Simple Summary
Due to adaptations to a completely subterranean lifestyle, blind mole rats have become an interesting research model for longevity and cancer resistance. The lesser blind mole rat from the genus Nannospalax is additionally characterized by extensive chromosomal changes, with 74 chromosomal forms described. As a result of these changes and of their morphological similarity, their taxonomy is unclear; consequently, many unrecognized species are endangered due to habitat fragmentation and reduction. Their official conservation status is still undefined due to insufficient data. Among the 25 chromosomal forms of N. leucodon in Europe, five have been identified as completely reproductively isolated and genetically divergent cryptic species in Serbia. The most endangered, N. l. syrmiensis, which was described 50 years ago as a group of populations endemic to Serbia, has been declared extinct in the literature. Using nucleotide comparison of two mitochondrial gene segments between old, archived and recently sampled material, we provide evidence that N. l. syrmiensis is not extinct. However, it has disappeared in a large part of its former range, mainly due to urbanization, invasive agriculture, and treatment as a pest. In order to preserve biodiversity, detailed monitoring, population-structure studies, risk assessment and appropriate conservation measures are required.
Abstract
Blind mole rats (genus Nannospalax) attract a great deal of attention because of their cancer resistance and longevity. Due to the high rate of chromosome rearrangements, 74 Nannospalax chromosomal forms have been discovered. The convergence of their external morphology complicates their taxonomy, and many cryptic species remain unrecognized. Thus, the European N. leucodon supersp. is listed in the IUCN Red List of Threatened Species with “Data Deficient” status. It is crucial for the conservation of biodiversity to clarify its taxonomy, to recognize each cryptic species, and assign to them the correct conservation status. Of the more than 20 chromosomal forms described within N. leucodon, five cryptic species occur in Serbia. The most threatened among them—N. l. syrmiensis, described and named 50 years ago in the regions of Srem, Belgrade and Mačva—has been declared extinct in the literature, which may have negative consequences for the conservation of wildlife genetic diversity. Through five years of fieldwork and comparison of 16SrRNA and MT-CYTB gene segments between old, archived teeth and recently collected material, we show that N. l. syrmiensis is not extinct. However, its habitat has been fragmented and reduced, owing primarily to anthropogenic impact. Therefore, detailed surveillance, population-structure studies, risk assessment, and appropriate conservation measures are needed.
1. Introduction
A clade of blind mole rats (BMRs) Spalacidae, Rodentia, draws constant attention in several scientific fields due to their numerous adaptations to a subterranean environment. They are small, solitary, exclusively subterranean, and highly specialized mammals with an extreme tolerance to hypoxia and hypercapnia, specific blood characteristics, high vessel density, and other physiological and morphological modifications. With a lifespan of about 20 years and a unique resistance to cancer [1,2,3,4,5], BMRs are the only long-lived and cancer-proof rodents in Europe.
These vastly unusual rodents are represented by two genera: the greater BMR, Spalax (Guldenstaedt 1770), and the lesser BMR, Nannospalax (Palmer 1903). The latter represents an excellent research model for chromosomal speciation studies, as the genus Nannospalax includes 74 groups with fixed chromosomal differences [6,7,8] with a wide range of diploid chromosomal number (2n), from 36 to 60. These chromosomal forms (CFs) are alternatively labelled as cytotypes, races, or sibling/good biological/cryptic species [8,9,10]. Another complicating circumstance in species delimitation in this genus is the species’ convergent morphology—reduced interspecific phenotypic variability that most likely developed as a result of stabilizing selection modeled by extreme environmental settings [11]. Biometric and/or morphological studies of skulls, jaws and teeth are a valuable source for distinguishing living mammal species in general, while morphological data, especially fossil teeth, can provide valuable insights into the evolutionary history and paleobiology of mammals. However, they are not reliable for species identification in morphologically convergent taxa such as the cryptic species of the lesser BMR. Therefore, the number of recognized species in the genus Nannospalax ranges from only one [12] to 14 [11]. The International Union for Conservation of Nature (IUCN) Red List of Threatened Species recognizes three morphospecies/superspecies, out of which the European species is classified as Data Deficient (DD) [13]: (1) the lesser BMR N. leucodon (Nordmann 1840) from parts of central and South-Eastern Europe (Figure 1a); the other two, classified as Least Concern (LC), are (2) the Anatolian BMR N. xanthodon (Nordmann 1845) (synonym: N. nehringi (Satunin 1898)), which occurs in Transcaucasia, most of Turkish Anatolia and certain East Aegean islands; and (3) the Palestine BMR N. ehrenbergi (Nehring 1898), which occurs in southeastern Anatolia in Turkey, Iraq, Syria, Lebanon, Israel, Jordan, and Egypt. Each of these three superspecies comprises more than 20 CFs, with several reproductively isolated and genetically well-differentiated cryptic species [8,14,15,16]. Some of them are already seriously endangered due to the loss and fragmentation of their natural habitats [6,10,17]. In most countries, there is no targeted species protection plan because of their uncertain conservation status, which results from insufficiently clarified taxonomy and cryptic speciation.
