E.; Froehlich, H.E.; Hodgson, E.E.; Koehn, L.E.; Oken, K.L.; Siple, M.C.; Stawitz, C.C. Fishing amplifies forage fish population collapses. Proc. Natl. Acad. Sci. USA 2015, 112, 6648–6652. [Google Scholar] [CrossRef]
- Queiroz, N.; Humphries, N.E.; Mucientes, G.; Hammerschlag, N.; Lima, F.P.; Scales, K.L.; Miller, P.I.; Sousa, L.L.; Seabra, R.; Sims, D.W. Ocean-wide tracking of pelagic sharks reveals extent of overlap with longline fishing hotspots. Proc. Natl. Acad. Sci. USA 2016, 113, 1582–1587. [Google Scholar] [CrossRef]
- Dario, F.D.; Alves, C.B.; Boos, H.; Frédou, F.L.; Lessa, R.P.; Mincarone, M.M.; Pinheiro, M.A.; Polaz, C.N.; Reis, R.E.; Rocha, L.A. A better way forward for Brazil’s fisheries. Science 2015, 347, 1079. [Google Scholar] [CrossRef]
- Camacho-Oliveira, R.B.; Daneluz, C.M.; do Prado, F.D.; Utsunomia, R.; Rodrigues Jr, C.E.; Foresti, F.; Porto-Foresti, F. DNA barcode reveals the illegal trade of rays commercialized in fishmongers in Brazil. Forensic Sci. Int. Synerg. 2020, 2, 95–97. [Google Scholar] [CrossRef]
- Bunholi, I.V.; da Silva Ferrette, B.L.; De Biasi, J.B.; de Oliveira Magalhães, C.; Rotundo, M.M.; Oliveira, C.; Foresti, F.; Mendonça, F.F. The fishing and illegal trade of the angelshark: DNA barcoding against misleading identifications. Fish. Res. 2018, 206, 193–197. [Google Scholar] [CrossRef]
- Dudgeon, C.; Blower, D.; Broderick, D.; Giles, J.; Holmes, B.; Kashiwagi, T.; Krueuck, N.; Morgan, J.; Tillett, B.; Ovenden, J. A review of the application of molecular genetics for fisheries management and conservation of sharks and rays. J. Fish Biol. 2012, 80, 1789–1843. [Google Scholar] [CrossRef]
- Pereyra, S.; Garcia, G.; Miller, P.; Oviedo, S.; Domingo, A. Low genetic diversity and population structure of the narrownose shark (Mustelus schmitti). Fish. Res. 2010, 106, 468–473. [Google Scholar] [CrossRef]
- Miranda, L.d.; Vooren, C. Captura e esforço da pesca de elasmobrânquios demersais no sul do Brasil nos anos de 1975 a 1997. Frente Marítimo 2003, 19, 217–231. [Google Scholar]
- Vooren, C.; Oddone, M. The diversity of the chondrichthyans of the far south of Brazil: The species, their origins, and their reproductive modes. In Ciencias Marino-Costeras en el Umbral del Siglo XXI. Desafios em Latinoamérica y el Caribe; AGT Editor: México, Mexico, 2019; pp. 173–214. [Google Scholar]
- Massa, A.; Hozbor, N.; Lucifora, L.; Colonello, J. Sugerencias de capturas para el año 2003 de gatuzo (Mustelus spp.), peces angel (Squatina spp.) y rayas costeras. In Informe Técnico Interno INIDEP; DNI; CONICET: Godoy Cruz, Argentina, 2003. [Google Scholar]
- Oddone, M.; Paesch, L.; Norbis, W. Reproductive biology and seasonal distribution of the patagonian smoothhound Mustelus schmitti (Elasmobranchii: Triakidae) in the Rio de La Plata oceanic front, South-Western Atlantic. J. Mar. Biol. Assoc. U. K. 2005, 85, 1193–1198. [Google Scholar] [CrossRef]
- Vooren, C.; Klippel, S.; Galina, A. Os elasmobrânquios das águas costeiras da Plataforma Sul. In Ações Para a Conservação de Tubarões e Raias No Sul do Brasil; Igaré: Porto Alegre, Brazil, 2005; pp. 113–120. [Google Scholar]
- Pereyra, I.; Orlando, L.; Norbis, W.; Paesch, L. Spatial and temporal variation of length and sex composition of the narrownose smooth-hound Mustelus schmitti Springer, 1939 in the trawl fishery off the oceanic coast of Uruguay during 2004. Rev. Biol. Mar. Oceanogr. 2008, 43, 159–166. [Google Scholar]
