Analysis of Genetic Diversity and Structure of Eight Populations of Nerita yoldii along the Coast of China Based on Mitochondrial COI Gene
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sampling
2.2. DNA Extraction and Polymerase Chain Reaction Amplification
2.3. Data Analysis
3. Results
3.1. Characteristics of the COI Sequence and the Genetic Diversity of N. yoldii
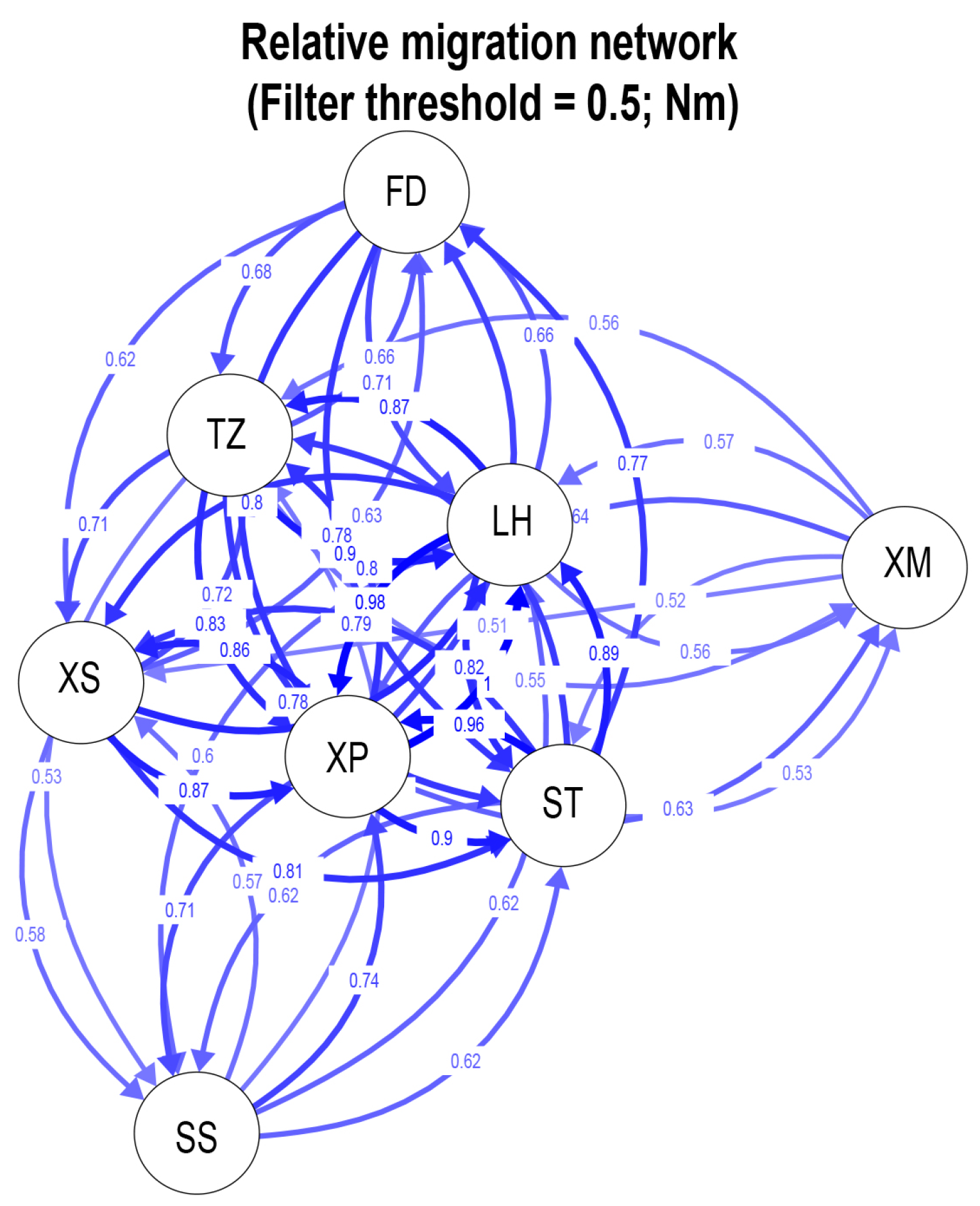
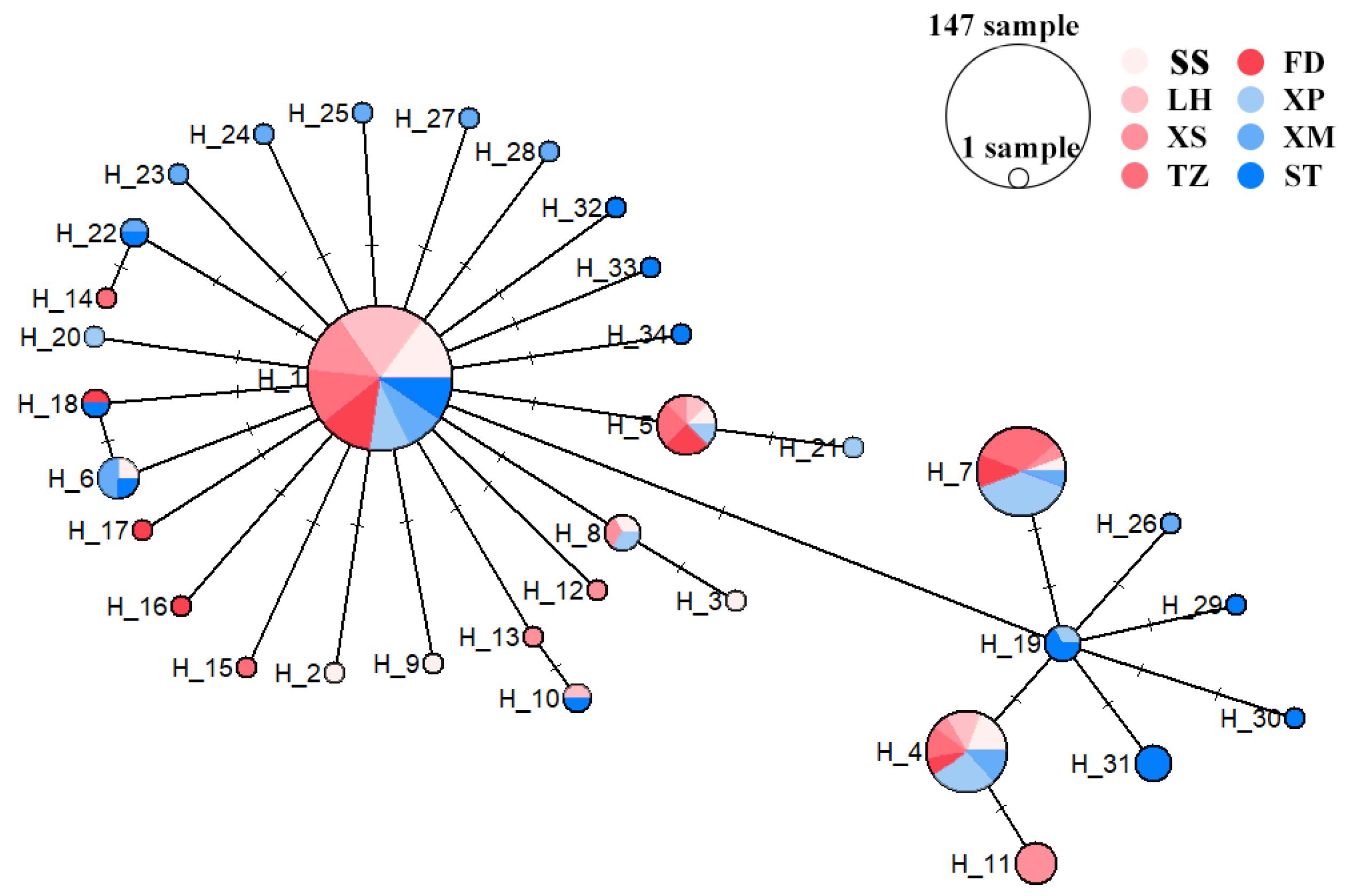
3.2. Population Genetic Structure of N. yoldii
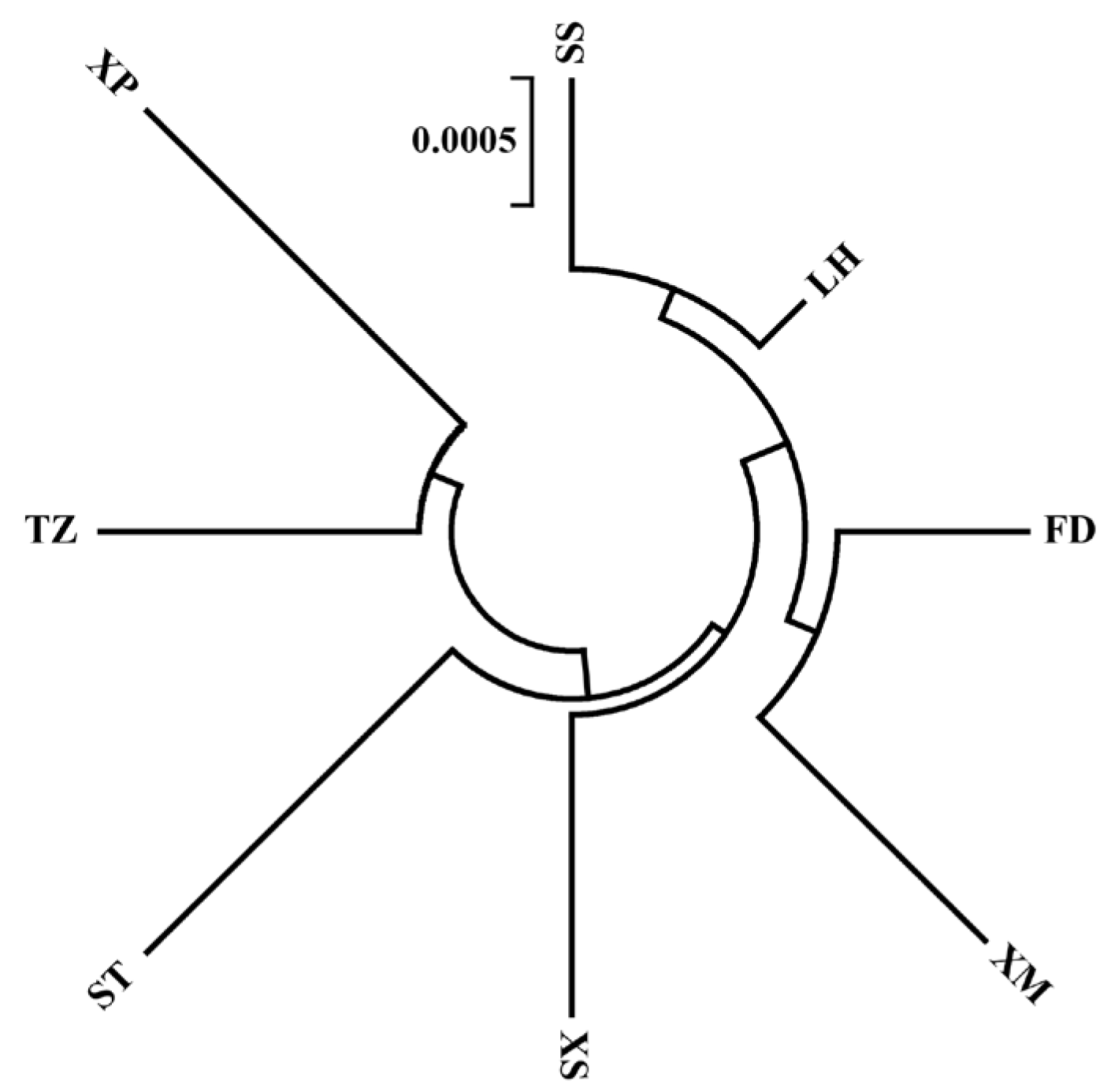
3.3. Population Phylogenetic Analysis
4. Discussion
4.1. Genetic Diversity of the Eight Populations of N. yoldii along the Coast of China
4.2. Population Differentiation of the Eight Populations of N. yoldii along the Coast of China
4.3. Phylogeny of the Eight Populations of N. yoldii along the Coast of China
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kano, Y.; Chiba, S.; Kase, T. Major adaptive radiation in neritopsine gastropods estimated from 28S rRNA sequences and fossil records. Proc. Biol. Sci. 2002, 269, 2457–2465. [Google Scholar] [CrossRef]
- Sasaki, T.; Ishikawa, H. The first occurrence of a neritopsine gastropod from a phreatic community. J. Molluscan Stud. 2002, 68, 286–288. [Google Scholar] [CrossRef]
- Waters, J.M.; King, T.M.; O’Loughlin, P.M.; Spencer, H.G. Phylogeographical disjunction in abundant high-dispersal littoral gastropods. Mol. Ecol. 2005, 14, 2789–2802. [Google Scholar] [CrossRef]
- Qi, L.; Xu, B.; Kong, L.; Li, Q. Improved phylogenetic resolution within Neritidae (Gastropoda, Nertimorpha) with implications for the evolution of shell traits and habitat. Zool. Scr. 2023, 52, 46–57. [Google Scholar] [CrossRef]
- Okutani, T. Marine Mollusks in Japan, 2nd ed.; Okutani, T., Ed.; Tokai University Press: Shibuya, Japan, 2017; pp. 259–1292. [Google Scholar]
- Zhang, X.; Qi, Z.; Zhang, F.; Ma, X. A preliminary study of the demarcation of marine Molluscan faunal regions of China and its adjacent waters. Oceanol. Limnol. Sin. 1963, 5, 124–138. [Google Scholar]
