Possible Spreading of SARS-CoV-2 from Humans to Captive Non-Human Primates in the Peruvian Amazon
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Collection
2.2. Viral Enrichment and RNA Extraction
2.3. Complementary DNA (cDNA) Synthesis and PCR for Coronavirus Detection
2.4. Metagenomic Sequencing and Bioinformatic Analysis
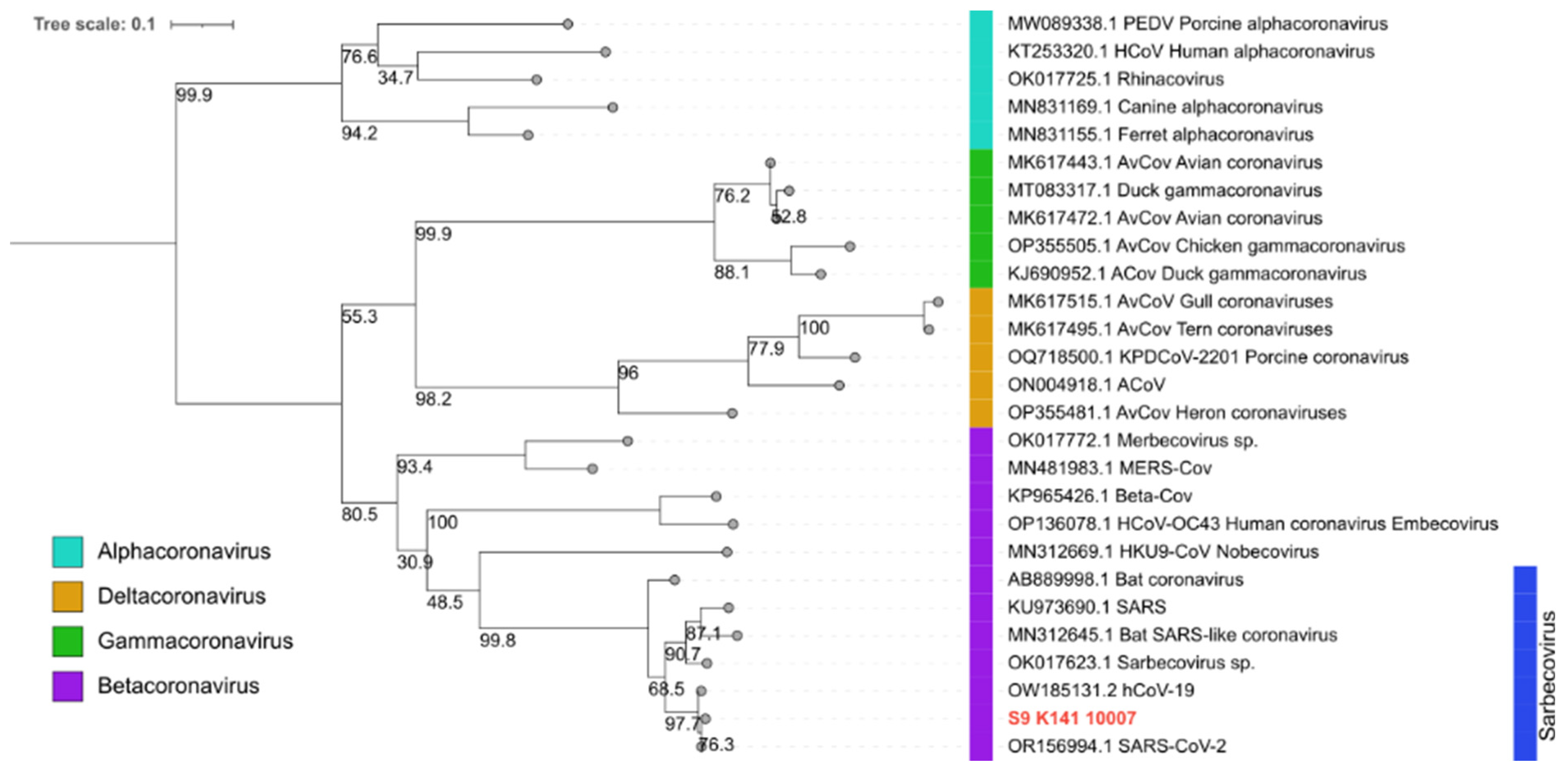
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Li, J.; Lai, S.; Gao, G.F.; Shi, W. The Emergence, Genomic Diversity and Global Spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef]
- Juscamayta-López, E.; Carhuaricra, D.; Tarazona, D.; Valdivia, F.; Rojas, N.; Maturrano, L.; Gavilán, R. Phylogenomics Reveals Multiple Introductions and Early Spread of SARS-CoV-2 into Peru. J. Med. Virol. 2021, 93, 5961–5968. [Google Scholar] [CrossRef]
- Ministry of Health of Peru Sala Situacional COVID-19 Peru. Available online: https://www.dge.gob.pe/covid19.html (accessed on 31 July 2023).