The lesser blind mole rat, Nannospalax leucodon (Nordmann 1840) superspecies, is endemic to Europe, with 25 CFs reported so far [6,8]. They are known as typical inhabitants of the steppe-like grasslands, mountain steppes, and sand steppes, and they avoid marshy areas and quicksand [18]. Breeding experiments that revealed the complete reproductive isolation of seven N. leucodon CFs [8,11,19], substantiated by the results of molecular phylogenetic investigations [14,15,16,20,21], strongly suggest their cryptic-species status. The extent of their evolutionary divergence resembles that recorded among clearly distinguishable Spalax species [14,21].
All five cryptic species of N. leucodon distributed in Serbia (Figure 1b), N. l. hungaricus (Nehring 1898); N. l. serbicus (Méhely 1909); N. l. montanoserbicus (Savić and Soldatović 1974); N. l. syrmiensis (Méhely 1909) and N. l. montanosyrmiensis (Savić and Soldatović 1974), which were documented 50–60 years ago [8], are reproductively completely isolated and meet the criteria to be classified as distinct species. Although N. leucodon has strictly protected status in Serbia, as proposed by Vasić et al. [22], the natural habitats of Serbian N. leucodon cryptic species have been increasingly reduced, mostly due to anthropogenic influences, i.e., extensive urbanization and invasive agriculture. Despite their national protected status, they are also treated as pests and killed. In a previous publication [21], we drew attention to two cryptic species with highly reduced and fragmented ranges, N. l. syrmiensis and N. l. montanosyrmiensis, categorized them as Potentially Endangered/Critically Endangered, and emphasized the need for detailed monitoring and population surveys.
Here, we report on our study of N. l. syrmiensis. During extensive cytogenetic mapping of BMRs in the 1970s, Soldatović [19,23] and Savić and Soldatović [24] described the BMR from the Srem district as a distinct CF, i.e., a group of seven populations with the diploid chromosome number 2n = 54 and the number of chromosomal arms NF = 90, and named it Spalax leucodon syrmiensis (Méhely 1909). Later, they accepted nomenclature with two genera and proposed a separate species with the name Nannospalax syrmiensis (Méhely 1909) on the basis of comparative morphometric, craniometric and karyotypic studies, evidence of reproductive isolation, and the character of surrounding ecogeographic formations [11]. It was widespread in the districts of Srem (locus typicus—Stara Pazova) and Mačva, as well as in the Belgrade region [11] (Figure 1c). After the investigations summarized in Savić and Soldatović [11], there have been no published data regarding distribution of N. l. syrmiensis since 1988 [25]. However, N. l. syrmiensis was declared probably extinct [26], and this statement was repeated [10,15]. Almost 35 years after the last data were published, Bugarski-Stanojević et al. [21] reported newly captured N. l. syrmiensis specimens in Serbia. Despite these results, in the latest comprehensive study, which included 22 N. leucodon CFs, Németh et al. [16] referred to N. l. syrmiensis as a missing taxon and “incertae sedis”.
For the conservation of biodiversity, including the genetic diversity of wildlife, declaring a particular cryptic species as probably extinct may have negative consequences. If N. l. syrmiensis is absent from the literature, or from part of its range, this does not mean that it is extinct. In this study, we conducted systematic fieldwork over several years/seasons, covering the entire known range of N. l. syrmiensis. For species confirmation, we compared the nucleotide sequences of two mitochondrial gene segments (16SrRNA and MT-CYTB) between newly captured animals and old archived tooth samples previously identified as N. l. syrmiensis by karyotyping. The tooth samples (collection Ivo Savić; Institute for Biological Research “Siniša Stanković”—National Institute of Republic of Serbia, University of Belgrade—IBISS) came from the locus typicus—Stara Pazova and several other localities in the Belgrade region. Another four cryptic N. leucodon species distributed in Serbia are newly sampled and included in this study together with tooth samples to avoid misinterpretation of species identity and account for the possible presence of hybrids. MtDNA genes were selected instead of nuclear genes to increase the success rate of sequencing from old archived material. The mentioned methods have already proven to be excellent for the identification of BMR cryptic species [14,15,16,21]. The 16SrRNA gene is regularly used to identify well-differentiated species, genera and distantly related taxa [27,28,29]. Polymorphism of the MT-CYTB gene has been widely used to identify species and phylogenetic relationships in BMRs [30,31,32]. In addition to proving that N. l. syrmiensis still occurs in Serbia, we determined its greatly reduced distribution range.