- Jie, W.; Yan, H.Y.; Cheng, Z.Y.; Huang, X.W.; Wei, W.; Ding, M.W.; Dong, Y.W. Recent northward range extension of Nerita yoldii (Gastropoda: Neritidae) on artificial rocky shores in China. J. Molluscan Stud. 2018, 84, 345–353. [Google Scholar]
- Wang, J.; Cheng, Z.Y.; Dong, Y.W. Demographic, physiological and genetic factors linked to the poleward range expansion of the snail Nerita yoldii along the shoreline of China. Mol. Ecol. 2022, 31, 4510–4526. [Google Scholar] [CrossRef] [PubMed]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, M.; Ge, S.; Wang, Q.; Yu, D.; Guan, S. Population structuring and historical demography of a common clam worm Perinereris aibuhitensis near the coasts of Shandong Peninsula. Biochem. Syst. Ecol. 2012, 44, 70–78. [Google Scholar] [CrossRef]
- Gary, S.F.; Fox, A.D.; Biastoch, A.; Roberts, J.M.; Cunningham, S.A. Larval behaviour, dispersal and population connectivity in the deep sea. Sci. Rep. 2020, 10, 10675. [Google Scholar] [CrossRef]
- Woodson, C.B.; McManus, M.A. Foraging behavior can influence dispersal of marine organisms. Limnol. Oceanogr. 2007, 52, 2701–2709. [Google Scholar] [CrossRef]
- Toonen, R.; Pawlik, J.R. Settlement of the gregarious tube worm Hydroides dianthus (Polychaeta: Serpulidae). I. Gregarious and nongregarious settlement. Mar. Ecol. Prog. Ser. 2001, 224, 103–114. [Google Scholar] [CrossRef]
- Addison, J.A.; Hart, M.W. Analysis of population genetic structure of the green sea urchin (Strongylocentrotus droebachiensis) using microsatellites. Mar. Biol. 2004, 144, 243–251. [Google Scholar] [CrossRef]
- Ye, Y.; Fu, Z.; Tian, Y.; Li, J.; Guo, B.; Lv, Z.; Wu, C. Pelagic larval dispersal habits influence the population genetic structure of clam Gomphina aequilatera in China. Genes Genom. 2018, 40, 1213–1223. [Google Scholar] [CrossRef] [PubMed]
- Timón-Gómez, A.; Nývltová, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef]
- Jiang, L.; Kang, L.; Wu, C.; Chen, M.; Lü, Z. A comprehensive description and evolutionary analysis of 9 Loliginidae mitochondrial genomes. Hydrobiologia 2018, 808, 115–124. [Google Scholar] [CrossRef]
- Dai, L.; Kausar, S.; Abbas, M.N.; Wang, T.-T. Complete sequence and characterization of the Ectropis oblique mitochondrial genome and its phylogenetic implications. Int. J. Biol. Macromol. 2018, 107 Pt A, 1142–1150. [Google Scholar] [CrossRef]