- Pacheco Torres, V.R.; Diaz, S.; Graham Angeles, L.A.; Flores-Quispe, M.; Calizaya-Mamani, G.; Ruelas, D.; Sánchez-Vendizú, P. Lista Actualizada de La Diversidad de Los Mamíferos Del Perú y Una Propuesta Para Su Actualización. Rev. Peru. Biol. 2021, 28, e21019. [Google Scholar] [CrossRef]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of Exposure to SARS-CoV-2 in Cats and Dogs from Households in Italy. Nat. Commun. 2020, 11, 6231. [Google Scholar] [CrossRef]
- Meisner, J.; Baszler, T.V.; Kuehl, K.E.; Ramirez, V.; Baines, A.; Frisbie, L.A.; Lofgren, E.T.; De Avila, D.M.; Wolking, R.M.; Bradway, D.S.; et al. Household Transmission of SARS-CoV-2 from Humans to Pets, Washington and Idaho, USA. Emerg. Infect. Dis. 2022, 28. [Google Scholar] [CrossRef]
- Račnik, J.; Kočevar, A.; Slavec, B.; Korva, M.; Rus, K.R.; Zakotnik, S.; Zorec, T.M.; Poljak, M.; Matko, M.; Rojs, O.Z.; et al. Transmission of SARS-CoV-2 from Human to Domestic Ferret. Emerg. Infect. Dis. 2021, 27, 2450–2453. [Google Scholar] [CrossRef] [PubMed]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2020, 11, e02220-20. [Google Scholar] [CrossRef] [PubMed]
- Aguiló-Gisbert, J.; Padilla-Blanco, M.; Lizana, V.; Maiques, E.; Muñoz-Baquero, M.; Chillida-Martínez, E.; Cardells, J.; Rubio-Guerri, C. First Description of SARS-CoV-2 Infection in Two Feral American Mink (Neovison vison) Caught in the Wild. Animals 2021, 11, 1422. [Google Scholar] [CrossRef]
- Padilla-Blanco, M.; Aguiló-Gisbert, J.; Rubio, V.; Lizana, V.; Chillida-Martínez, E.; Cardells, J.; Maiques, E.; Rubio-Guerri, C. The Finding of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) in a Wild Eurasian River Otter (Lutra lutra) Highlights the Need for Viral Surveillance in Wild Mustelids. Front. Vet. Sci. 2022, 9, 826991. [Google Scholar] [CrossRef]
- Nagy, A.; Stará, M.; Vodička, R.; Černíková, L.; Jiřincová, H.; Křivda, V.; Sedlák, K. Reverse-Zoonotic Transmission of SARS-CoV-2 Lineage Alpha (B.1.1.7) to Great Apes and Exotic Felids in a Zoo in the Czech Republic. Arch. Virol. 2022, 167, 1681–1685. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Sikkema, R.S.; Velkers, F.C.; Nieuwenhuijse, D.F.; Fischer, E.A.J.; Meijer, P.A.; Bouwmeester-Vincken, N.; Rietveld, A.; Wegdam-Blans, M.C.A.; Tolsma, P.; et al. Adaptation, Spread and Transmission of SARS-CoV-2 in Farmed Minks and Associated Humans in the Netherlands. Nat. Commun. 