Improving the Efficiency of CRISPR Ribonucleoprotein-Mediated Precise Gene Editing by Small Molecules in Porcine Fibroblasts
Abstract
:Simple Summary
Abstract
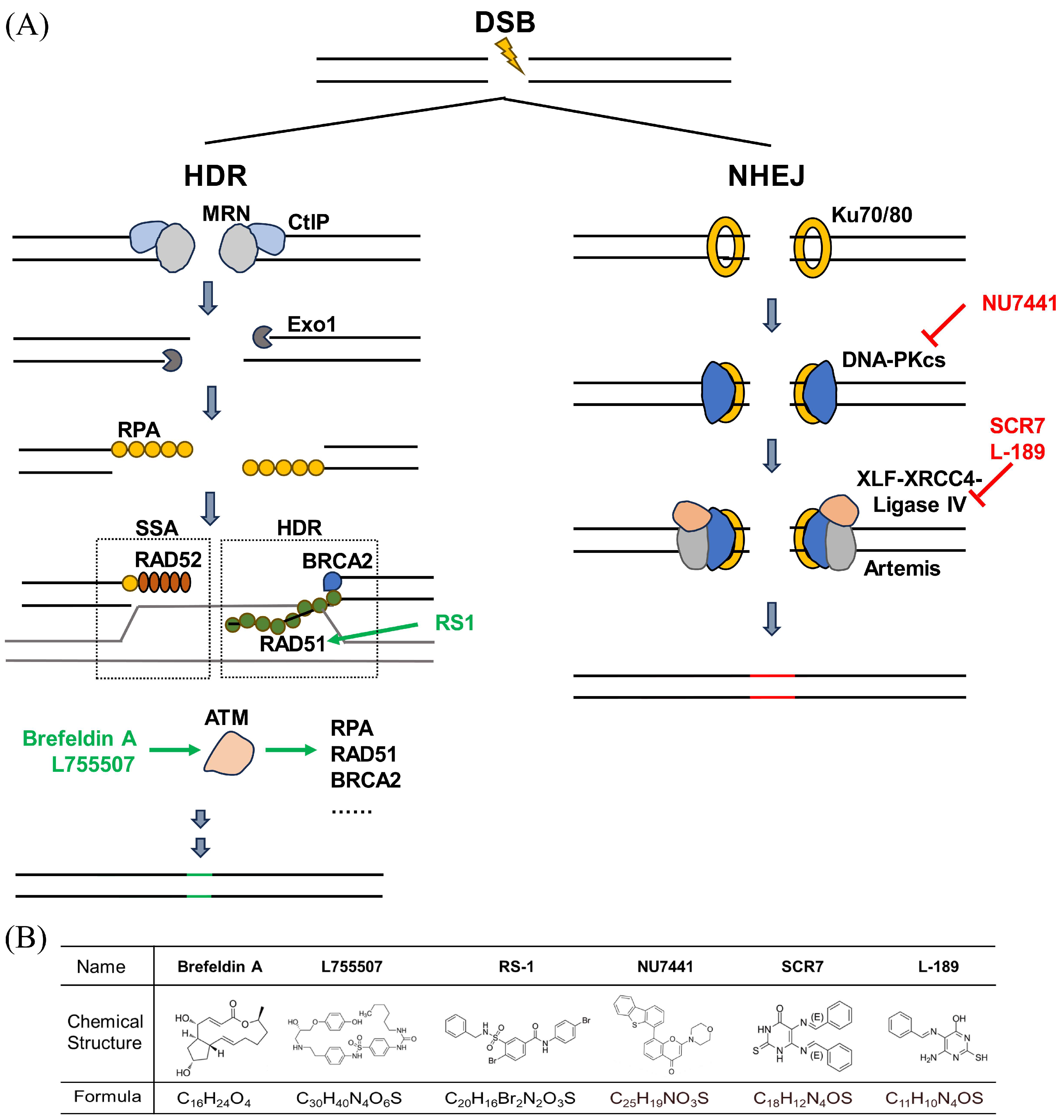
1. Introduction
2. Materials and Methods
2.1. Cell Culture
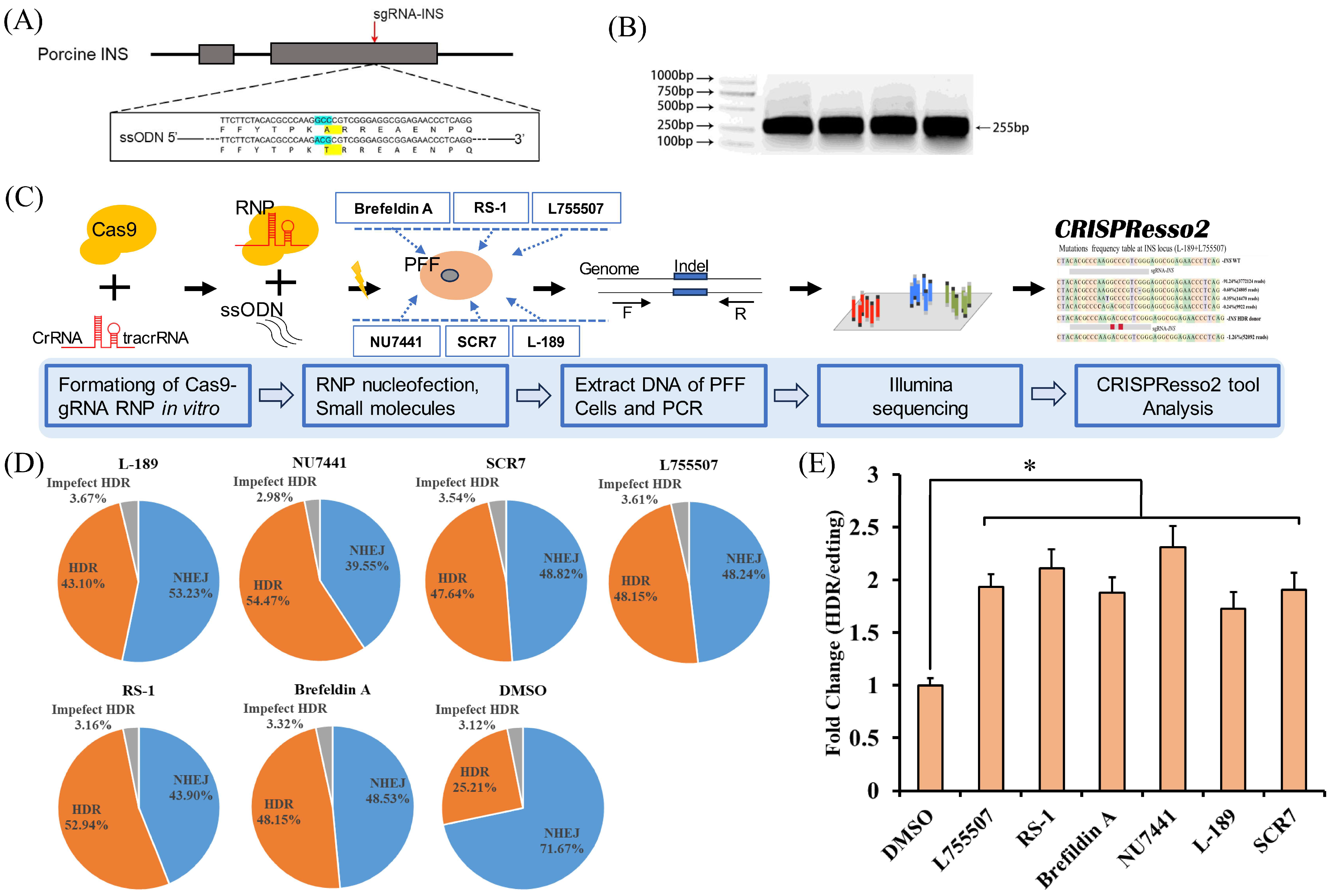
2.2. Design of gRNAs and ssODNs
2.3. Oligonucleotide and Ribonucleoprotein Electroporation
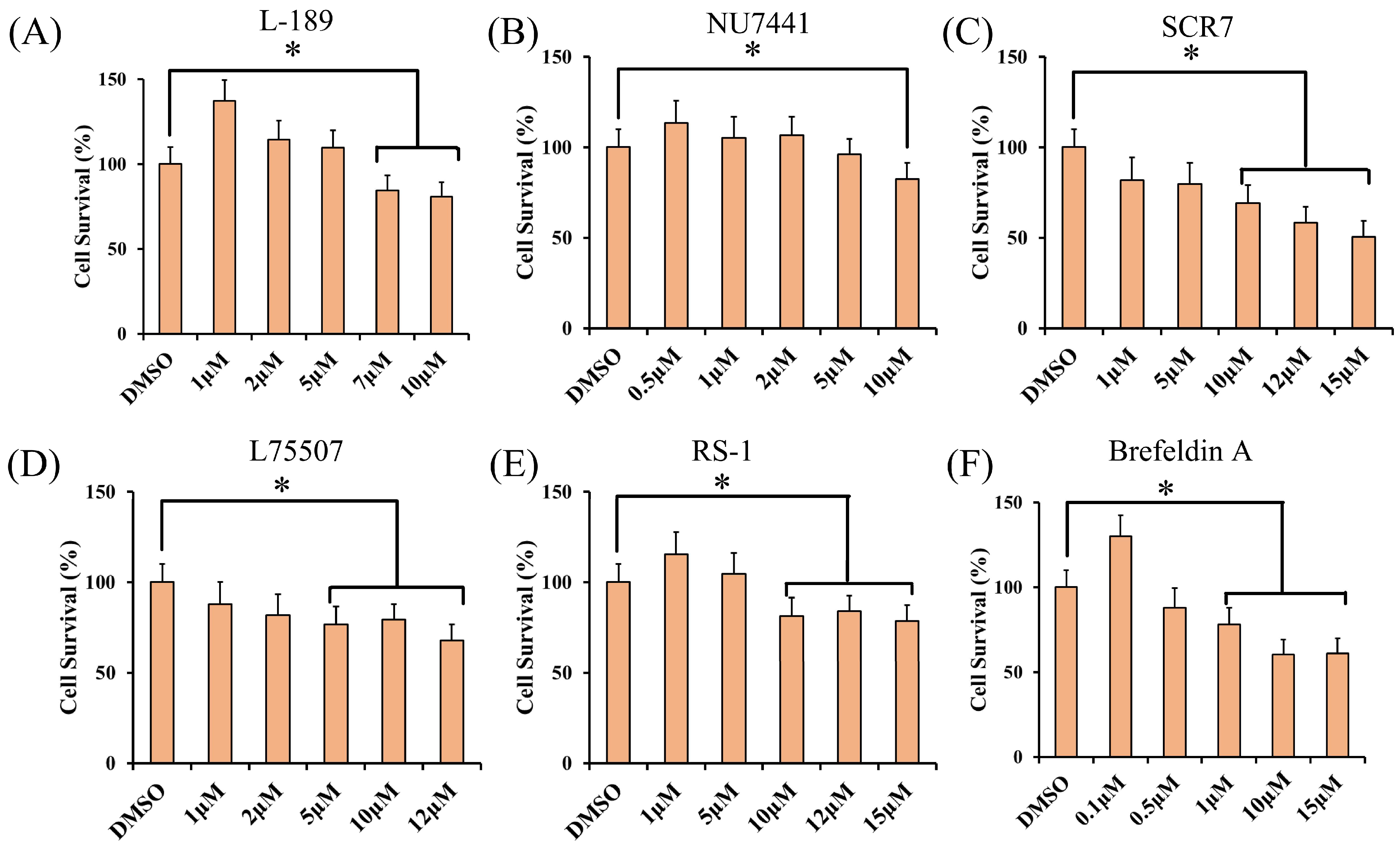
2.4. Cell Viability Assay
2.5. DNA Extraction and PCR
2.6. Deep Sequencing (Deep-Seq) and Analysis
2.7. Statistical Analysis
3. Results
3.1. The Effect of Different Concentrations of Small Molecules on Cell Viability
3.2. The Effect of Individual Small Molecules on the Efficiency of CRISPR RNP Mediated Precise Genome Editing
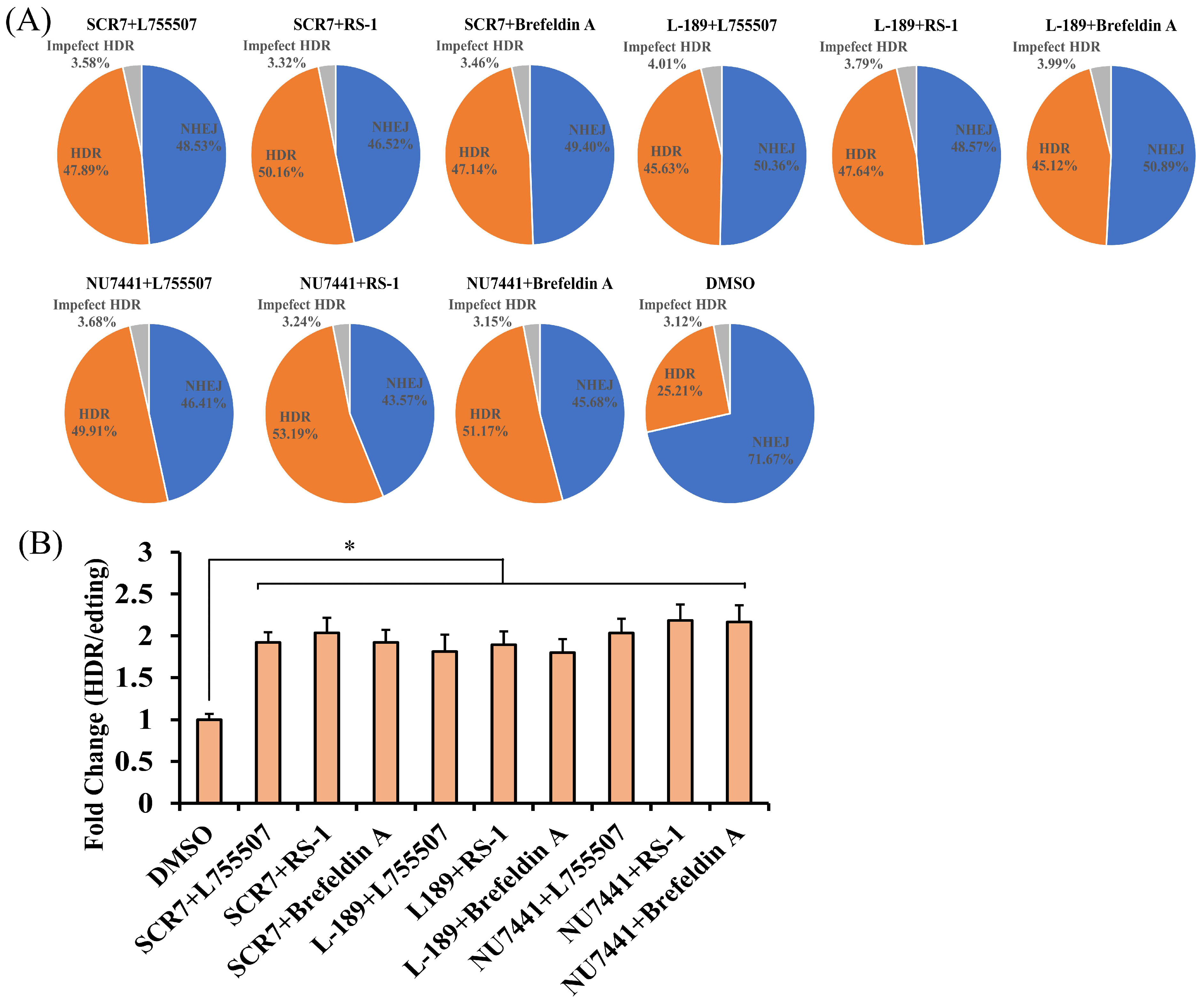
3.3. The Effect of Combining of Two Small Molecules on the Efficiency of CRISPR RNP Mediated Precise Genome Editing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, H.; Wu, Z. Genome Editing of Pigs for Agriculture and Biomedicine. Front. Genet. 2018, 9, 360. [Google Scholar] [CrossRef]
- Song, R.G.; Wang, Y.; Zheng, Q.T.; Yao, J.; Cao, C.W.; Wang, Y.F.; Zhao, J.G. One-step base editing in multiple genes by direct embryo injection for pig trait improvement. Sci. China-Life Sci. 2022, 65, 739–752. [Google Scholar] [CrossRef]
- Xu, K.; Zhou, Y.R.; Mu, Y.L.; Liu, Z.G.; Hou, S.H.; Xiong, Y.J.; Fang, L.R.; Ge, C.L.; Wei, Y.H.; Zhang, X.L.; et al. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance. eLife 2020, 9, e57132. [Google Scholar] [CrossRef]