- Feng, J.; Guo, Y.; Yan, C.; Ye, Y.; Li, J.; Guo, B.; Lü, Z. Sequence comparison of the mitochondrial genomes in two species of the genus Nerita (Gastropoda: Neritimorpha: Neritidae): Phylogenetic implications and divergence time estimation for Neritimorpha. Mol. Biol. Rep. 2020, 47, 7903–7916. [Google Scholar] [CrossRef]
- Devassy, A.; Kumar, R.; Shajitha, P.P.; John, R.; Padmakumar, K.G.; Basheer, V.S.; Gopalakrishnan, A.; Mathew, L. Genetic identification and phylogenetic relationships of Indian clariids based on mitochondrial COI sequences. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 3777–3780. [Google Scholar] [CrossRef]
- Toda, S.; Osakabe, M.; Komazaki, S. Interspecific diversity of mitochondrial COI sequences in Japanese Panonychus species (Acari: Tetranychidae). Exp. Appl. Acarol. 2000, 24, 821–829. [Google Scholar] [CrossRef]
- Dumidae, A.; Janthu, P.; Subkrasae, C.; Pumidonming, W.; Dekumyoy, P.; Thanwisai, A.; Vitta, A. Genetic analysis of Cryptozona siamensis (Stylommatophora, Ariophantidae) populations in Thailand using the mitochondrial 16S rRNA and COI sequences. PLoS ONE 2020, 15, e0239264. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, K.; Deng, D.; Zhang, Y.N.; Peng, S.; Xu, X. Genetic Diversity of Daphnia pulex in the Middle and Lower Reaches of the Yangtze River. PLoS ONE 2016, 11, e0152436. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Zhang, J.; Huang, J.; Wang, L.; Li, Y.; Hafeez, M.; Lu, Y. Population Genetic Diversity and Structure of Thrips tabaci (Thysanoptera: Thripidae) on Allium Hosts in China, Inferred From Mitochondrial COI Gene Sequences. J. Econ. Entomol. 2020, 113, 1426–1435. [Google Scholar] [CrossRef]
- Qasim, M.; Baohua, W.; Zou, H.; Lin, Y.; Dash, C.K.; Bamisile, B.S.; Hussain, M.; Zhiwen, Z.; Wang, L. Phylogenetic relationship and genetic diversity of citrus psyllid populations from China and Pakistan and their associated Candidatus bacterium. Mol. Phylogenet. Evol. 2018, 126, 173–180. [Google Scholar] [CrossRef]