2021, 12, 6802. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 Infection in Free-Ranging White-Tailed Deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Italiya, J.; Vacek, V.; Matějů, P.; Dering, C.; Celina, S.S.; Ndiaye, A.; Černý, J. First Detection of SARS-CoV-2 in White Rhinoceros during a Small-Scale Coronavirus Surveillance in the Bandia Reserve, Senegal. Animals 2023, 13, 2593. [Google Scholar] [CrossRef]
- Tan, C.C.S.; Lam, S.D.; Richard, D.; Owen, C.J.; Berchtold, D.; Orengo, C.; Nair, M.S.; Kuchipudi, S.V.; Kapur, V.; Van Dorp, L.; et al. Transmission of SARS-CoV-2 from Humans to Animals and Potential Host Adaptation. Nat. Commun. 2022, 13, 2988. [Google Scholar] [CrossRef]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Salguero, F.J.; White, A.D.; Slack, G.S.; Fotheringham, S.A.; Bewley, K.R.; Gooch, K.E.; Longet, S.; Humphries, H.E.; Watson, R.J.; Hunter, L.; et al. Comparison of Rhesus and Cynomolgus Macaques as an Infection Model for COVID-19. Nat. Commun. 2021, 12, 1260. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.H.; Vasconcelos, A.L.; Silva, V.L.; Nogueira, B.S.; Silva, A.C.; Pacheco, R.C.; Souza, M.A.; Colodel, E.M.; Ubiali, D.G.; Biondo, A.W.; et al. Natural SARS-CoV-2 Infection in a Free-Ranging Black-Tailed Marmoset (Mico melanurus) from an Urban Area in Mid-West Brazil. J. Comp. Pathol. 2022, 194, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Qian, X.K.; Hong, M.H.; Zhang, J.L.; Li, D.Y.; Wang, T.H.; Yang, Z.M.; Zhang, L.Y.; Wang, Z.M.; Nie, H.J.; et al. Global View on Virus Infection in Non-Human Primates and Implications for Public Health and Wildlife Conservation. Zool. Res. 2021, 42, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Djikeng, A.; Halpin, R.; Kuzmickas, R.; Depasse, J.; Feldblyum, J.; Sengamalay, N.; Afonso, C.; Zhang, X.; Anderson, N.G.; Ghedin, E.; et al. Viral genome sequencing by random priming methods. BMC Genom. 2008, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.; Leung, C.Y.; Gilbert, M.; Joyner, P.H.; Ng, E.M.; Tse, T.M.; Guan, Y.; Peiris, J.S.; Poon, L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011, 85, 12815–12820. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- NCBI Resource Coordinators; Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Gaudreault, N.N.; Richt, J.A. Natural and Experimental SARS-CoV-2 Infection in Domestic and Wild Animals. Viruses 2021, 13, 1993. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Dahlen, A.; Hedlin, H.; Park, R.M.; Han, A.; Schmidtke, D.T.; Verma, R.; et al. Gastrointestinal Symptoms and Fecal Shedding of SARS-CoV-2 RNA Suggest Prolonged Gastrointestinal Infection. Clin. Adv. 2022, 3, 371–387.e9. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E.; et al. Wastewater Sequencing Reveals Early Cryptic SARS-CoV-2 Variant Transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, e02703-20. [Google Scholar] [CrossRef]
- Joshi, M.; Mohandas, S.; Prasad, S.; Shinde, M.; Chavan, N.; Yadav, P.D.; Lavania, M. Lack of Evidence of Viability and Infectivity of SARS-CoV-2 in the Fecal Specimens of COVID-19 Patients. Front. Public Health 2022, 10, 1030249. [Google Scholar] [CrossRef]
- Cerrada-Romero, C.; Berastegui-Cabrera, J.; Camacho-Martínez, P.; Goikoetxea-Aguirre, J.; Pérez-Palacios, P.; Santibáñez, S.; Blanco-Vidal, M.J.; Valiente, A.; Alba, J.; Rodríguez-Álvarez, R.; et al. Excretion and viability of SARS-CoV-2 in feces and its association with the clinical outcome of COVID-19. Sci. Rep. 2022, 12, 7397. [Google Scholar] [CrossRef]
- Abdalhamid, B.; Bilder, C.R.; McCutchen, E.L.; Hinrichs, S.H.; Koepsell, S.A.; Iwen, P.C. Assessment of Specimen Pooling to Conserve SARS CoV-2 Testing Resources. Am. J. Clin. Pathol. 2020, 153, 715–718. [Google Scholar] [CrossRef]
- Álvarez-Antonio, C.; Meza-Sánchez, G.; Calampa, C.; Casanova, W.; Carey, C.; Alava, F.; Rodríguez-Ferrucci, H.; Quispe, A.M. Seroprevalence of Anti-SARS-CoV-2 Antibodies in Iquitos, Peru in July and August, 2020: A Population-Based Study. Lancet Glob. Health 2021, 9, e925–e931. [Google Scholar] [CrossRef]
- Vargas-Herrera, N.; Araujo-Castillo, R.V.; Mestanza, O.; Galarza, M.; Rojas-Serrano, N.; Solari-Zerpa, L. SARS-CoV-2 Lambda and Gamma Variants Competition in Peru, a Country with High Seroprevalence. Lancet Reg. Health-Am. 2022, 6, 100112. [Google Scholar] [CrossRef]