- Xiang, G.H.; Ren, J.L.; Hai, T.; Fu, R.; Yu, D.W.; Wang, J.; Li, W.; Wang, H.Y.; Zhou, Q. Editing porcine regulatory element improved meat production in Chinese Bama pigs. Cell Mol. Life Sci. 2018, 75, 4619–4628. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Xue, C.Y.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021, 37, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Saikot, F.K.; Rahman, M.S.; Jamal, M.A.M.; Rahman, S.M.K.; Islam, S.M.R.; Kim, K.H. Programmable Molecular Scissors: Applications of a New Tool for Genome Editing in Biotech. Mol. Ther.-Nucleic Acids 2019, 14, 212–238. [Google Scholar] [CrossRef] [PubMed]
- Remy, S.; Chenouard, V.; Tesson, L.; Usal, C.; Menoret, S.; Brusselle, L.; Heslan, J.M.; Nguyen, T.H.; Bellien, J.; Merot, J.; et al. Generation of gene-edited rats by delivery of CRISPR/Cas9 protein and donor DNA into intact zygotes using electroporation. Sci. Rep. 2017, 7, 16554. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.Q.; Pruett-Miller, S.M.; Huang, Y.P.; Gjoka, M.; Duda, K.; Taunton, J.; Collingwood, T.N.; Frodin, M.; Davis, G.D. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 2011, 8, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Di Stazio, M.; Foschi, N.; Athanasakis, E.; Gasparini, P.; d’Adamo, A.P. Systematic analysis of factors that improve homologous direct repair (HDR) efficiency in CRISPR/Cas9 technique. PLoS ONE 2021, 16, e0247603. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.W.; Dai, X.Y.; Wang, W.T.; Yang, Z.X.; Zhao, J.J.; Zhang, J.P.; Wen, W.; Zhang, F.; Oberg, K.C.; Zhang, L.; et al. Dynamics and competition of CRISPR-Cas9 ribonucleoproteins and AAV donor-mediated NHEJ, MMEJ and HDR editing. Nucleic Acids Res. 2021, 49, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, D.N.; Haber, J.E. Single-strand template repair: Key insights to increase the efficiency of gene editing. Curr. Genet. 