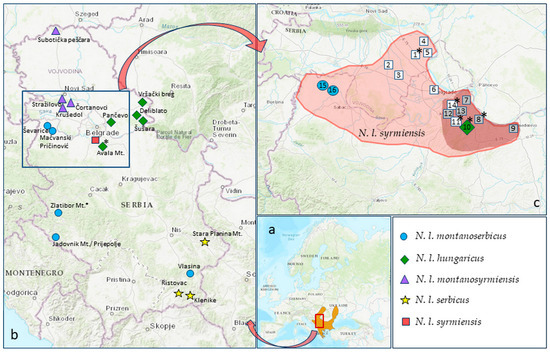
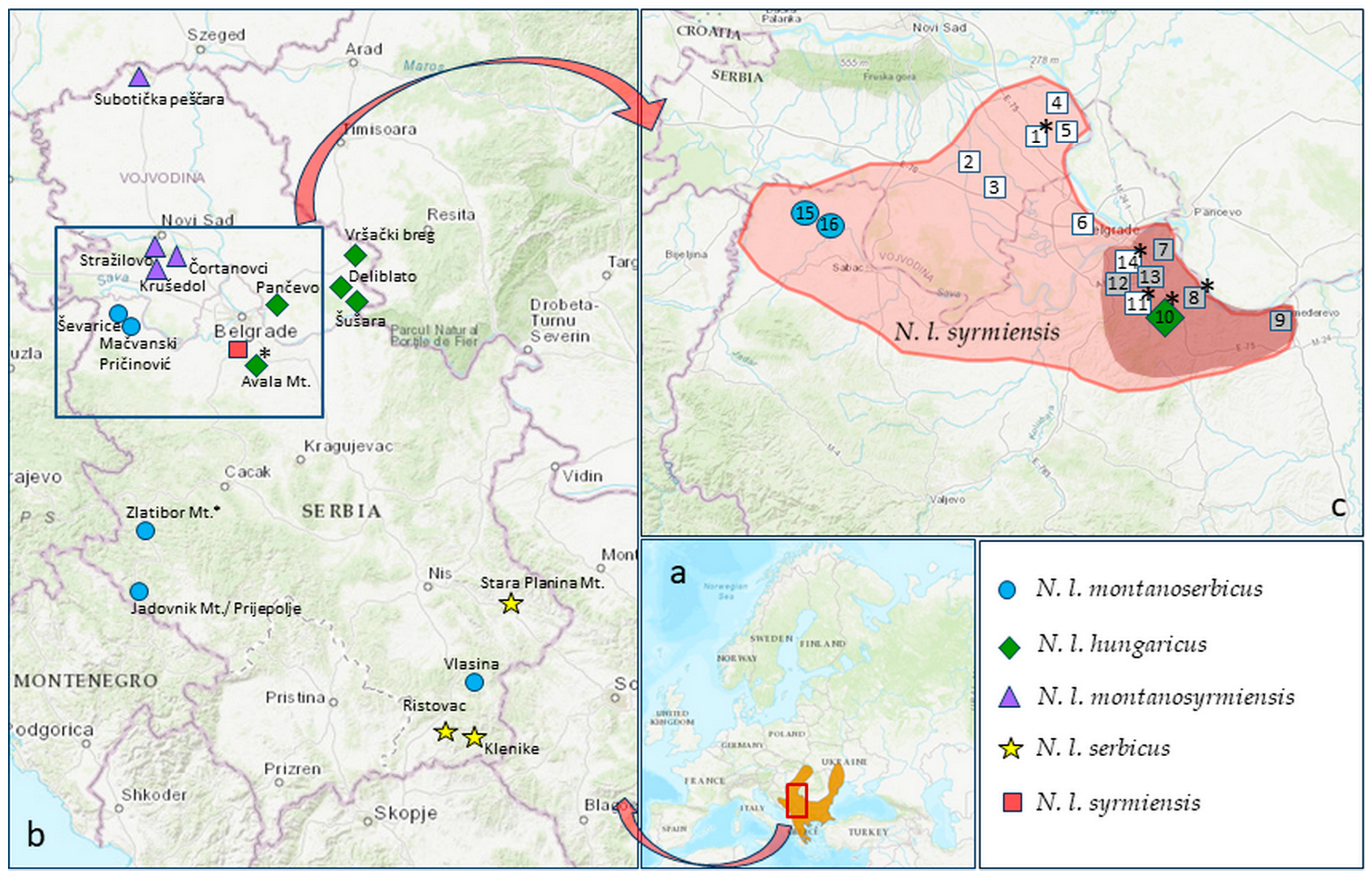
Figure 1.
Geographical distribution of five N. leucodon cryptic species from Serbia. (a) Distribution area of N. leucodon supersp. (b) Capturing localities in Serbia, including our previously published data (listed in Table S1): N. l. syrmiensis—square; N. l. montanosyrmiensis—triangle; N. l. hungaricus—rhomb; N. l. montanoserbicus—circle; N. l. serbicus—star. (c) Distributional area of N. l. syrmiensis according to [11,19]—light red; distribution according to our research—dark red. Localities: 1—Stara Pazova, 2—Donji Petrovac, 3—Krnješevci, 4—Slankamen, 5—Stari Banovci, 6—Zemun Polje, 7—Višnjica, 8—Vinča, 9—Udovice (Smederevo), 10—Avala, 11—Jajinci, 12—Banjica, 13—Košutnjak, 14—Banovo Brdo, 15—Mačvanski Pričinović (Bogatić), 16—Ševarice (Bogatić); empty squares—BMR not present; filled squares—fresh samples; *—old, archived samples. Source of the map: IUCN (International Union for Conservation of Nature) 2011. Nannospalax leucodon. The IUCN Red List of Threatened Species. Version 2023-1. Accessed on 8 February 2024.