- Chiaramonte, G.E.; Pettovello, A.D. The biology of Mustelus schmitti in southern Patagonia, Argentina. J. Fish Biol. 2000, 57, 930–942. [Google Scholar] [CrossRef]
- Begg, G.A.; Friedland, K.D.; Pearce, J.B. Stock identification and its role in stock assessment and fisheries management: An overview. Fish. Res. 1999, 43, 1–8. [Google Scholar] [CrossRef]
- Pimiento, C.; Albouy, C.; Silvestro, D.; Mouton, T.L.; Velez, L.; Mouillot, D.; Judah, A.B.; Griffin, J.N.; Leprieur, F. Functional diversity of sharks and rays is highly vulnerable and supported by unique species and locations worldwide. Nat. Commun. 2023, 14, 7691. [Google Scholar] [CrossRef]
- Reiss, H.; Hoarau, G.; Dickey-Collas, M.; Wolff, W.J. Genetic population structure of marine fish: Mismatch between biological and fisheries management units. Fish Fish. 2009, 10, 361–395. [Google Scholar] [CrossRef]
- López, J.A.; Ryburn, J.A.; Fedrigo, O.; Naylor, G.J. Phylogeny of sharks of the family Triakidae (Carcharhiniformes) and its implications for the evolution of carcharhiniform placental viviparity. Mol. Phylogenetics Evol. 2006, 40, 50–60. [Google Scholar] [CrossRef]
- Cortés, F. Sustentabilidad de la Explotación del Gatuzo Mustelus schmitti, en el Ecosistema Costero Bonaerense (34–42 S). Bachelor Thesis, Universidad Nacional de Mar del Plata, Mar del Plata, Argentina, 2007. [Google Scholar]
- Lim, D.D.; Motta, P.; Mara, K.; Martin, A.P. Phylogeny of hammerhead sharks (Family Sphyrnidae) inferred from mitochondrial and nuclear genes. Mol. Phylogenetics Evol. 2010, 55, 572–579. [Google Scholar] [CrossRef]
- Dosay-Akbulut, M. The phylogenetic relationship within the genus Carcharhinus. Comptes Rendus Biol. 2008, 331, 500–509. [Google Scholar] [CrossRef]
- Cunha, D.B.; Silva Rodrigues-Filho, L.F.; Luna Sales, J.B. A Review of the Mitogenomic Phylogeny of the Chondrichthyes. In Chondrichthyes-Multidisciplinary Approach; InTech: London, UK, 2017. [Google Scholar]
- Naylor, G.J.; Ryburn, J.; Fedrigo, O.; Lopez, J. Phylogenetic relationships among the major lineages of modern elasmobranchs. Reprod. Biol. Phylogeny 2005, 3, 25. [Google Scholar]
- Maisey, J.; Naylor, G.; Ward, D.; Arratia, G.; Tintori, A. Mesozoic elasmobranchs, neoselachian phylogeny and the rise of modern elasmobranch diversity. Mesoz. Fishes 3-Syst. Paleoenviron. Biodivers. 2004, 3, 17–56. [Google Scholar]
- McEachran, J.D.; Aschliman, N. Phylogeny of Batoidea; CRC Press: Boca Raton, FL, USA, 2004; pp. 79–113. [Google Scholar]
- Renz, A.J.; Meyer, A.; Kuraku, S. Revealing less derived nature of cartilaginous fish genomes with their evolutionary time scale inferred with nuclear genes. PLoS ONE 2013, 8, e66400. [Google Scholar] [CrossRef]
- Kousteni, V.; Mazzoleni, S.; Vasileiadou, K.; Rovatsos, M. Complete mitochondrial DNA genome of nine species of sharks and rays and their phylogenetic placement among modern elasmobranchs. Genes 2021, 12, 324. [Google Scholar] [CrossRef]
- Iglésias, S.P.; Lecointre, G.; Sellos, D.Y. Extensive paraphylies within sharks of the order Carcharhiniformes inferred from nuclear and mitochondrial genes. Mol. Phylogenetics Evol. 2005, 34, 569–583. [Google Scholar] [CrossRef]