2021, 67, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.D.; Simoes-Pires, C.A.; Suzuki, M.; Greally, J.M. High-efficiency genomic editing in Epstein-Barr virus-transformed lymphoblastoid B cells using a single-stranded donor oligonucleotide strategy. Commun. Biol. 2019, 2, 312. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Mintier, G.; Ma-Edmonds, M.; Storton, D.; Wang, X.; Xiao, X.; Kienzle, B.; Zhao, D.; Feder, J.N. ‘Cold shock’ increases the frequency of homology directed repair gene editing in induced pluripotent stem cells. Sci. Rep. 2018, 8, 2080. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Amaishi, Y.; Maki, I.; Enoki, T.; Mineno, J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci. Rep. 2019, 9, 4811. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.S.; Thommandru, B.; Woodley, J.; Turk, R.; Yan, S.; Kurgan, G.; McNeill, M.S.; Rettig, G.R. Optimized design parameters for CRISPR Cas9 and Cas12a homology-directed repair. Sci. Rep. 2021, 11, 19482. [Google Scholar] [CrossRef] [PubMed]
- Shams, F.; Bayat, H.; Mohammadian, O.; Mahboudi, S.; Vahidnezhad, H.; Soosanabadi, M.; Rahimpour, A. Advance trends in targeting homology-directed repair for accurate gene editing: An inclusive review of small molecules and modified CRISPR-Cas9 systems. Bioimpacts 2022, 12, 371–391. [Google Scholar] [CrossRef]