- Woolley, V.C.; Tembo, Y.L.; Ndakidemi, B.; Obanyi, J.N.; Arnold, S.E.; Belmain, S.R.; Ndakidemi, P.A.; Ogendo, J.O.; Stevenson, P.C. The diversity of aphid parasitoids in East Africa and implications for biological control. Pest Manag. Sci. 2022, 78, 1109–1116. [Google Scholar] [CrossRef]
- Mao, Y.; Gao, T.; Yanagimoto, T.; Xiao, Y. Molecular phylogeography of Ruditapes philippinarum in the Northwestern Pacific Ocean based on COI gene. J. Exp. Mar. Biol. Ecol. 2011, 407, 171–181. [Google Scholar] [CrossRef]
- Hsu, T.-H.; Gwo, J.-C. Genetic diversity and stock identification of small abalone (Haliotis diversicolor) in Taiwan and Japan. PLoS ONE 2017, 12, e0179818. [Google Scholar] [CrossRef]
- Huang, X.C.; Su, J.H.; Ouyang, J.X.; Ouyang, S.; Zhou, C.H.; Wu, X.P. Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida): Species delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns. Mol. Phylogenet. Evol. 2019, 130, 45–59. [Google Scholar] [CrossRef]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Sundqvist, L.; Keenan, K.; Zackrisson, M.; Prodöhl, P.; Kleinhans, D. Directional genetic differentiation and relative migration. Ecol. Evol. 2016, 6, 3461–3475. [Google Scholar] [CrossRef] [PubMed]
- Grant, W.; Bowen, B. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Yan, C.; Miao, J.; Feng, J.; Xia, L.; Ye, Y.; Li, J.; Guo, B. Pelagic Larval Dispersal Habits Shape the Weak Population Structure of Thais clavigera in the Coastal Areas of China Sea. Pak. J. Zool. 2023. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [PubMed]
- Feng, J.T.; Xia, L.P.; Yan, C.R.; Miao, J.; Ye, Y.Y.; Li, J.J.; Guo, B.Y.; Lü, Z.M. Characterization of four mitochondrial genomes of family Neritidae (Gastropoda: Neritimorpha) and insight into its phylogenetic relationships. Sci. Rep. 2021, 11, 11748. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-X.; Gao, T.-X.; Yokogawa, K.; Zhang, Y.-P. Differential population structuring and demographic history of two closely related fish species, Japanese sea bass (Lateolabrax japonicus) and spotted sea bass (Lateolabrax maculatus) in Northwestern Pacific. Mol. Phylogenet. Evol. 2006, 39, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Ni, G.; Li, Q.; Kong, L.; Zheng, X. Phylogeography of bivalve Cyclina sinensis: Testing the historical glaciations and Changjiang River outflow hypotheses in northwestern Pacific. PLoS ONE 2012, 7, e49487. [Google Scholar] [CrossRef]
