Serological and Molecular Characterization of Small Ruminant Lentiviruses in Morocco
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection and Serological Analysis
2.2. DNA Extraction and SRLV Proviral Amplification
2.3. Amplicon Sequencing
2.4. Data Analysis
3. Results
3.1. Serology
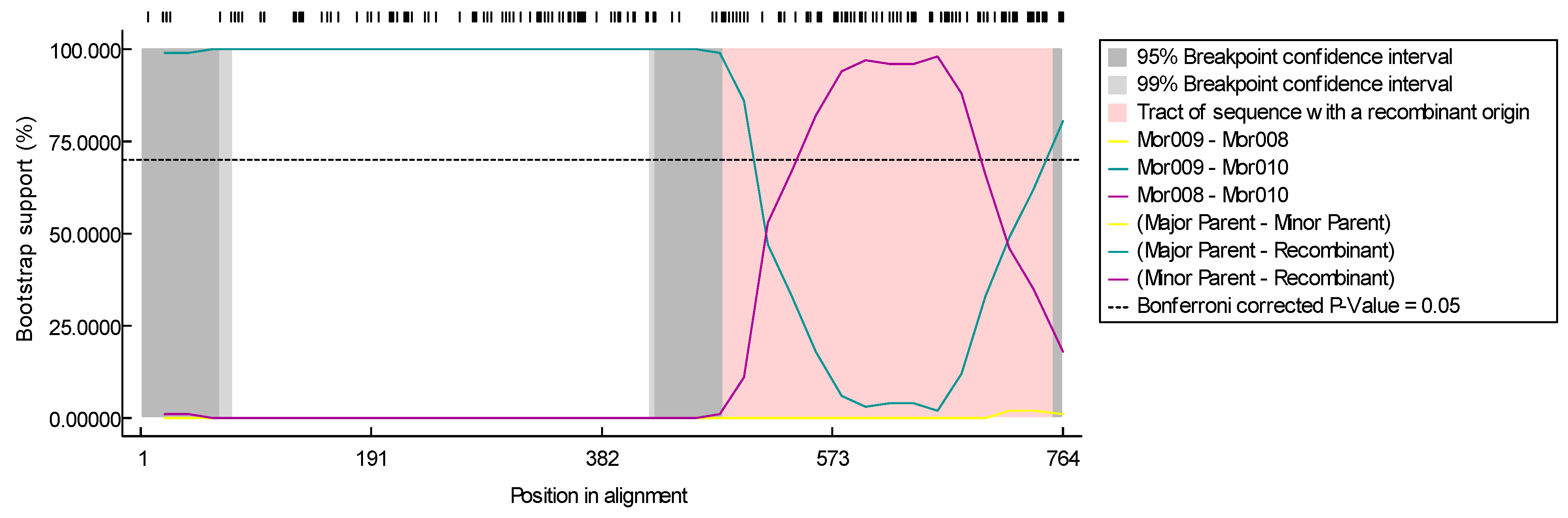
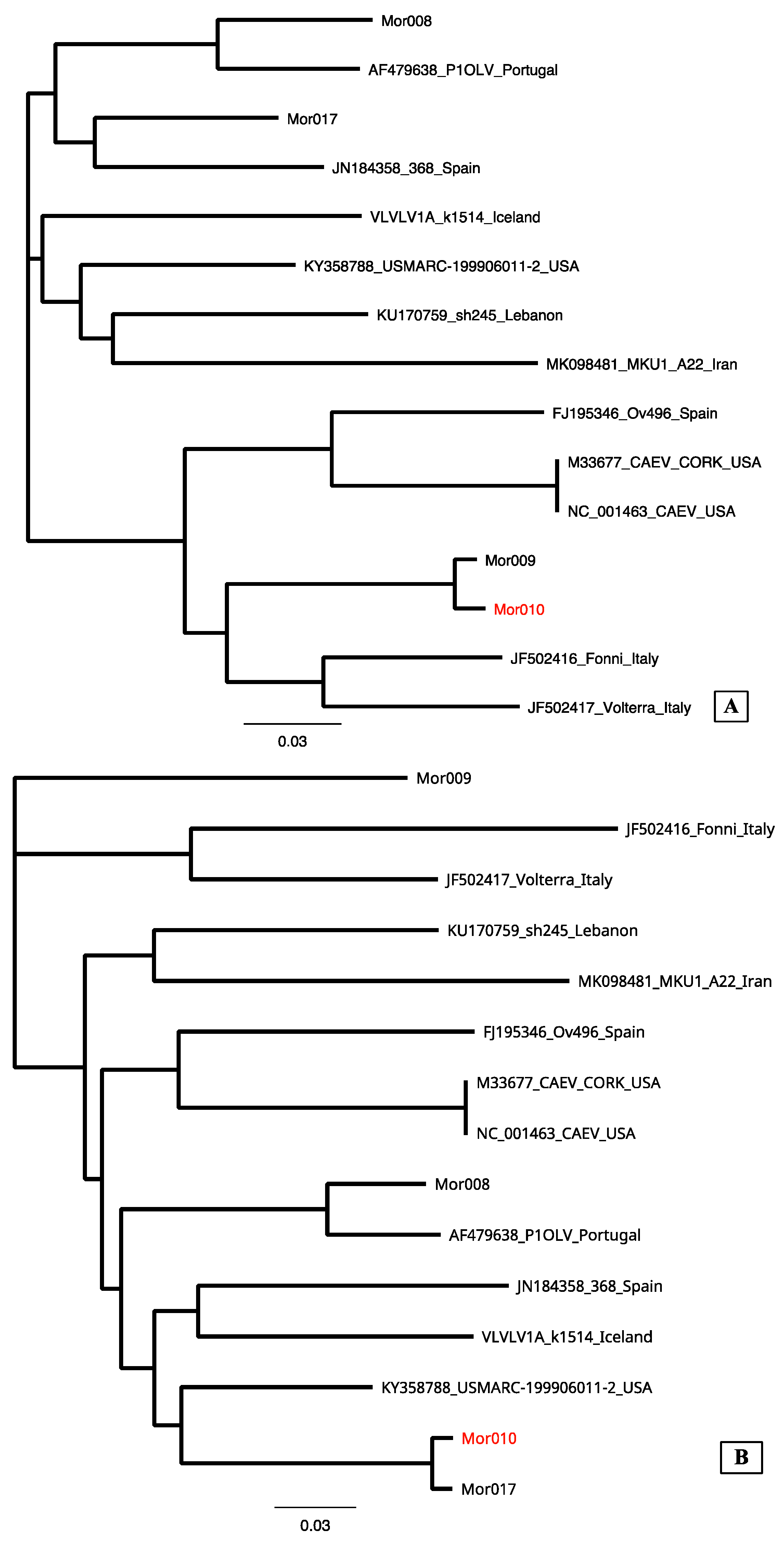
3.2. Sequencing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iniguez, L. Characterization of Small Ruminant Breeds in West Asia and North Africa; ICARDA: Aleppo, Syria, 2005. [Google Scholar]
- de Miguel, R.; Arrieta, M.; Rodríguez-largo, A.; Echeverría, I.; Resendiz, R.; Pérez, E.; Ruiz, H.; Pérez, M.; de Andrés, D.; Reina, R.; et al. Worldwide Prevalence of Small Ruminant Lentiviruses in Sheep: A Systematic Review and Meta-Analysis. Animals 2021, 11, 784. [Google Scholar] [CrossRef] [PubMed]
- Minguijón, E.; Reina, R.; Pérez, M.; Polledo, L.; Villoria, M.; Ramírez, H.; Leginagoikoa, I.; Badiola, J.J.; García-Marín, J.F.; de Andrés, D.; et al. Small Ruminant Lentivirus Infections and Diseases. Vet. Microbiol. 2015, 181, 75–89. [Google Scholar] [CrossRef]
- Blacklaws, B.A. Small Ruminant Lentiviruses: Immunopathogenesis of Visna-Maedi and Caprine Arthritis and Encephalitis Virus. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Bazzucchi, M.; Pierini, I.; Gobbi, P.; Pirani, S.; Torresi, C.; Iscaro, C.; Feliziani, F.; Giammarioli, M. Genomic Epidemiology and Heterogeneity of SRLV in Italy from 1998 to 2019. Viruses 2021, 13, 2338. [Google Scholar] [CrossRef] [PubMed]
- Olech, M. The Genetic Variability of Small-Ruminant Lentiviruses and Its Impact on Tropism, the Development of Diagnostic Tests and Vaccines and the Effectiveness of Control Programmes. J. Vet. Res. 2023, 67, 479–502. [Google Scholar] [CrossRef]
- Olech, M.; Rachid, A.; Croisé, B.; Kuźmak, J.; Valas, S. Genetic and Antigenic Characterization of Small Ruminant Lentiviruses Circulating in Poland. Virus Res. 2012, 163, 528–536. [Google Scholar] [CrossRef]