Figure 1.
Geographical distribution of five N. leucodon cryptic species from Serbia. (a) Distribution area of N. leucodon supersp. (b) Capturing localities in Serbia, including our previously published data (listed in Table S1): N. l. syrmiensis—square; N. l. montanosyrmiensis—triangle; N. l. hungaricus—rhomb; N. l. montanoserbicus—circle; N. l. serbicus—star. (c) Distributional area of N. l. syrmiensis according to [11,19]—light red; distribution according to our research—dark red. Localities: 1—Stara Pazova, 2—Donji Petrovac, 3—Krnješevci, 4—Slankamen, 5—Stari Banovci, 6—Zemun Polje, 7—Višnjica, 8—Vinča, 9—Udovice (Smederevo), 10—Avala, 11—Jajinci, 12—Banjica, 13—Košutnjak, 14—Banovo Brdo, 15—Mačvanski Pričinović (Bogatić), 16—Ševarice (Bogatić); empty squares—BMR not present; filled squares—fresh samples; *—old, archived samples. Source of the map: IUCN (International Union for Conservation of Nature) 2011. Nannospalax leucodon. The IUCN Red List of Threatened Species. Version 2023-1. Accessed on 8 February 2024.
2. Materials and Methods
Field work—Localities previously reported for the occurrence of N. l. syrmiensis [11], as well as a wide surrounding area, were surveyed from 2019–2023. During the same period, another four cryptic N. leucodon species distributed in Serbia were sampled in eight localities, so that a total of 18 animals were newly collected and sampled from their natural habitat for this study (Figure 1b, Table S1). Our fieldwork included the following: (1) detection of active burrows, which involved noting their distribution/shape; a detailed inspection to distinguish BMRs from moles, e.g., examining the structure of underground tunnel walls and measuring of their diameters, which should be about 5–7 cm [33]; and searching for imprinted animal tracks in the soil and the presence of food stores; (2) capture of live animals by opening their tunnel systems; (3) collection of specimens accidentally killed during capture or by local residents during agricultural activities; and (4) conduction of interviews of citizens. All animal-sampling protocols complied with the current laws of the Republic of Serbia. The Ministry of Environmental Protection of the Republic of Serbia issued annual permits (numbers listed in Institutional Review Board Statement) that allowed the capture and sampling of a limited number of animals per location.
Data collection—After capture, the live animals were transported in separate cages to IBISS and treated by veterinarian with isoflurane inhalation anaesthesia. A fingertip from a hind foot was removed and stored at −80 °C for DNA extraction. After a few days of recovery, each animal was released back into its own underground tunnel system. Liver and muscle tissue from dead animals was stored in absolute ethanol or frozen at −20 °C. Live animals were treated in accordance with Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. All animal procedures were approved by the Ethical Committee for the Use of Laboratory Animals of the Institute for Biological Research “Siniša Stanković”, University of Belgrade (IBISS) and the Veterinary Directorate of the Ministry of Agriculture, Forestry, and Water Economy of the Republic of Serbia (Licence No. 323-07-11307).
The teeth (molars) were extracted from four old, archived skull specimens (Collection Ivo Savić, IBISS). They were karyotyped between 1963 and 1977 [11]. Three were identified as N. l. syrmiensis; one of these animals was captured in the Srem district (locus typicus—Stara Pazova) in 1969, while the other two were from the Belgrade region—Vinča and Jajinci—and were captured in 1963 and 1977, respectively (Figure 1c, Table S1). A specimen caught in sympatry with N. l. syrmiensis from the locality Avala Mt. (1969) was identified as N. l. hungaricus.
Total DNA was extracted from finger parts of living animals or from liver and muscle tissue using the DNeasy Blood and Tissue Kit, Qiagen. DNA isolation from teeth was carried out at the Institute of Forensic Medicine according to the procedures described in [14]. Universal primers [34] and seminested PCR were used for the amplification of ~600 bp long 16SrRNA gene fragments, as described in [14]. To obtain longer sequences of DNA extracted from old teeth, MT-CYTB gene parts were amplified with several primers that had been newly designed and modified according to [35,36]. Primer sequences and the reaction conditions are those given in [21]. Sequencing was performed in both directions by a third party.
Sequence analysis—Our analysis generated 22 new 16SrRNA and 20 MT-CYTB sequences from five cryptic N. leucodon species. They were all visually examined using the FinchTV 1.4.0 chromatogram viewer (Geospiza Inc. Seattle WA, USA), analysed using BioEdit Ver. 7.2.5 [37], and checked for the presence of stop codons/chimeric sequences. Additional sets of 40 16SrRNA and 42 MT-CYTB sequences were compared using the Basic Local Alignment Search Tool (BLAST) and obtained from GenBank (Table S1). The final 16SrRNA dataset of 62 individual sequences included a total of 46 individuals from five cryptic N. leucodon species, five N. xanthodon, eight N. ehrenbergi, and three Spalax sp. sequences, which were used as an outgroup. The final MT-CYTB dataset comprised 62 individual sequences, 45 N. leucodon, four N. xanthodon, eight N. ehrenbergi, and five Spalax sp. for the outgroup. They were subsequently aligned using ClustalW in MEGA Ver. X software [38].