- Zhou, N.; Shen, H.; Chen, C.; Sun, B.; Zheng, P.; Wang, C. Genetic structure of Onchidium “struma” (Mollusca: Gastropoda: Eupulmonata) from the coastal area of China based on mtCO I. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 1319–1323. [Google Scholar]
- Wright, S. Evolution in Mendelian Populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef]
- Zhan, A.; Hu, J.; Hu, X.; Zhou, Z.; Hui, M.; Wang, S.; Peng, W.; Wang, M.; Bao, Z. Fine-Scale Population Genetic Structure of Zhikong Scallop (Chlamys farreri): Do Local Marine Currents Drive Geographical Differentiation? Mar. Biotechnol. 2009, 11, 223–235. [Google Scholar] [CrossRef]
- Je Lee, H.; Boulding, E.G. Spatial and temporal population genetic structure of four northeastern Pacific littorinid gastropods: The effect of mode of larval development on variation at one mitochondrial and two nuclear DNA markers. Mol. Ecol. 2009, 18, 2165–2184. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Liu, Y.; Xu, Z.; Yin, Y.; Hou, Y. Bidirectional Volume Exchange between Kuroshio and East China Sea Shelf Water Based on a Whole-Region Passive-Tracing Method. J. Geophys. Res. Oceans 2020, 125, e2019JC015528. [Google Scholar] [CrossRef]
- Yuan, D.; Zhu, J.; Li, C.; Hu, D. Cross-shelf circulation in the Yellow and East China Seas indicated by MODIS satellite observations. J. Mar. Syst. 2008, 70, 134–149. [Google Scholar] [CrossRef]
- Li, G.; Qiao, L.; Dong, P.; Ma, Y.; Xu, J.; Liu, S.; Liu, Y.; Li, J.; Li, P.; Ding, D.; et al. Hydrodynamic condition and suspended sediment diffusion in the Yellow Sea and East China Sea. J. Geophys. Res. Oceans 2016, 121, 6204–6222. [Google Scholar] [CrossRef]
- Jilan, S. Overview of the South China Sea circulation and its influence on the coastal physical oceanography outside the Pearl River Estuary. Cont. Shelf Res. 2004, 24, 1745–1760. [Google Scholar] [CrossRef]