- Braz, G.F.; Heinemann, M.B.; Reis, J.K.P.; Teixeira, B.M.; Cruz, J.C.M.; Rajão, D.S.; Oliveira, F.G.; Alves, F.; Castro, R.S.; Leite, R.C.; et al. Genetic and Antigenic Characterization of Brazilian SRLV Strains: Natural Small Ruminant Interspecies Transmission from Mixed Herds. Infect. Genet. Evol. 2022, 103, 105322. [Google Scholar] [CrossRef]
- Olech, M.; Hodor, D.; Toma, C.; Negoescu, A.; Taulescu, M. First Molecular Characterization of Small Ruminant Lentiviruses Detected in Romania. Animals 2023, 13, 3718. [Google Scholar] [CrossRef]
- Olech, M.; Murawski, M.; Kuźmak, J. Molecular Analysis of Small-Ruminant Lentiviruses in Polish Flocks Reveals the Existence of a Novel Subtype in Sheep. Arch. Virol. 2019, 164, 1193–1198. [Google Scholar] [CrossRef]
- Santry, L.A.; de Jong, J.; Gold, A.C.; Walsh, S.R.; Menzies, P.I.; Wootton, S.K. Genetic Characterization of Small Ruminant Lentiviruses Circulating in Naturally Infected Sheep and Goats in Ontario, Canada. Virus Res. 2013, 175, 30–44. [Google Scholar] [CrossRef]
- De la Luz-Armendáriz, J.; Ducoing-Watty, A.E.; Ramírez-Mendoza, H.; Gómez-Núñez, L.; Tufiño-Loza, C.; Cabrera-Domínguez, E.M.; Díaz-Aparicio, E.; Rivera-Benítez, J.F. Prevalence, Molecular Detection, and Pathological Characterization of Small Ruminant Lentiviruses in Goats from Mexico. Small Rumin. Res. 2021, 202, 106474. [Google Scholar] [CrossRef]
- Laamanen, I.; Jakava-Viljanen, M.; Sihvonen, L. Genetic Characterization of Maedi-Visna Virus (MVV) Detected in Finland. Vet. Microbiol. 2007, 122, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Michiels, R.; Adjadj, N.R.; De Regge, N. Phylogenetic Analysis of Belgian Small Ruminant Lentiviruses Supports Cross Species Virus Transmission and Identifies New Subtype B5 Strains. Pathogens 2020, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Kalogianni, A.I.; Stavropoulos, I.; Chaintoutis, S.C.; Bossis, I.; Gelasakis, A.I. Serological, Molecular and Culture-based Diagnosis of Lentiviral Infections in Small Ruminants. Viruses 2021, 13, 1711. [Google Scholar] [CrossRef] [PubMed]