- Kiser, H.; Skufca, K.; Bemis, K.E.; Baeza, J.A. Comparative analysis of the mitochondrial genomes of Smoothhound sharks provide insight into the phylogenetic relationships within the family Triakidae. Gene Rep. 2024, 36, 101957. [Google Scholar] [CrossRef]
- Winn, J.C.; Maduna, S.N.; Bester-van der Merwe, A.E. A comprehensive phylogenomic study unveils evolutionary patterns and challenges in the mitochondrial genomes of Carcharhiniformes: A focus on Triakidae. Genomics 2024, 116, 110771. [Google Scholar] [CrossRef]
- Wang, C.; Lai, T.; Ye, P.; Yan, Y.; Feutry, P.; He, B.; Huang, Z.; Zhu, T.; Wang, J.; Chen, X. Novel duplication remnant in the first complete mitogenome of Hemitriakis japanica and the unique phylogenetic position of family Triakidae. Gene 2022, 820, 146232. [Google Scholar] [CrossRef] [PubMed]
- Heinicke, M.; Naylor, G.; Hedges, S. Cartilaginous fishes (Chondrichthyes). Timetree Life 2009, 9, 320–327. [Google Scholar]
- Maduna, S.N.; Rossouw, C.; Da Silva, C.; Soekoe, M.; Bester-van der Merwe, A.E. Species identification and comparative population genetics of four coastal houndsharks based on novel NGS-mined microsatellites. Ecol. Evol. 2017, 7, 1462–1486. [Google Scholar] [CrossRef]
- Nickum, J.; Bart, H., Jr.; Bowser, P.; Greer, I.; Hubbs, C.; Jenkins, J.; MacMillan, J.; Rachlin, J.; Rose, J.; Sorensen, P. Guidelines for the use of fishes in research. Am. Fish. Soc. 2004, 29, 26. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Mikheenko, A.; Valin, G.; Prjibelski, A.; Saveliev, V.; Gurevich, A. Icarus: Visualizer for de novo assembly evaluation. Bioinformatics 2016, 32, 3321–3323. [Google Scholar] [CrossRef]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. TRNAscan-SE: Searching for TRNA Genes in Genomic Sequences; Springer: New York, NY, USA, 2019. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Griffin, A.M.; Griffin, H.G.; Higgins, D.G. CLUSTAL V: Multiple alignment of DNA and protein sequences. Comput. Anal. Seq. Data Part II 1994, 307–318. [Google Scholar]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Alvarenga, M.; Bunholi, I.V.; de Brito, G.R.; Siqueira, M.V.B.M.; Domingues, R.R.; Charvet, P.; Foresti, F.; Solé-Cava, A.M.; da Cruz, V.P. Fifteen years of elasmobranchs trade unveiled by DNA tools: Lessons for enhanced monitoring and conservation actions. Biol. Conserv. 2024, 292, 110543. [Google Scholar] [CrossRef]
- Kenchington, E.L. The Effects of Fishing on Species and Genetic Diversity. Responsible Fish. Mar. Ecosyst. 2003. [Google Scholar] [CrossRef]
- Villate-Moreno, M.; Cubillos-M, J.C.; Stibor, H.; Crawford, A.J.; Straube, N. Molecular identification and first demographic insights of sharks based on artisanal fisheries bycatch in the Pacific Coast of Colombia: Implications for conservation. PeerJ 2022, 10, e13478. [Google Scholar] [CrossRef]
- Douady, C.J.; Dosay, M.; Shivji, M.S.; Stanhope, M.J. Molecular phylogenetic evidence refuting the hypothesis of Batoidea (rays and skates) as derived sharks. Mol. Phylogenetics Evol. 2003, 26, 215–221. [Google Scholar] [CrossRef]
- Martin, A. The phylogenetic placement of Chondrichthyes: Inferences from analysis of multiple genes and implications for comparative studies. Genetica 2001, 111, 349–357. [Google Scholar] [CrossRef]
- Kamal, S.A.; Baeza, J.A. Detailed characterization of the complete mitochondrial genome of the oceanic whitetip shark Carcharhinus longimanus (Poey, 1861). Mol. Biol. Rep. 2024, 51, 826. [Google Scholar] [CrossRef]