- Chen, R.; Li, J. Large-scale ecological study on planktonic ostracoda in China’s seas and adjacent waters Ⅱ. Correlation between planktonic ostracoda and water systems. Acta Oceanol. Sin. (Chin. Ed.) 1998, 20, 96–101. [Google Scholar]
- Xue, C.; Zhang, Y. Sediment transportation of longshore current and coastal current in China littoral zone. Mar. Geol. Quat. Geol. 2010, 30, 1–8. [Google Scholar] [CrossRef]
- Wu, J.; Chen, J.; Wang, C.; Yan, X.; Yin, X.; Liu, Q. Constraining the origin of sedimentary organic matter in the eastern Guangdong coast of China using δ13C and δ15N. Front. Mar. Sci. 2023, 10, 1234116. [Google Scholar] [CrossRef]
- Briggs, J.C. Marine centres of origin as evolutionary engines. J. Biogeogr. 2003, 30, 1–18. [Google Scholar] [CrossRef]
- Pinxian, W.; Xiangjun, S. Last glacial maximum in China: Comparison between land and sea. CATENA 1994, 23, 341–353. [Google Scholar] [CrossRef]
- Voris, H.K. Maps of Pleistocene sea levels in Southeast Asia: Shorelines, river systems and time durations. J. Biogeogr. 2000, 27, 1153–1167. [Google Scholar] [CrossRef]
- Wang, L.; Yu, H.; Li, Q. Development of microsatellite markers and analysis of genetic diversity of Barbatia virescens in the southern coasts of China. Genes Genom. 2019, 41, 407–416. [Google Scholar] [CrossRef]
- Marko, P.B. Fossil calibration of molecular clocks and the divergence times of geminate species pairs separated by the isthmus of panama. Mol. Biol. Evol. 2002, 19, 2005–2021. [Google Scholar] [CrossRef]



| Sampling Site | Abbreviation | Coordinates | Sampling Date | Sample Size |
|---|---|---|---|---|
| Shengsi | SS | 30°44′ N 122°28′ E | 2023.03 | 32 |
| Liuheng | LH | 29°40′ N 122°12′ E | 2021.01 | 33 |
| Xiangshan | XS | 29°26′ N 121°58′ E | 2021.01 | 30 |
| Taizhou | TZ | 28°13′ N 121°22′ E | 2023.05 | 30 |
| Fuding | FD | 27°09′ N 120°25′ E | 2019.08 | 26 |
| Xiapu | XP | 26°52′ N 120°04′ E | 2018.11 | 30 |
| Xiamen | XM | 24°26′ N 118°04′ E | 2021.04 | 24 |
| Shantou | ST | 23°09′ N 116°38′ E | 2020.12 | 28 |
| Total | 233 |
| Population | S | h | Hd | K | Pi | PiJC |
|---|---|---|---|---|---|---|
| SS | 10 | 9 | 0.5282 | 0.9597 | 0.0019 | 0.0020 |