- Arcangeli, C.; Torricelli, M.; Sebastiani, C.; Lucarelli, D.; Ciullo, M.; Passamonti, F.; Giammarioli, M.; Biagetti, M. Genetic Characterization of Small Ruminant Lentiviruses (SRLVs) Circulating in Naturally Infected Sheep in Central Italy. Viruses 2022, 14, 686. [Google Scholar] [CrossRef] [PubMed]
- Colitti, B.; Coradduzza, E.; Puggioni, G.; Capucchio, M.T.M.T.; Reina, R.; Bertolotti, L.; Rosati, S. A New Approach for Small Ruminant Lentivirus Full Genome Characterization Revealed the Circulation of Divergent Strains. PLoS ONE 2019, 14, e0212585. [Google Scholar] [CrossRef] [PubMed]
- Olech, M.; Bomba, A.; Kuźmak, J. Quasispecies Composition of Small Ruminant Lentiviruses Found in Blood Leukocytes and Milk Epithelial Cells. Viruses 2021, 13, 2497. [Google Scholar] [CrossRef] [PubMed]
- Gifford, R.J. Viral Evolution in Deep Time: Lentiviruses and Mammals. Trends Genet. 2012, 28, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Tebit, D.M.; Arts, E.J. Tracking a Century of Global Expansion and Evolution of HIV to Drive Understanding and to Combat Disease. Lancet Infect. Dis. 2011, 11, 45–56. [Google Scholar] [CrossRef]
- Katzourakis, A.; Gifford, R.J.; Tristem, M.; Thomas, M.; Gilbert, P.; Pybus, O.G. Macroevolution of Complex Retroviruses. Science 2009, 325, 1512. [Google Scholar] [CrossRef]
- Shah, C.; Böni, J.; Huder, J.B.; Vogt, H.R.; Mühlherr, J.; Zanoni, R.; Miserez, R.; Lutz, H.; Schüpbach, J. Phylogenetic Analysis and Reclassification of Caprine and Ovine Lentiviruses Based on 104 New Isolates: Evidence for Regular Sheep-to-Goat Transmission and Worldwide Propagation through Livestock Trade. Virology 2004, 319, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Straub, O.C. Maedi-Visna Virus Infection in Sheep. History and Present Knowledge. Comp. Immunol. Microbiol. Infect. Dis. 2004, 27, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Carrozza, M.L.; Niewiadomska, A.M.; Mazzei, M.; Abi-Said, M.R.; Hué, S.; Hughes, J.; Gatseva, A.; Gifford, R.J. Emergence and Pandemic Spread of Small Ruminant Lentiviruses. Virus Evol. 2023, 9, vead005. [Google Scholar] [CrossRef] [PubMed]