- Inoue, J.G.; Miya, M.; Lam, K.; Tay, B.-H.; Danks, J.A.; Bell, J.; Walker, T.I.; Venkatesh, B. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): A mitogenomic perspective. Mol. Biol. Evol. 2010, 27, 2576–2586. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.-C.; Liang, Y.-Y.; Wu, N.; Guo, H.-Y.; Zhang, N.; Jiang, S.-G.; Zhang, D.-C. Sequencing and characterization of the complete mitochondrial genome of Japanese Swellshark (Cephalloscyllium umbratile). Sci. Rep. 2017, 7, 15299. [Google Scholar] [CrossRef]
- Palacios-Barreto, P.; Mar-Silva, A.F.; Bayona-Vasquez, N.J.; Adams, D.H.; Díaz-Jaimes, P. Characterization of the complete mitochondrial genome of the Brazilian cownose ray Rhinoptera brasiliensis (Myliobatiformes, Rhinopteridae) in the western Atlantic and its phylogenetic implications. Mol. Biol. Rep. 2023, 50, 4083–4095. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Takeshima, H.; Endo, H.; Ishiguro, N.B.; Inoue, J.G.; Mukai, T.; Satoh, T.P.; Yamaguchi, M.; Kawaguchi, A.; Mabuchi, K. Major patterns of higher teleostean phylogenies: A new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenetics Evol. 2003, 26, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Condamine, F.L.; Romieu, J.; Guinot, G. Climate cooling and clade competition likely drove the decline of lamniform sharks. Proc. Natl. Acad. Sci. USA 2019, 116, 20584–20590. [Google Scholar] [CrossRef]
- Klein, J.D.; Maduna, S.N.; Dicken, M.L.; da Silva, C.; Soekoe, M.; McCord, M.E.; Potts, W.M.; Hagen, S.B.; Bester-van der Merwe, A.E. Local adaptation with gene flow in a highly dispersive shark. Evol. Appl. 2024, 17, e13628. [Google Scholar] [CrossRef]
- Sabadin, D.E.; Lucifora, L.O.; Barbini, S.A.; Figueroa, D.E.; Kittlein, M. Towards regionalization of the chondrichthyan fauna of the Southwest Atlantic: A spatial framework for conservation planning. ICES J. Mar. Sci. 2020, 77, 1893–1905. [Google Scholar] [CrossRef]
- Kraft, D.W.; Conklin, E.E.; Barba, E.W.; Hutchinson, M.; Toonen, R.J.; Forsman, Z.H.; Bowen, B.W. Genomics versus mtDNA for resolving stock structure in the silky shark (Carcharhinus falciformis). PeerJ 2020, 8, e10186. [Google Scholar] [CrossRef]
- Domingues, R.R.; Hilsdorf, A.W.S.; Gadig, O.B.F. The importance of considering genetic diversity in shark and ray conservation policies. Conserv. Genet. 2018, 19, 501–525. [Google Scholar] [CrossRef]
- Hara, Y.; Yamaguchi, K.; Onimaru, K.; Kadota, M.; Koyanagi, M.; Keeley, S.D.; Tatsumi, K.; Tanaka, K.; Motone, F.; Kageyama, Y. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol. 2018, 2, 1761–1771. [Google Scholar] [CrossRef]
- Read, T.D.; Petit, R.A.; Joseph, S.J.; Alam, M.T.; Weil, M.R.; Ahmad, M.; Bhimani, R.; Vuong, J.S.; Haase, C.P.; Webb, D.H. Draft sequencing and assembly of the genome of the world’s largest fish, the whale shark: Rhincodon typus Smith 1828. BMC Genom. 2017, 18, 532. [Google Scholar]
- Kuraku, S. Shark and ray genomics for disentangling their morphological diversity and vertebrate evolution. Dev. Biol. 2021, 477, 262–272. [Google Scholar] [CrossRef]
- Ho, S.Y.; Lanfear, R.; Bromham, L.; Phillips, M.J.; Soubrier, J.; Rodrigo, A.G.; Cooper, A. Time-dependent rates of molecular evolution. Mol. Ecol. 2011, 20, 3087–3101. [Google Scholar] [CrossRef]