- Hu, J.H.; Davis, K.M.; Liu, D.R. Chemical Biology Approaches to Genome Editing: Understanding, Controlling, and Delivering Programmable Nucleases. Cell Chem. Biol. 2016, 23, 57–73. [Google Scholar] [CrossRef]
- Chen, S.W.; Chen, D.; Liu, B.; Haisma, H.J. Modulating CRISPR/Cas9 genome-editing activity by small molecules. Drug Discov. Today 2022, 27, 951–966. [Google Scholar] [CrossRef]
- Ma, X.J.; Kong, L.H.; Zhu, S.Y. Reprogramming cell fates by small molecules. Protein Cell 2017, 8, 328–348. [Google Scholar] [CrossRef]
- Baumgard, L.H.; Hausman, G.J.; Sanz Fernandez, M.V. Insulin: Pancreatic secretion and adipocyte regulation. Domest. Anim. Endocrinol. 2016, 54, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Kleinwort, K.J.H.; Amann, B.; Hauck, S.M.; Hirmer, S.; Blutke, A.; Renner, S.; Uhl, P.B.; Lutterberg, K.; Sekundo, W.; Wolf, E.; et al. Retinopathy with central oedema in an INSC94Y transgenic pig model of long-term diabetes. Diabetologia 2017, 60, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Wang, J.; Liu, S.; Wang, J.; Xue, B.; Li, J.; Wei, R.; Zhao, Y.; Liu, Z. Positive correlation between the efficiency of induced pluripotent stem cells and the development rate of nuclear transfer embryos when the same porcine embryonic fibroblast lines are used as donor cells. Cell Reprogram 2014, 16, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019, 37, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Kruger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL bioactivity database: An update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Milanowska, K.; Krwawicz, J.; Papaj, G.; Kosinski, J.; Poleszak, K.; Lesiak, J.; Osinska, E.; Rother, K.; Bujnicki, J.M. REPAIRtoire—A database of DNA repair pathways. FEBS J. 2013, 280, 56. [Google Scholar] [CrossRef]