Species identification—Estimates of evolutionary divergence over sequence pairs were performed between eight groups, i.e., five different cryptic species of the N. leucodon species complex, N. xanthodon, N. ehrenbergi and the outgroup (Spalax sp.), for each mtDNA gene separately in MEGA X, using the maximum composite likelihood model between groups with 10,000 bootstraps. Analyses were performed using the Kimura two-parameter model [39]. Rate variation between sites was modelled with a gamma distribution (shape parameter = 1). The two final data sets analysed comprised 62 nucleotide sequences, with a total of 492 and 425 positions for 16srRNA and MT-CYTB, respectively. The codon positions included were 1st + 2nd + 3rd + Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option).
We applied two different phylogenetic analysis methods to confirm the strength of the tree topology: maximum likelihood (ML) in PhyML [40] and Bayesian analysis in MrBayes [41]. Phylogenetic trees were drawn using FigTree Ver. 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/) (accessed on 6 February 2024) and MEGA X. Prior to ML phylogenetic analysis, a best-fit substitution model in aligned sequences with the Akaike Information Criterion corrected for small sample sizes (AICc) [42] was established using jModelTest v.2.1.4. [40,43]. BI analyses originated with random starting trees and were run for 1 × 106 generations, sampling every 100th generation, with the burn-in value set to 500. Combined trees from the various runs produced a 50% majority-rule consensus tree with the Bayesian posterior probability values of the relevant branches.
3. Results
3.1. Cryptic Species Identification
3.1.1. 16srRNA Gene Polymorphism
The obtained sequences are deposited in the GenBank database under the accession numbers PP349436–PP349461 (Table S1). The total data set contains 22 sequences that are free of stop codons, insertions, or deletions. In the 492 bp data set (485 excluding sites with gaps/missing data), there were 105 variable (polymorphic) sites, with 88 being parsimony informative, and a total of 135 mutations. A summary of the findings follows: total number of haplotypes, h = 34; nucleotide diversity, Pi: 0.05207; haplotype diversity, Hd = 0.9513 (variance of Hd = 0.0002, SD = 0.015).
The evolutionary divergence values calculated in MEGA X (Table 1) between the genera Spalax/Nannospalax ranged from 0.0939 to 0.1429; among three Nannospalax superspecies, N. leucodon was closer to N. xanthodon (0.063 to 0.0854) and N. xanthodon was closer to N. ehrenbergi (0.1024 to 0.1172). Among the five N. leucodon cryptic species distributed in Serbia, the lowest divergence (0.0140) was that between N. l. syrmiensis and N. l. serbicus and the highest (0.0537) was that between N. l. hungaricus and N. l. montanosyrmiensis. The lowest values of evolutionary divergence were found in N. ehrenbergi supersp.: that of N. golani and N. galili was 0.0052, and that of N. judaei and N. carmeli was 0.0136.