- Díaz-Jaimes, P.; Bayona-Vásquez, N.J.; Adams, D.H.; Uribe-Alcocer, M. Complete mitochondrial DNA genome of bonnethead shark, Sphyrna tiburo, and phylogenetic relationships among main superorders of modern elasmobranchs. Meta Gene 2016, 7, 48–55. [Google Scholar] [CrossRef]
- Heist, E.J. Genetics of sharks, skates and rays. In Biology of Sharks and Their Relatives; CRC Press: Boca Raton, FL, USA, 2012; 18p, ISBN 9780429106545. [Google Scholar]
- Stingo, V.; Capriglione, T.; Rocco, L.; Improta, R.; Morescalchi, A. Genome size and AT rich DNA in selachians. Genetica 1989, 79, 197–205. [Google Scholar] [CrossRef]
- Huang, X.; Yu, J.; Chen, H.; Chen, X.; Wang, J. Complete mitochondrial genome and the phylogenetic position of the snaggletooth shark Hemipristis elongata (Carcharhiniformes: Hemigaleidae). Mitochondrial DNA Part B 2016, 1, 538–539. [Google Scholar] [CrossRef]
- Brée, B.; Condamine, F.L.; Guinot, G. Combining palaeontological and neontological data shows a delayed diversification burst of carcharhiniform sharks likely mediated by environmental change. Sci. Rep. 2022, 12, 21906. [Google Scholar] [CrossRef]
- Dudgeon, C.L.; Corrigan, S.; Yang, L.; Allen, G.R.; Erdmann, M.V.; Sugeha, H.Y.; White, W.T.; Naylor, G.J. Walking, swimming or hitching a ride? Phylogenetics and biogeography of the walking shark genus Hemiscyllium. Mar. Freshw. Res. 2020, 71, 1107–1117. [Google Scholar] [CrossRef]
- Sayyari, E.; Mirarab, S. Testing for polytomies in phylogenetic species trees using quartet frequencies. Genes 2018, 9, 132. [Google Scholar] [CrossRef]
- McLay, T.G.; Fowler, R.M.; Fahey, P.S.; Murphy, D.J.; Udovicic, F.; Cantrill, D.J.; Bayly, M.J. Phylogenomics reveals extreme gene tree discordance in a lineage of dominant trees: Hybridization, introgression, and incomplete lineage sorting blur deep evolutionary relationships despite clear species groupings in Eucalyptus subgenus Eudesmia. Mol. Phylogenetics Evol. 2023, 187, 107869. [Google Scholar] [CrossRef]
- Jeiter, J.; Smets, E. Integrating comparative morphology and development into evolutionary research. Taxon 2023, 72, 724–732. [Google Scholar] [CrossRef]
- Lücking, R.; Leavitt, S.D.; Hawksworth, D.L. Species in lichen-forming fungi: Balancing between conceptual and practical considerations, and between phenotype and phylogenomics. Fungal Divers. 2021, 109, 99–154. [Google Scholar] [CrossRef]
- Steenwyk, J.L.; Li, Y.; Zhou, X.; Shen, X.-X.; Rokas, A. Incongruence in the phylogenomics era. Nat. Rev. Genet. 2023, 24, 834–850. [Google Scholar] [CrossRef]
- DeSalle, R.; Goldstein, P. Review and interpretation of trends in DNA barcoding. Front. Ecol. Evol. 2019, 7, 302. [Google Scholar] [CrossRef]
- Guo, M.; Yuan, C.; Tao, L.; Cai, Y.; Zhang, W. Life barcoded by DNA barcodes. Conserv. Genet. Resour. 2022, 14, 351–365. [Google Scholar] [CrossRef]
- Human, B.A.; Owen, E.P.; Compagno, L.J.; Harley, E.H. Testing morphologically based phylogenetic theories within the cartilaginous fishes with molecular data, with special reference to the catshark family (Chondrichthyes; Scyliorhinidae) and the interrelationships within them. Mol. Phylogenetics Evol. 2006, 39, 384–391. [Google Scholar] [CrossRef]
- Corrigan, S.; Beheregaray, L.B. A recent shark radiation: Molecular phylogeny, biogeography and speciation of wobbegong sharks (family: Orectolobidae). Mol. Phylogenetics Evol. 2009, 52, 205–216. [Google Scholar] [CrossRef]