- Alberto, F.J.; Boyer, F.; Orozco-Terwengel, P.; Streeter, I.; Servin, B.; De Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent Genomic Signatures of Domestication in Sheep and Goats. Nat. Commun. 2018, 9, 813. [Google Scholar] [CrossRef]
- Mahin, L.; Chadli, M.; Houwers, D.J. A Preliminary Report on the Occurrence of Maedi-visna in Sheep in Morocco. Vet. Q. 1984, 6, 104. [Google Scholar] [CrossRef] [PubMed]
- Bouljihad, M.; Leipold, H.W. Ovine Lentiviral Infection (Maedi/Visna) in Morocco: A Serologic and Postmortem Survey. J. Vet. Med. Ser. A 1994, 41, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Daif, S.; El Berbri, I.; Lhor, Y.; Fassi Fihri, O. Serological and Molecular Prevalence Study of Bluetongue Virus in Small Domestic Ruminants in Morocco. Sci. Rep. 2022, 12, 19448. [Google Scholar] [CrossRef]
- Rosati, S.; Profiti, M.; Lorenzetti, R.; Bandecchi, P.; Mannelli, A.; Ortoffi, M.; Tolari, F.; Ciabatti, I.M. Development of Recombinant Capsid Antigen/Transmembrane Epitope Fusion Proteins for Serological Diagnosis of Animal Lentivirus Infections. J. Virol. Methods 2004, 121, 73–78. [Google Scholar] [CrossRef]
- Grego, E.; Bertolotti, L.L.; Quasso, A.; Profiti, M.; Lacerenza, D.; Muz, D.; Rosati, S. Genetic Characterization of Small Ruminant Lentivirus in Italian Mixed Flocks: Evidence for a Novel Genotype Circulating in a Local Goat Population. J. Gen. Virol. 2007, 88, 3423–3427. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F.; Seconds-Pichon, A.; Biggins, F.; Wingett, S. FastQC. A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics. Babraham Inst. 2015, 1, 1. [Google Scholar]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Datamonkey: Rapid Detection of Selective Pressure on Individual Sites of Codon Alignments. Bioinformatics 2005, 21, 2531–2533. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, H.; Reina, R.; Amorena, B.; Andrés, D.; Martínez, H. Small Ruminant Lentiviruses: Genetic Variability, Tropism and Diagnosis. Viruses 2013, 5, 1175–1207. [Google Scholar] [CrossRef]