Table 1.
Estimates of evolutionary divergence over sequence pairs between groups using 16SrRNA gene polymorphism dataset. Standard error estimates are shown above the diagonal.
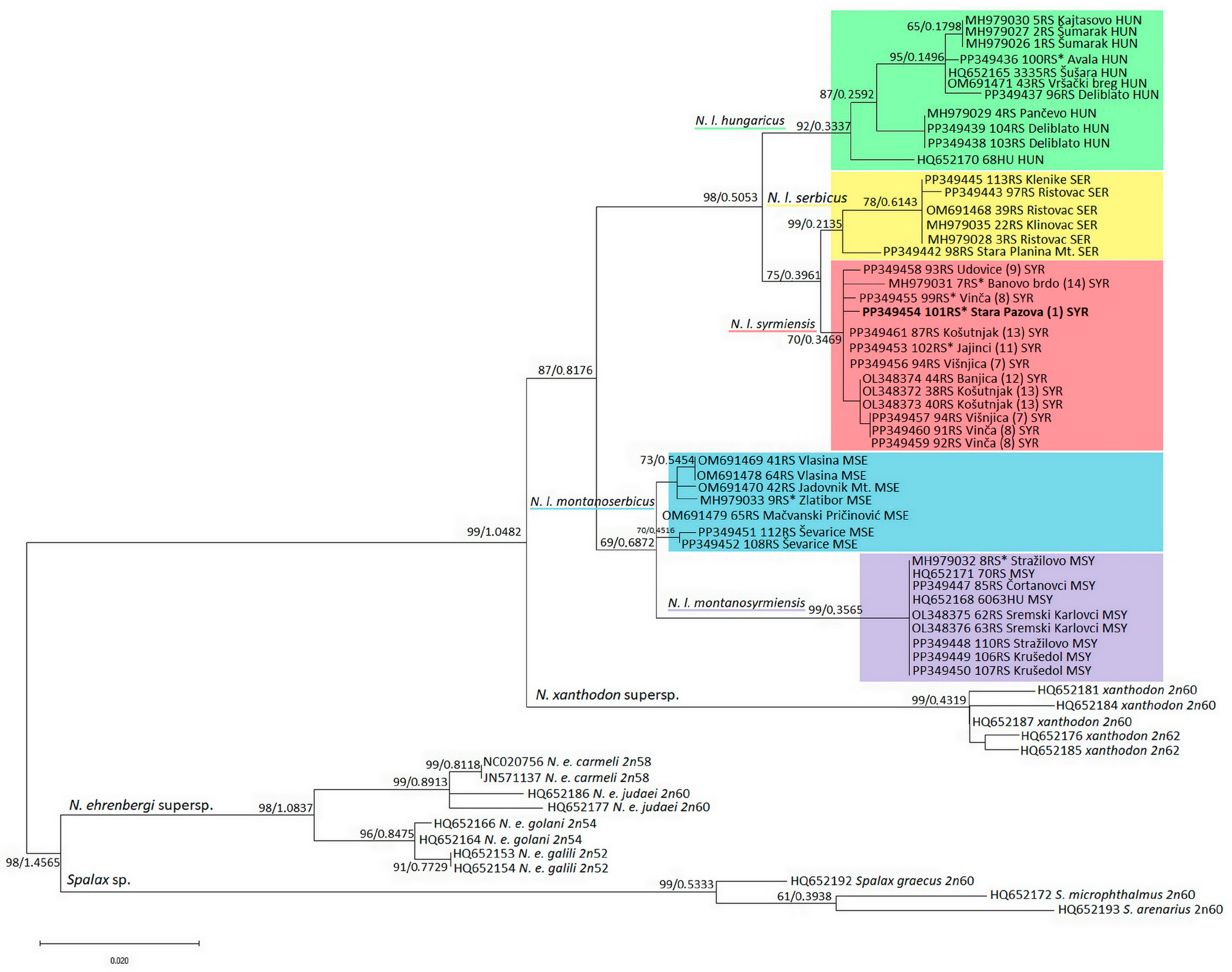
The topologies of the two trees, the ML phylogenetic tree generated using the Maximum Likelihood method with the TRN + I + G model suggested by jModelTest and the BI tree, are identical; therefore, only the ML tree with nodes supporting values from both methods is shown (Figure 2). The tree is rooted in the genus Spalax and shows a strongly supported division into two groups, the first being N. ehrenbergi, which is more closely related to Spalax sp., and the second being the sister position of N. xanthodon and N. leucodon, which are grouped into one clade. Each of the five N. leucodon cryptic species forms a distinct clade with high significance. There are two main clades, the first containing the phylogenetically older N. l. montanosyrmiensis and N. l. montanoserbus in the basal position, and the second containing N. l. syrmiensis, N. l. serbicus, and the youngest branch, N. l. hungaricus. All 18 sequences of newly collected individuals are grouped together with old, archived tooth-sample sequences of the same cryptic species. The tooth sample of a specimen from Avala Mt. (10 in Figure 1c), identified as N. l. hungaricus in the past, is clustered with other recent N. l. hungaricus specimens. Four tooth samples (1, 8, 11, and 14 in Figure 1c) from specimens previously determined by karyotyping to be N. l. syrmiensis, including the sample from the locus typicus Stara Pazova (1 in Figure 1c), are grouped with recently collected animals from the Belgrade (7, 8, 12, and 13 in Figure 1c) and Smederevo (9 in Figure 1c) regions. Our new samples (15 and 16 in Figure 1c) from the Mačva district (Ševarice near Bogatić) are grouped with other N. l. montanoserbicus samples from recently collected animals, but also with a tooth sample from a specimen captured in 1965 from Zlatibor Mt. The tooth sample of an individual from Stražilovo (on the slopes of Fruška Gora), N. l. montanosyrmiensis, is grouped with all other fresh samples from nearby localities, as well as from Kelebia. This cryptic species is the most homogeneous clade of N. leucodon analysed here.
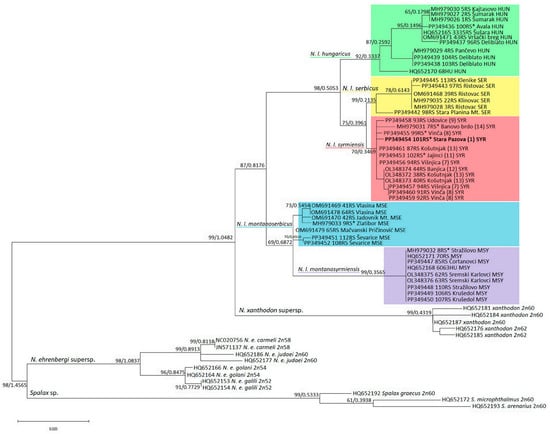
Figure 2.
ML phylogenetic tree constructed using 16SrRNA segment nucleotide polymorphism analysis. Similar topologies were inferred using the ML and BI methods; therefore, the support values were placed at the tree nodes. The sequences with asterisks are old, archived karyotyped samples (Collection Ivo Savić, IBISS). The numbers in brackets for N. l. syrmiensis correspond to the locality numbers in Figure 1c.