- Compagno, L.J. Systematics and body form. In Sharks, Skates and Rays: The Biology of Elasmobranch Fishes; Johns Hopkins University Press: Baltimore, MD, USA, 1999; pp. 1–42. [Google Scholar]
- Rasmussen, A.S.; Arnason, U. Phylogenetic studies of complete mitochondrial DNA molecules place cartilaginous fishes within the tree of bony fishes. J. Mol. Evol. 1999, 48, 118–123. [Google Scholar] [CrossRef]
- Boomer, J.J.; Harcourt, R.G.; Francis, M.P.; Stow, A.J. Genetic divergence, speciation and biogeography of Mustelus (sharks) in the central Indo-Pacific and Australasia. Mol. Phylogenetics Evol. 2012, 64, 697–703. [Google Scholar] [CrossRef]
- Maduna, S.N. Unravelling the Mystery of the Shark Genus Mustelus in Southern Africa using a Multidisciplinary Approach. Ph.D. Thesis, Stellenbosch University, Stellenbosch, South Africa, 2017. [Google Scholar]
- Sandoval-Castillo, J.; Beheregaray, L.B. Oceanographic heterogeneity influences an ecological radiation in elasmobranchs. J. Biogeogr. 2020, 47, 1599–1611. [Google Scholar] [CrossRef]
- Da Silva Rodrigues-Filho, L.F.; da Costa Nogueira, P.; Sodré, D.; da Silva Leal, J.R.; Nunes, J.L.S.; Rincon, G.; Lessa, R.P.T.; Sampaio, I.; Vallinoto, M.; Ready, J.S. Evolutionary history and taxonomic reclassification of the critically endangered daggernose shark, a species endemic to the Western Atlantic. J. Zool. Syst. Evol. Res. 2023, 2023, 4798805. [Google Scholar] [CrossRef]
- Sternes, P.C.; Schmitz, L.; Higham, T.E. The rise of pelagic sharks and adaptive evolution of pectoral fin morphology during the Cretaceous. Curr. Biol. 2024, 34, 2764–2772.e3. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.R.; Karl, S.A.; Horn, R.L.; Bernard, A.M.; Lea, J.S.; Hazin, F.H.; Prodöhl, P.A.; Shivji, M.S. Global mitochondrial DNA phylogeography and population structure of the silky shark, Carcharhinus falciformis. Mar. Biol. 2015, 162, 945–955. [Google Scholar] [CrossRef]
- Karl, S.; Castro, A.; Lopez, J.; Charvet, P.; Burgess, G. Phylogeography and conservation of the bull shark (Carcharhinus leucas) inferred from mitochondrial and microsatellite DNA. Conserv. Genet. 2011, 12, 371–382. [Google Scholar] [CrossRef]
- Harnlett, W.C. Placentatrophy in Sharks. In Reproductive Biology and Phylogeny of Chondrichthyes; CRC Press: Boca Raton, FL, USA, 2011; pp. 473–477. [Google Scholar]

| Family | Species | Size (bp) | AT% | GenBank |
|---|---|---|---|---|
| Carcharhinidae: | Carcharhinus acronotus | 16,719 | 61.6 | NC_024055 |
| Carcharhinus albimarginatus | 16,706 | 61.4 | NC_047239 | |
| Carcharhinus amblyrhynchoides | 16,705 | 61.8 | NC_023948 | |
| Carcharhinus amblyrhynchos | 16,705 | 61.6 | NC_047238 | |
| Carcharhinus amboinensis | 16,704 | 62.0 | NC_026696 | |
| Carcharhinus brachyurus | 16,704 | 61.7 | NC_057525 | |
| Carcharhinus brevipinna | 16,706 | 61.4 | NC_027081 | |
| Carcharhinus falciformis | 16,677 | 61.4 | NC_042256 | |
| Carcharhinus leucas | 16,704 | 62.6 | NC_023522 | |
| Carcharhinus limbatus | 16,705 | 61.7 | NC_057057 | |
| Carcharhinus longimanus | 16,706 | 61.5 | NC_025520 | |
| Carcharhinus macloti | 16,701 | 60.8 | NC_024862 | |
| Carcharhinus melanopterus | 16,706 | 61.4 | NC_024284 | |
| Carcharhinus obscurus | 16,706 | 61.5 | NC_020611 | |