- Molaee, V.; Bazzucchi, M.; De Mia, G.M.; Otarod, V.; Abdollahi, D.; Rosati, S.; Lühken, G. Phylogenetic Analysis of Small Ruminant Lentiviruses in Germany and Iran Suggests Their Expansion with Domestic Sheep. Sci. Rep. 2020, 10, 2243. [Google Scholar] [CrossRef]
- Peterhans, E.; Greenland, T.; Badiola, J.; Harkiss, G.; Bertoni, G.; Amorena, B.; Eliaszewicz, M.; Juste, R.A.; Kraßnig, R.; Lafont, J.-P.; et al. Routes of Transmission and Consequences of Small Ruminant Lentiviruses (SRLVs) Infection and Eradication Schemes. Vet. Res. 2004, 35, 257–274. [Google Scholar] [CrossRef]
- Gjerset, B. Genetic Diversity of Small-Ruminant Lentiviruses: Characterization of Norwegian Isolates of Caprine Arthritis Encephalitis Virus. J. Gen. Virol. 2006, 87, 573–580. [Google Scholar] [CrossRef]
- Muigai, A.W.T.; Hanotte, O. The Origin of African Sheep: Archaeological and Genetic Perspectives. Afr. Archaeol. Rev. 2013, 30, 39–50. [Google Scholar] [CrossRef]
- de Andrés, X.; Ramírez, H.; Bertolotti, L.; San Román, B.; Glaria, I.; Crespo, H.; Jáuregui, P.; Minguijón, E.; Juste, R.; Leginagoikoa, I.; et al. An Insight into a Combination of ELISA Strategies to Diagnose Small Ruminant Lentivirus Infections. Vet. Immunol. Immunopathol. 2013, 152, 277–288. [Google Scholar] [CrossRef]
- Cardinaux, L.; Zahno, M.L.; Deubelbeiss, M.; Zanoni, R.; Vogt, H.R.; Bertoni, G. Virological and Phylogenetic Characterization of Attenuated Small Ruminant Lentivirus Isolates Eluding Efficient Serological Detection. Vet. Microbiol. 2013, 162, 572–581. [Google Scholar] [CrossRef]