- Frazzini, S.; Amadori, M.; Turin, L.; Riva, F. SARS CoV-2 Infections in Animals, Two Years into the Pandemic. Arch. Virol. 2022, 167, 2503–2517. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.-P.; Pfenning, A.R.; Zhao, H.; et al. Broad Host Range of SARS-CoV-2 Predicted by Comparative and Structural Analysis of ACE2 in Vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Melin, A.D.; Janiak, M.C.; Marrone, F.; Arora, P.S.; Higham, J.P. Comparative ACE2 Variation and Primate COVID-19 Risk. Commun. Biol. 2020, 3, 641. [Google Scholar] [CrossRef] [PubMed]
- De Abreu, F.V.S.; Macedo, M.V.; Da Silva, A.J.J.; De Oliveira, C.H.; De Ottone, V.O.; De Almeida, M.A.B.; Dos Santos, E.; Da Cardoso, J.C.; Campos, A.S.; Da Silva, C.M.D.; et al. No Evidence of SARS-CoV-2 Infection in Neotropical Primates Sampled During COVID-19 Pandemic in Minas Gerais and Rio Grande Do Sul, Brazil. EcoHealth 2021, 18, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Sacchetto, L.; Chaves, B.A.; Costa, E.R.; De Menezes Medeiros, A.S.; Gordo, M.; Araújo, D.B.; Oliveira, D.B.L.; Da Silva, A.P.B.; Negri, A.F.; Durigon, E.L.; et al. Lack of Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spillover in Free-Living Neotropical Non-Human Primates, Brazil. Viruses 2021, 13, 1933. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Bartrand, T.A.; Weir, M.H.; Omura, T.; Haas, C.N. Development of a dose-response model for SARS coronavirus. Risk Anal. 2010, 30, 1129–1138. [Google Scholar] [CrossRef]
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol. 2023, 21, 147–161. [Google Scholar] [CrossRef]
- Devaux, C.A.; Mediannikov, O.; Medkour, H.; Raoult, D. Infectious Disease Risk Across the Growing Human-Non-Human Primate Interface: A Review of the Evidence. Front. Public Health 2019, 7, 305. [Google Scholar] [CrossRef]
- Esposito, M.M.; Turku, S.; Lehrfield, L.; Shoman, A. The Impact of Human Activities on Zoonotic Infection Transmissions. Animals 2023, 13, 1646. [Google Scholar] [CrossRef]
- Delahay, R.J.; De La Fuente, J.; Smith, G.C.; Sharun, K.; Snary, E.L.; Flores Girón, L.; Nziza, J.; Fooks, A.R.; Brookes, S.M.; Lean, F.Z.X.; et al. Assessing the Risks of SARS-CoV-2 in Wildlife. One Health Outlook 2021, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Gryseels, S.; De Bruyn, L.; Gyselings, R.; Calvignac-Spencer, S.; Leendertz, F.H.; Leirs, H. Risk of Human-to-wildlife Transmission of SARS-CoV-2. Mammal Rev. 2021, 51, 272–292. [Google Scholar] [CrossRef] [PubMed]