| Carcharhinus perezii | 16,709 | 61.5 | MW528216 | |
| Carcharhinus plumbeus | 16,706 | 61.2 | NC_024596 | |
| Carcharhinus sorrah | 16,707 | 61.0 | NC_023521 | |
| Carcharhinus tjutjot | 16,705 | 60.6 | NC_026871 | |
| Glyphis fowlerae | 16,704 | 60.6 | NC_028342 | |
| Glyphis garricki | 16,702 | 60.8 | NC_023361 | |
| Glyphis glyphis | 16,701 | 61.0 | NC_021768 | |
| Lamiopsis temminckii | 16,708 | 61.1 | NC_028341 | |
| Lamiopsis tephrodes | 16,705 | 61.2 | NC_028340 | |
| Loxodon macrorhinus | 16,702 | 61.1 | NC_029843 | |
| Prionace glauca | 16,705 | 62.5 | NC_022819 | |
| Rhizoprionodon acutus | 16,693 | 63.0 | NC_046016 | |
| Scoliodon laticaudus | 16,695 | 63.1 | NC_042504 | |
| Scoliodon macrorhynchos | 16,693 | 63.1 | NC_018052 | |
| Triaenodon obesus | 16,700 | 61.1 | NC_026287 | |
| Galeocerdidae: | Galeocerdo cuvier | 16,703 | 63.1 | NC_022193 |
| Hemigaleidae: | Hemigaleus microstoma | 16,701 | 60.1 | NC_029400 |
| Hemipristis elongata | 16,691 | 63.0 | NC_032065 | |
| Proscylliidae: | Proscyllium habereri | 16,708 | 62.1 | NC_030216 |
| Pseudotriakidae: | Pseudotriakis microdon | 16,700 | 63.6 | NC_022735 |
| Scyliorhinidae: | Cephaloscyllium fasciatum | 16,703 | 61.9 | MZ424309 |
| Cephaloscyllium umbratile | 16,698 | 62.1 | NC_029399 | |
| Galeus melastomus | 16,706 | 63.2 | NC_049881 | |
| Halaelurus buergeri | 19,100 | 61.1 | NC_031811 | |
| Parmaturus melanobranchus | 16,687 | 62.5 | NC_056784 | |
| Poroderma pantherinum | 16,686 | 61.1 | NC_043830 | |
| Scyliorhinus canicula | 16,697 | 62.0 | NC_001950 | |
| Scyliorhinus torazame | 17,861 | 61.8 | AP019520 | |
| Sphyrnidae: | Eusphyra blochii | 16,727 | 61.3 | NC_031812 |
| Sphyrna lewini | 16,726 | 60.5 | NC_022679 | |
| Sphyrna mokarran | 16,719 | 61.4 | NC_035491 | |
| Sphyrna tiburo | 16,723 | 60.7 | NC_028508 | |
| Sphyrna zygaena | 16,731 | 61.7 | NC_025778 | |
| Triakidae: | Galeorhinus galeus | 17,488 | 62.0 | ON652874 |
| Hemitriakis japanica | 17,301 | 60.0 | KJ617039 | |
| Mustelus asterias * | 16,708 | 61.5 | ON652873 | |
| Mustelus canis | 16,758 | 60.8 | OP056805 | |
| Mustelus griseus * | 16,754 | 61.0 | NC_023527 | |
| Mustelus manazo * | 16,707 | 61.8 | NC_000890 | |
| Mustelus mosis | 16,755 | 60.7 | ON075077 | |
| Mustelus mustelus * | 16,755 | 60.8 | NC_039629 | |
| Mustelus norrisi | 16,769 | 61.2 | OP056963 | |
| Mustelus palumbes * | 16,708 | 61.5 | NC_077463 | |
| Mustelus schmitti ** | 16,764 | 61.4 | PQ182775 | |
| Triakis megalopterus | 16,746 | 61.3 | ON075075 | |
| Triakis semifasciata | 16,613 | 61.2 | NC_077588 | |
| Hemiscylliidae | Chiloscyllium griseum | 16,755 | 63.9 | NC_017882 |
| Lamnidae | Lamna ditropis | 16,702 | 61.1 | NC_024269 |
| Name | Codon Start | Codon Stop | Anti-Codon | AT% | CG% | Type | Position from | Position to | Length | Strand |
|---|---|---|---|---|---|---|---|---|---|---|
| D-loop | 63.9 | 35.1 | D-loop | 15641 | 16763 | 1123 | H | |||
| tRNA-Pro | TGG | 43.5 | 56.5 | tRNA | 15572 | 15640 | 69 | L | ||
| tRNA-Thr | TGT | 57.0 | 43.0 | tRNA | 15498 | 15569 | 72 | H | ||
| Cytb | ATG | TAG | 59.3 | 40.7 | gene | 14353 | 15497 | 1145 | H | |
| tRNA-Glu | TTC | 68.6 | 31.4 | tRNA | 14281 | 14350 | 70 | L | ||
| ND6 | CTA | CAT | 61.5 | 38.1 | gene | 13759 | 14280 | 522 | L | |
| ND5 | ATG | TAA | 63.4 | 36.5 | gene | 11934 | 13763 | 1830 | H | |