- Denes, C.E.; Cole, A.J.; Aksoy, Y.A.; Li, G.; Neely, G.G.; Hesselson, D. Approaches to Enhance Precise CRISPR/Cas9-Mediated Genome Editing. Int. J. Mol. Sci. 2021, 22, 8571. [Google Scholar] [CrossRef]
- Yang, D.; Scavuzzo, M.A.; Chmielowiec, J.; Sharp, R.; Bajic, A.; Borowiak, M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 2016, 6, 21264. [Google Scholar] [CrossRef]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks. Int. J. Mol. Sci. 2020, 21, 6461. [Google Scholar] [CrossRef]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 538–542. [Google Scholar] [CrossRef]
- Chen, X.; Zhong, S.J.; Zhu, Y.; Dziegielewska, B.; Ellenberger, T.; Wilson, G.M.; MacKerell, A.D.; Tomkinson, A.E. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res. 2008, 68, 3169–3177. [Google Scholar] [CrossRef]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef]
- Li, G.; Zhang, X.; Zhong, C.; Mo, J.; Quan, R.; Yang, J.; Liu, D.; Li, Z.; Yang, H.; Wu, Z. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci. Rep. 2017, 7, 8943. [Google Scholar] [CrossRef]
- Paek, S.M. Recent Synthesis and Discovery of Brefeldin A Analogs. Mar Drugs 2018, 16, 133. [Google Scholar] [CrossRef]
- Richardson, C.D.; Kazane, K.R.; Feng, S.J.; Zelin, E.; Bray, N.L.; Schafer, A.J.; Floor, S.N.; Corn, J.E. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 2018, 50, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, G.L.; D’Andrea, A.D. FANCD2 hurdles the DNA interstrand crosslink. Cell 2009, 139, 1222–1224. [Google Scholar] [CrossRef]