- Glaria, I.; Reina, R.; Ramírez, H.; de Andrés, X.; Crespo, H.; Jauregui, P.; Salazar, E.; Luján, L.; Pérez, M.M.; Benavides, J.; et al. Visna/Maedi Virus Genetic Characterization and Serological Diagnosis of Infection in Sheep from a Neurological Outbreak. Vet. Microbiol. 2012, 155, 137–146. [Google Scholar] [CrossRef]
- Kryazhimskiy, S.; Plotkin, J.B. The Population Genetics of DN/DS. PLoS Genet. 2008, 4, 1000304. [Google Scholar] [CrossRef] [PubMed]
- Olech, M.; Kuzmak, J. Compartmentalization of Subtype A17 of Small Ruminant Lentiviruses between Blood and Colostrum in Infected Goats Is Not Exclusively Associated to the Env Gene. Viruses 2019, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, H.; Reina, R.; Bertolotti, L.; Cenoz, A.; Hernández, M.M.; San Román, B.; Glaria, I.; de Andrés, X.; Crespo, H.; Jáuregui, P.; et al. Study of Compartmentalization in the Visna Clinical Form of Small Ruminant Lentivirus Infection in Sheep. BMC Vet. Res. 2012, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
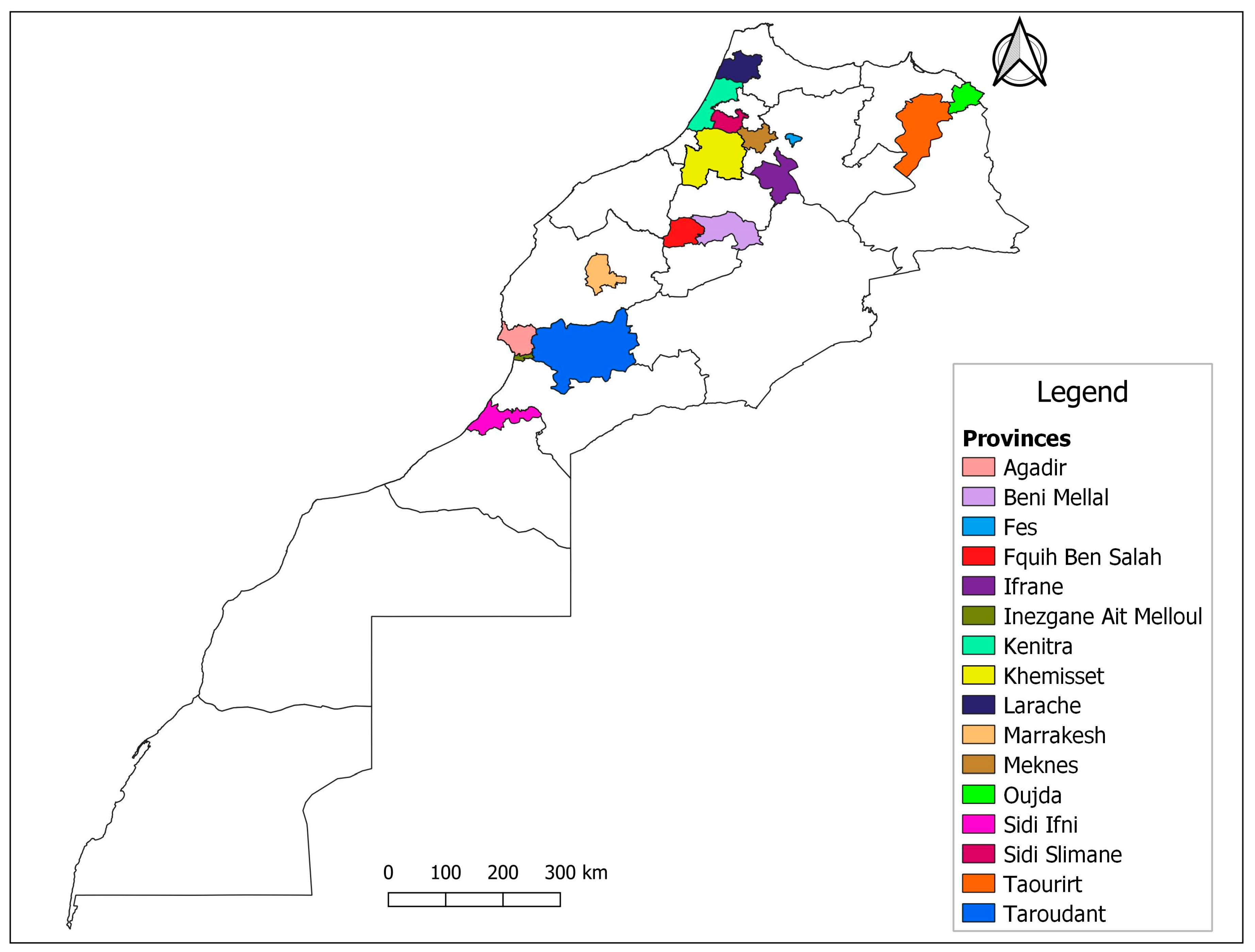


| Region | Species | Number of Flocks Tested | Number of Animals | Number of Tested Animals | Screening Positive | Screening Doubt | Genotyping A | Genotyping B | Genotyping E | Genotyping Indeterminate | Genotyping Negative |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Agadir | Sheep | 9 | 1896 | 151 | 3 | 6 | 0 | 3 | 0 | 2 | 4 |
| Goat | 7 | 358 | 86 | 1 | 6 | 0 | 2 | 2 | 1 | 2 | |
| Marrachech | Sheep | 8 | 515 | 91 | 1 | 2 | 0 | 1 | 0 | 2 | 0 |
| Goat | 2 | 22 | 7 | 0 | 0 | - | - | - | - | - | |
| Taourirt | Sheep | 2 | 700 | 58 | 8 | 1 | 1 | 1 | 0 | 4 | 3 |
| Goat | 2 | 20 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Kenitra | Sheep | 3 | 140 | 65 | 0 | 0 | - | - | - | - | - |
| Laraache | Sheep | 3 | 210 | 43 | 7 | 0 | 0 | 0 | 1 | 3 | 3 |
| Kcer Lkbir | Sheep | 2 | 100 | 38 | 7 | 0 | 3 | 0 | 3 | 0 | 1 |
| Fes | Sheep | 1 | 50 | 18 | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
| Ifrane | Sheep | 2 | 110 | 29 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Fquih Ben Salem | Sheep | 2 | 220 | 20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Goat | 2 | 40 | 12 | 0 | - | - | - | - | - | - | |