| tRNA-Leu2 | TAG | 59.7 | 40.2 | tRNA | 11862 | 11933 | 72 | H | ||
| tRNA-Ser2 | GCT | 49.3 | 50.8 | tRNA | 11795 | 11861 | 67 | H | ||
| tRNA-His | GTG | 79.7 | 20.3 | tRNA | 11726 | 11794 | 69 | H | ||
| ND4 | ATG | T- | 62.4 | 37.6 | gene | 10345 | 11725 | 1381 | H | |
| ND4L | ATG | TAA | 58.3 | 41.7 | gene | 10055 | 10351 | 297 | H | |
| tRNA-Arg | TCG | 67.1 | 32.9 | tRNA | 9985 | 10054 | 70 | H | ||
| ND3 | ATG | TAG | 56.4 | 43.5 | gene | 9636 | 9984 | 349 | H | |
| tRNA-Gly | TCC | 70.0 | 30.0 | tRNA | 9566 | 9635 | 70 | H | ||
| COXIII | ATG | TAA | 57.9 | 42.1 | gene | 8778 | 9563 | 786 | H | |
| ATPase6 | ATG | TAA | 64.2 | 35.7 | gene | 8095 | 8777 | 683 | H | |
| ATPase8 | ATG | TAA | 72.0 | 27.9 | gene | 7937 | 8104 | 168 | H | |
| tRNA-Lys | TTT | 60.8 | 39.2 | tRNA | 7862 | 7935 | 74 | H | ||
| COXII | ATG | T- | 61.8 | 38.2 | gene | 7171 | 7861 | 691 | H | |
| tRNA-Asp | GTC | 62.9 | 37.2 | tRNA | 7094 | 7163 | 70 | H | ||
| tRNA-Ser1 | TGA | 53.5 | 46.5 | tRNA | 7020 | 7090 | 71 | L | ||
| COXI | GTG | TAA | 61.5 | 38.5 | gene | 5462 | 7018 | 1557 | H | |
| tRNA-Tyr | GTA | 47.2 | 50.0 | tRNA | 5391 | 5460 | 70 | L | ||
| tRNA-Cys | GCA | 49.2 | 50.7 | tRNA | 5321 | 5389 | 69 | L | ||
| tRNA-Asn | GTT | 61.6 | 38.4 | tRNA | 5213 | 5285 | 73 | L | ||
| tRNA-Ala | TGC | 66.6 | 33.3 | tRNA | 5144 | 5212 | 69 | L | ||
| tRNA-Trp | CCA | 67.6 | 32.4 | tRNA | 5072 | 5142 | 71 | H | ||
| ND2 | ATG | TAG | 62.2 | 37.8 | gene | 4027 | 5071 | 1045 | H | |
| tRNA-Met | CAT | 57.9 | 42.0 | tRNA | 3958 | 4026 | 69 | H | ||
| tRNA-Gln | TTG | 65.3 | 34.7 | tRNA | 3886 | 3957 | 72 | L | ||
| tRNA-Ile | GAT | 54.3 | 45.7 | tRNA | 3815 | 3884 | 70 | H | ||
| ND1 | ATG | TAA | 60.3 | 39.7 | gene | 2840 | 3814 | 975 | H | |
| tRNA-Leu1 | TAA | 54.7 | 45.3 | tRNA | 2765 | 2839 | 75 | H | ||
| 16S rRNA | 62.5 | 37.4 | rRNA | 1095 | 2764 | 1670 | H | |||
| tRNA-Val | TAV | 56.9 | 43.0 | tRNA | 1023 | 1094 | 72 | H | ||
| 12S rRNA | 58.2 | 41.8 | rRNA | 70 | 1022 | 953 | H | |||
| tRNA-Phe | GAA | 60.9 | 39.1 | tRNA | 1 | 69 | 69 | H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisa-Castro-Neto, W.; Wagner, P.G.C.; Kipper, D.; da Silveira, V.P.; Fonseca, A.S.K.; Ikuta, N.; Lunge, V.R. Mitochondrial Genome and Phylogenetic Analysis of the Narrownose Smooth-Hound Shark Mustelus schmitti Springer, 1939. Animals 2024, 14, 3396. https://doi.org/10.3390/ani14233396
Nisa-Castro-Neto W, Wagner PGC, Kipper D, da Silveira VP, Fonseca ASK, Ikuta N, Lunge VR. Mitochondrial Genome and Phylogenetic Analysis of the Narrownose Smooth-Hound Shark Mustelus schmitti Springer, 1939. Animals. 2024; 14(23):3396. https://doi.org/10.3390/ani14233396
Chicago/Turabian StyleNisa-Castro-Neto, Walter, Paulo Guilherme Carniel Wagner, Diéssy Kipper, Vinicius Proença da Silveira, André Salvador Kazantzi Fonseca, Nilo Ikuta, and Vagner Ricardo Lunge. 2024. "Mitochondrial Genome and Phylogenetic Analysis of the Narrownose Smooth-Hound Shark Mustelus schmitti Springer, 1939" Animals 14, no. 23: 3396. https://doi.org/10.3390/ani14233396
APA StyleNisa-Castro-Neto, W., Wagner, P. G. C., Kipper, D., da Silveira, V. P., Fonseca, A. S. K., Ikuta, N., & Lunge, V. R. (2024). Mitochondrial Genome and Phylogenetic Analysis of the Narrownose Smooth-Hound Shark Mustelus schmitti Springer, 1939. Animals, 14(23), 3396. https://doi.org/10.3390/ani14233396