| LH | 5 | 4 | 0.2292 | 0.4167 | 0.0008 | 0.0009 |
| XS | 9 | 8 | 0.5494 | 1.2575 | 0.0026 | 0.0026 |
| TZ | 8 | 6 | 0.6092 | 1.5816 | 0.0032 | 0.0032 |
| FD | 8 | 7 | 0.5231 | 0.9631 | 0.0020 | 0.0020 |
| XP | 8 | 8 | 0.7287 | 1.8046 | 0.0037 | 0.0037 |
| XM | 13 | 11 | 0.7536 | 1.4420 | 0.0029 | 0.0029 |
| ST | 13 | 12 | 0.7487 | 1.4418 | 0.0029 | 0.0029 |
| Total | 34 | 34 | 0.5838 | 1.2334 | 0.0025 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among populations | 7 | 8.593 | 0.02127 Va | 3.38 |
| Within populations | 225 | 136.961 | 0.60872 Vb | 96.62 |
| Total | 232 | 145.554 | 0.62999 |
| Statistics | SS | LH | XS | TZ | FD | XP | XM | ST | Mean | s.d. |
|---|---|---|---|---|---|---|---|---|---|---|
| Tajima’s D | −1.92 | −1.76 | −1.39 | −0.66 | −1.70 | −0.32 | −2.03 | −1.90 | −1.46 | 0.63 |
| Tajima’s D p-value | 0.01 | 0.01 | 0.07 | 0.28 | 0.03 | 0.42 | 0.01 | 0.01 | 0.10 | 0.16 |
| FS | −5.40 | −1.55 | −3.00 | −0.30 | −3.14 | −1.65 | −7.16 | −8.07 | −3.78 | 2.80 |
| FS p-value | 0.00 | 0.06 | 0.03 | 0.46 | 0.01 | 0.19 | 0.00 | 0.00 | 0.09 | 0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Li, Z.; Li, J.; Xu, K.; Ye, Y. Analysis of Genetic Diversity and Structure of Eight Populations of Nerita yoldii along the Coast of China Based on Mitochondrial COI Gene. Animals 2024, 14, 718. https://doi.org/10.3390/ani14050718
Jiang S, Li Z, Li J, Xu K, Ye Y. Analysis of Genetic Diversity and Structure of Eight Populations of Nerita yoldii along the Coast of China Based on Mitochondrial COI Gene. Animals. 2024; 14(5):718. https://doi.org/10.3390/ani14050718
Chicago/Turabian StyleJiang, Senping, Zhenhua Li, Jiji Li, Kaida Xu, and Yingying Ye. 2024. "Analysis of Genetic Diversity and Structure of Eight Populations of Nerita yoldii along the Coast of China Based on Mitochondrial COI Gene" Animals 14, no. 5: 718. https://doi.org/10.3390/ani14050718
APA StyleJiang, S., Li, Z., Li, J., Xu, K., & Ye, Y. (2024). Analysis of Genetic Diversity and Structure of Eight Populations of Nerita yoldii along the Coast of China Based on Mitochondrial COI Gene. Animals, 14(5), 718. https://doi.org/10.3390/ani14050718