- Killian, T.; Dickopf, S.; Haas, A.K.; Kirstenpfad, C.; Mayer, K.; Brinkmann, U. Disruption of diphthamide synthesis genes and resulting toxin resistance as a robust technology for quantifying and optimizing CRISPR/Cas9-mediated gene editing. Sci. Rep. 2017, 7, 15480. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, Y.A.; Nguyen, D.T.; Chow, S.; Chung, R.S.; Guillemin, G.J.; Cole, N.J.; Hesselson, D. Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos. Commun. Biol. 2019, 2, 198. [Google Scholar] [CrossRef] [PubMed]
- Wienert, B.; Nguyen, D.N.; Guenther, A.; Feng, S.J.; Locke, M.N.; Wyman, S.K.; Shin, J.; Kazane, K.R.; Gregory, G.L.; Carter, M.A.M.; et al. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nat. Commun. 2020, 11, 2109. [Google Scholar] [CrossRef]
- Liu, B.; Chen, S.W.; La Rose, A.; Chen, D.; Cao, F.Y.; Zwinderman, M.; Kiemel, D.; Aissi, M.; Dekker, F.J.; Haisma, H.J. Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res. 2020, 48, 517–532. [Google Scholar] [CrossRef]
- Kagita, A.; Lung, M.S.Y.; Xu, H.G.; Kita, Y.; Sasakawa, N.; Iguchi, T.; Ono, M.; Wang, X.H.; Gee, P.; Hotta, A. Efficient ssODN-Mediated Targeting by Avoiding Cellular Inhibitory RNAs through Precomplexed CRISPR-Cas9/sgRNA Ribonucleoprotein. Stem Cell Rep. 2021, 16, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, M.P.; Krishnakumar, R.; Timlin, J.A.; Carney, J.P.; Butler, K.S. Gene editing and CRISPR in the clinic: Current and future perspectives. Biosci. Rep. 2020, 40, BSR20200127. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Lu, H.; Wang, H.; Li, S.; Truong, L.; Li, J.; Liu, S.; Xiang, R.; Wu, X. Break-induced replication plays a prominent role in long-range repeat-mediated deletion. EMBO J. 2019, 38, e101751. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chen, X.; Jin, Y.; Ge, W.; Wang, W.; Kong, L.; Ji, J.; Guo, X.; Huang, J.; Feng, X.H.; et al. Small molecules promote CRISPR-Cpf1-mediated genome editing in human pluripotent stem cells. Nat. Commun. 2018, 9, 1303. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Li, X.; Liu, C.; Jiang, C.; Guo, X.; Xu, Q.; Yin, Z.; Liu, Z.; Mu, Y. Improving the Efficiency of CRISPR Ribonucleoprotein-Mediated Precise Gene Editing by Small Molecules in Porcine Fibroblasts. Animals 2024, 14, 719. https://doi.org/10.3390/ani14050719
Zhao Y, Li X, Liu C, Jiang C, Guo X, Xu Q, Yin Z, Liu Z, Mu Y. Improving the Efficiency of CRISPR Ribonucleoprotein-Mediated Precise Gene Editing by Small Molecules in Porcine Fibroblasts. Animals. 2024; 14(5):719. https://doi.org/10.3390/ani14050719
Chicago/Turabian StyleZhao, Yunjing, Xinyu Li, Chang Liu, Chaoqian Jiang, Xiaochen Guo, Qianqian Xu, Zhi Yin, Zhonghua Liu, and Yanshuang Mu. 2024. "Improving the Efficiency of CRISPR Ribonucleoprotein-Mediated Precise Gene Editing by Small Molecules in Porcine Fibroblasts" Animals 14, no. 5: 719. https://doi.org/10.3390/ani14050719
APA StyleZhao, Y., Li, X., Liu, C., Jiang, C., Guo, X., Xu, Q., Yin, Z., Liu, Z., & Mu, Y. (2024). Improving the Efficiency of CRISPR Ribonucleoprotein-Mediated Precise Gene Editing by Small Molecules in Porcine Fibroblasts. Animals, 14(5), 719. https://doi.org/10.3390/ani14050719