3.1.2. MT-CYTB Gene Polymorphism
The obtained sequences were deposited in the GenBank database under the accession numbers PP378178–PP378201 (Table S1). The total dataset comprises 20 sequences that are free of stop codons, insertions, or deletions. In the 425 bp dataset (424 excluding sites with gaps/missing data), there were 138 variable (polymorphic) sites, of which 126 were parsimony informative, and a total of 180 mutations were found. A summary of the findings follows: total number of haplotypes, h = 37; nucleotide diversity, Pi: 0.08913; haplotype diversity, Hd = 0.975 (variance of Hd = 0.00007, Sd = 0.008).
The values of evolutionary divergence generated from MT-CYTB polymorphism analysis (Table 2) are about twice as high as those derived from the 16SrRNA gene, but the same relationships were found between the groups. Values of divergence among the genera Spalax/Nannospalax ranged from 0.2042 to 0.2380; N. leucodon was again found to be closely related to N. xanthodon (0.1020 to 0.1269) and N. xanthodon to N. ehrenbergi (0.1326 to 0.1416). The lowest divergence value, 0.0314, was found between N. l. syrmiensis and N. l. serbicus, but the highest value, 0.1073, was between N. l. syrmiensis and N. l. montanosyrmiensis. The lowest values of evolutionary divergence were again found in the N. ehrenbergi supersp.: N. golani and N. galili, 0.0183 and N. judaei and N. carmeli, 0.0195.

Table 2.
Estimates of evolutionary divergence over sequence pairs between groups using the MT-CYTB gene polymorphism dataset. Standard error estimates are shown above the diagonal.
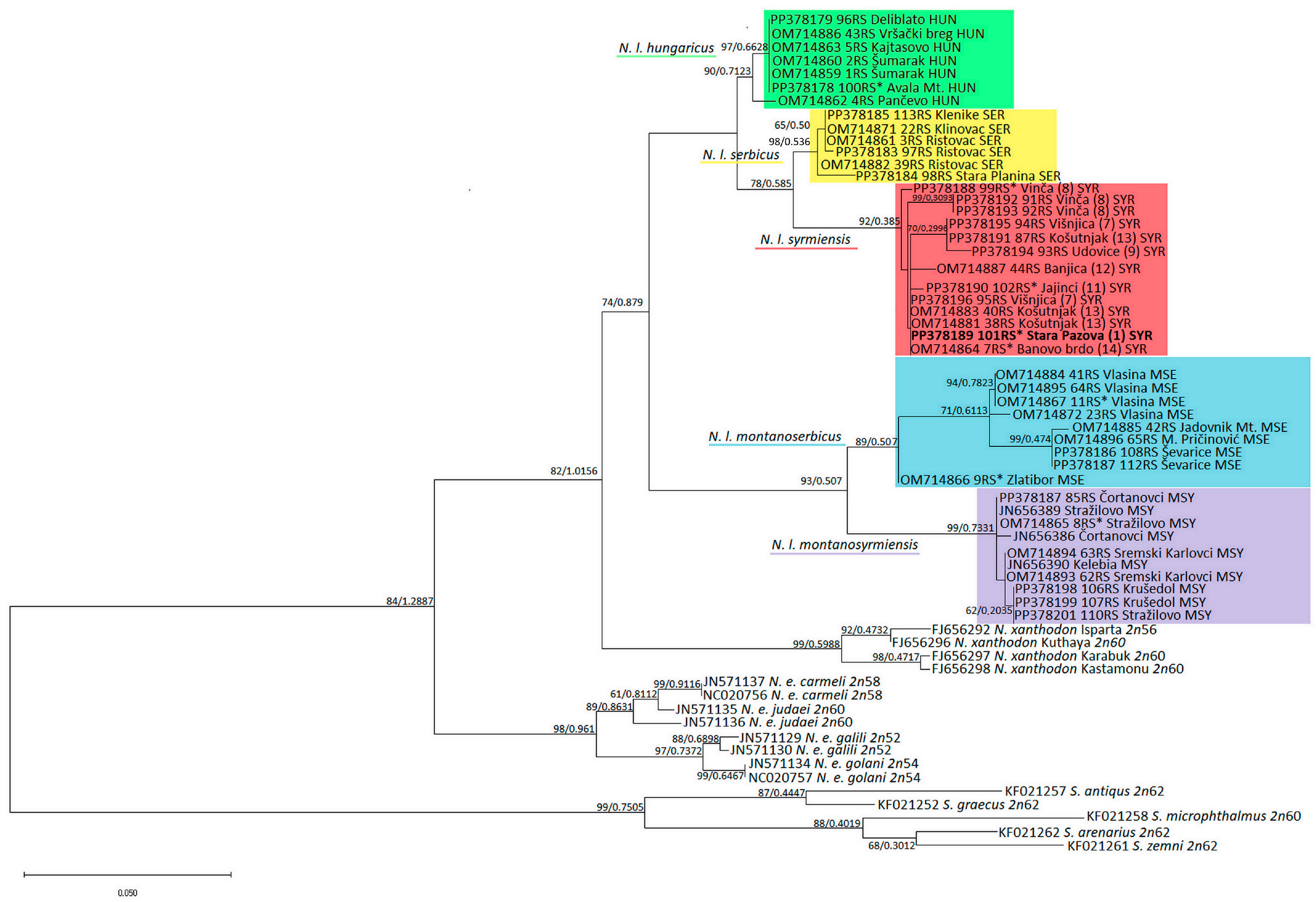
Similarly, the topologies of the ML phylogenetic tree generated using the HKY + I + G model from jModelTest and the BI tree are identical, so the support values of both methods were placed at the tree nodes in the ML tree (Figure 3). The topology of the MT-CYTB tree corresponds to 16SrRNA rooted in Spalax sp. All five N. leucodon cryptic species, are divided into five clusters with high significance. The phylogenetically older N. l. montanosyrmiensis and N. l. montanoserbicus are in the basal position of the N. leucodon group. N. l. syrmiensis, N. l. serbicus, and N. l. hungaricus are closer to each other. An old, archived tooth sample from a specimen from Avala Mt. (10 in Figure 1c) is clustered with N. l. hungaricus. The sequences from four tooth samples (1, 8, 11, and 14 in Figure 1c) from specimens identified as N. l. syrmiensis by karyotyping in the past, including the sample from the locus typicus Stara Pazova (1 in Figure 1c), are clustered with samples from recently captured animals from the Belgrade (7, 8, 12, and 13 in Figure 1c) and Smederevo (9 in Figure 1c) regions. Fresh samples from localities near Bogatić (15 and 16 in Figure 1c) are clustered with the tooth sample of an animal from Vlasina and other newly collected N. l. montanoserbicus samples. The old, archived sample from Stražilovo (Fruška Gora Mt.), which was determined by karyotyping to be N. l. montanosyrmiensis, is clustered with newer samples from nearby localities and from Kelebia.