| Rommani | Sheep | 4 | 74 | 48 | 3 | 0 | 1 | 0 | 0 | 2 | 0 |
| Goat | 1 | 5 | 1 | 0 | - | - | - | - | - | - | |
| Meknes | Sheep | 1 | 75 | 9 | 0 | - | - | - | - | - | - |
| Oujda | Sheep | 3 | 140 | 25 | 0 | 2 | 0 | 2 | 0 | 0 | 0 |
| Goat | 1 | 2 | 2 | 0 | - | - | - | - | - | - | |
| Sidi Ifni | Goat | 3 | 87 | 21 | 0 | - | - | - | - | - | - |
| Benimellal | Goat | 1 | 17 | 10 | 0 | - | - | - | - | - | - |
| Sidi Slimane | Goat | 1 | 15 | 5 | 0 | - | - | - | - | - | - |
| Berkane | Sheep | 1 | 20 | 7 | 0 | - | - | - | - | - | - |
| Sequence | Subtype | Alignment |
|---|---|---|
| VLVLV1A_k1514_Iceland | A1 |  |
| Mor019 |  | |
| Mor023 |  | |
| Mor026 |  | |
| Mor033 |  | |
| Mor034 |  | |
| AY101611_85-34_USA | A2 |  |
| AY454175_SNCR5560_Switzerland | A5 |  |
| AY454176_5561_Switzerland | A3 |  |
| AY454208_5692_Switzerland | A7 |  |
| MH374287_It0038_2017_Italy | A19 |  |
| MH374291_ItVda_2017_Italy | A8 |  |
| MK098481_MKU1_Iran | A22 |  |
| MG554409_It0009_2017_Italy | A20 |  |
| Mor017 |  | |
| AM084209_Finland | A |  |
| Mor008 |  | |
| Mor018 |  | |
| Mor020 |  | |
| Mor021 |  | |
| Mor022 |  | |
| AY445885_G4668 _Switzerland | A4 |  |
| AF479638_P1OLV_Portugal | A2 |  |
| M33677_CAEV_CORK_USA | B1 |  |
| Mor038 | 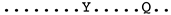 | |
| Mor009 | 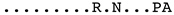 | |
| Mor010 | 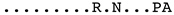 | |
| JF502416_Fonni_Italy | B3 | 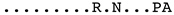 |
| GU120138_Shanxi _China | B1 |  |
| KT214469_Sichuan_China | B1 |  |
| HM210570_FESC-752_Spain | B1 |  |
| LC002526_Philippine | B |  |
| Mor035 |  | |
| Mor037 |  | |
| FJ195346_Ov496_Spain | B2 |  |
| AF015181_CA680_France | B1 | 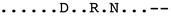 |
| EU293537_Roccaverano _Italy | E1 | 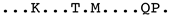 |
| GQ381130_Seui _Italy | E2 | 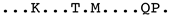 |
| AF322109_1GA_Norway | C |  |