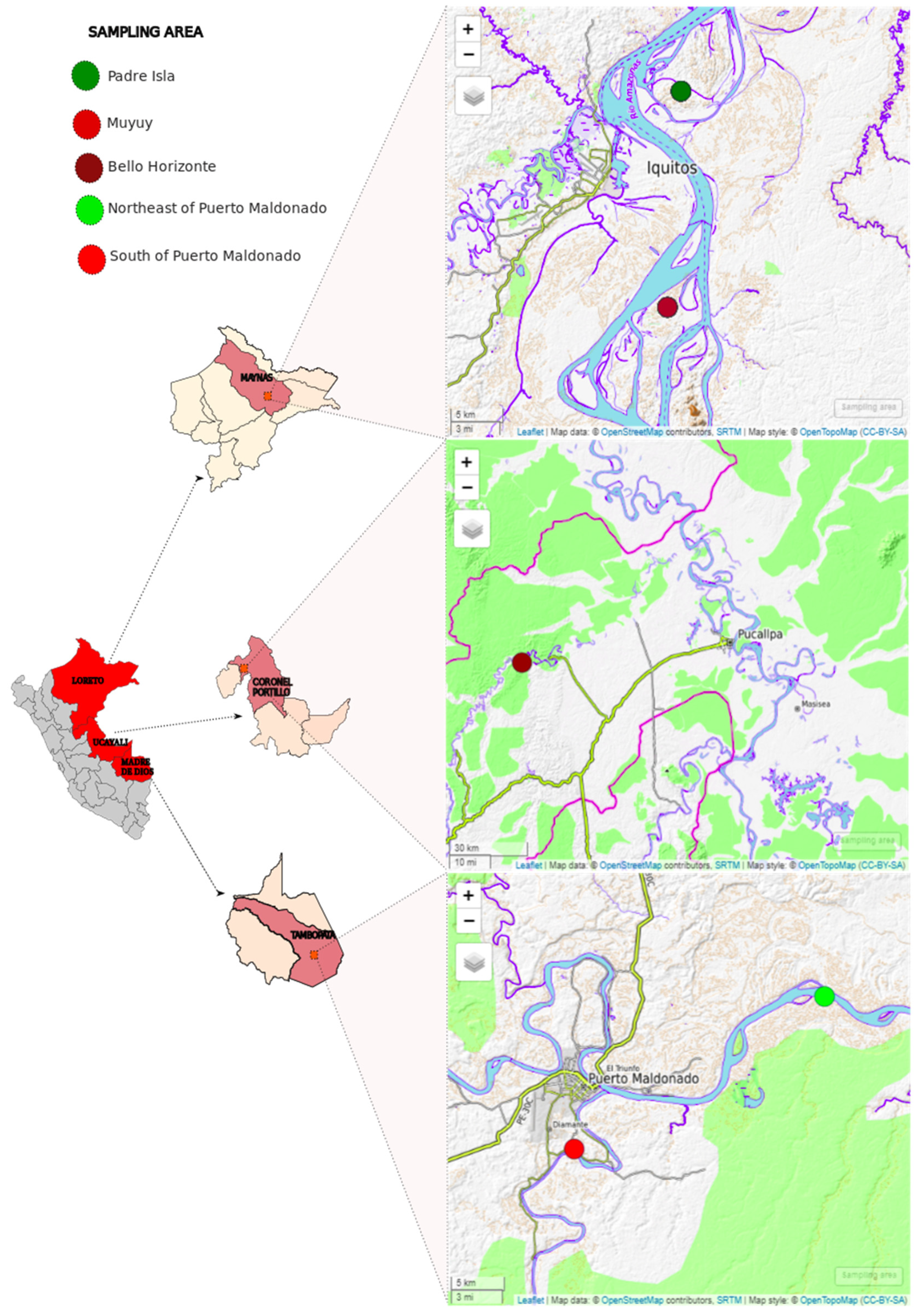
| Region | Sampling Area | NHP Species | Pooled Samples | # Pool |
|---|---|---|---|---|
| Madre de Dios | South of Puerto Maldonado | Red howler monkey (Alouatta seniculus) | 7 | Pool 1 |
| Madre de Dios | Northeast of Puerto Maldonado | Black spider monkey (Ateles chamek) | 7 | Pool 2 |
| Madre de Dios | Northeast of Puerto Maldonado | Red howler monkey (Alouatta seniculus) | 5 | Pool 3 |
| Madre de Dios | South of Puerto Maldonado | White fronted capuchin (Cebus unicolor) | 7 | Pool 4 |
| Ucayali | Bello Horizonte | Black spider monkey (Ateles chamek) | 11 | Pool 5 |
| Loreto | Muyuy | White lipped tamarin (Saguinus labiatus) | 9 | Pool 6 |
| Loreto | Padre Isla | Moustached tamarin (Saguinus mystax) | 8 | Pool 7 |
| Madre de Dios | Northeast of Puerto Maldonado | Black spider monkey (Ateles chamek) | 7 | Pool 8 |
| Ucayali | Bello Horizonte | White fronted capuchin (Cebus unicolor) | 9 | Pool 9 |
| Madre de Dios | South of Puerto Maldonado | Red howler monkey (Alouatta seniculus) | 6 | Pool 10 |
| Total samples | 76 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavera Gonzales, A.; Bazalar Gonzales, J.; Silvestre Espejo, T.; Leiva Galarza, M.; Rodríguez Cueva, C.; Carhuaricra Huamán, D.; Luna Espinoza, L.; Maturrano Hernández, A. Possible Spreading of SARS-CoV-2 from Humans to Captive Non-Human Primates in the Peruvian Amazon. Animals 2024, 14, 732. https://doi.org/10.3390/ani14050732
Tavera Gonzales A, Bazalar Gonzales J, Silvestre Espejo T, Leiva Galarza M, Rodríguez Cueva C, Carhuaricra Huamán D, Luna Espinoza L, Maturrano Hernández A. Possible Spreading of SARS-CoV-2 from Humans to Captive Non-Human Primates in the Peruvian Amazon. Animals. 2024; 14(5):732. https://doi.org/10.3390/ani14050732
Chicago/Turabian StyleTavera Gonzales, Andrea, Jhonathan Bazalar Gonzales, Thalía Silvestre Espejo, Milagros Leiva Galarza, Carmen Rodríguez Cueva, Dennis Carhuaricra Huamán, Luis Luna Espinoza, and Abelardo Maturrano Hernández. 2024. "Possible Spreading of SARS-CoV-2 from Humans to Captive Non-Human Primates in the Peruvian Amazon" Animals 14, no. 5: 732. https://doi.org/10.3390/ani14050732