| Sample | Region | Specie | ELISA Screening | ELISA Genotyping | PCR | Subtype |
|---|---|---|---|---|---|---|
| Mor008 | Taourirt | Ovine | Doubt | Indeterminate | Positive | A2 |
| Mor009 | Taourirt | Ovine | Positive | B | Positive | B6 |
| Mor010 | Taourirt | Ovine | Doubt | Indeterminate | Positive | B6 |
| Mor017 | Agadir | Caprine | Positive | Indeterminate | Positive | A |
| Mor018 | Agadir | Caprine | Doubt | Indeterminate | Positive | A2 |
| Mor019 | Rommani | Ovine | Doubt | A | Doubt | A2 |
| Mor020 | Laraache | Ovine | Doubt | Negative | Doubt | A2 |
| Mor021 | Laraache | Ovine | Doubt | Negative | Doubt | A2 |
| Mor022 | Laraache | Ovine | Doubt | Negative | Doubt | A2 |
| Mor023 | Kcer Lkbir | Ovine | Positive | E | Doubt | A2 |
| Mor026 | Fes | Ovine | Positive | Negative | Positive | A2 |
| Mor033 | Kcer Lkbir | Ovine | Positive | Negative | Positive | A2 |
| Mor034 | Kcer Lkbir | Ovine | Positive | A | Positive | A2 |
| Mor035 | Agadir | Caprine | Negative | B | Doubt | B2 |
| Mor037 | Rommani | Ovine | Doubt | Indeterminate | Positive | B2 |
| Mor038 | Taourirt | Ovine | Positive | E | Positive | A2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colitti, B.; Daif, S.; Choukri, I.; Scalas, D.; Jerre, A.; El Berbri, I.; Fassi Fihri, O.; Rosati, S. Serological and Molecular Characterization of Small Ruminant Lentiviruses in Morocco. Animals 2024, 14, 550. https://doi.org/10.3390/ani14040550
Colitti B, Daif S, Choukri I, Scalas D, Jerre A, El Berbri I, Fassi Fihri O, Rosati S. Serological and Molecular Characterization of Small Ruminant Lentiviruses in Morocco. Animals. 2024; 14(4):550. https://doi.org/10.3390/ani14040550
Chicago/Turabian StyleColitti, Barbara, Soukaina Daif, Imane Choukri, Daniela Scalas, Anniken Jerre, Ikhlass El Berbri, Ouafaa Fassi Fihri, and Sergio Rosati. 2024. "Serological and Molecular Characterization of Small Ruminant Lentiviruses in Morocco" Animals 14, no. 4: 550. https://doi.org/10.3390/ani14040550