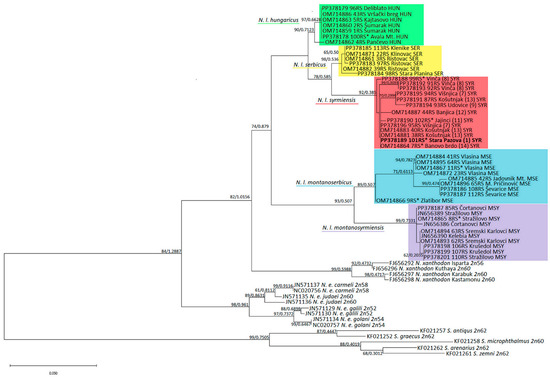
Figure 3.
ML phylogenetic tree constructed using nucleotide polymorphism analysis of MT-CYTB gene sequences. Similar topologies were inferred using ML and BI methods; therefore, the support values were placed at the tree nodes. Sequences with asterisks are old, archived karyotyped samples (Collection Ivo Savić, IBISS). The numbers in brackets for N. l. syrmiensis correspond to the locality numbers in Figure 1c.
3.2. Presence and Distribution of N. l. syrmiensis
As a result of the five-year survey of localities attributed to N. l. syrmiensis (Stara Pazova (locus typicus), Višnjica, Banovo Brdo, Košutnjak, Avala, Jajinci, Smederevo, Vinča, and Bogatić [11]) and nearby areas, we report habitat loss, i.e., a greatly reduced distribution range of N. l. syrmiensis (Figure 1c). Nannospalax l. syrmiensis is no longer present at the Stara Pazova locality (locus typicus) (locality 1 in Figure 1c) or in the wide surroundings in the district of Srem (Vojvodina province) (localities 2–6 in Figure 1c) due to intensive anthropogenic activities. After extensive field work in cooperation with citizens, only moles were found in the entire area. On the slopes of Mount Avala (locality 10 in Figure 1c), the site of Beli Potok, where N. l. syrmiensis and N. l. hungaricus lived sympatrically in the same area (more precisely, in the same meadow), has now been destroyed by the massive construction of a highway.
We were able to confirm the occurrence of N. l. syrmiensis only in the Belgrade region, i.e., in Višnjica, Vinča, Banjica, and Košutnjak (7, 8, 12, and 13 in Figure 1c) and at the easternmost limit of its geographical range—Udovice, near Smederevo (locality 9 in Figure 1c). However, the westernmost part of its range in the Mačva district (Mačvanski Pričinović and Ševarice, both near Bogatić; 15 and 16 in Figure 1c) is now inhabited by another cryptic species, N. l. montanoserbicus.
4. Discussion
Here, we conducted five years of systematic field work, covering the entire known distribution range of N. l. syrmiensis [11,19]. For species identification, we compared the nucleotide sequences of two mitochondrial gene segments (16SrRNA and MT-CYTB) between newly collected animals and 46–61-year-old archived tooth samples that had been previously identified by karyotyping. For the species identity of N. l. syrmiensis, we used the tooth sample from the locus typicus—Stara Pazova (1 in Figure 1c)—together with three tooth samples from the Belgrade region—Vinča, Jajinci, and Banovo Brdo (8, 11, and 14 in Figure 1c).
Both genes yielded almost identical topologies using two different molecular phylogenetic methods (BI and ML), a result that is consistent with formerly published phylogenies [15,16,20,21]. The estimates of evolutionary divergence confirmed the relationships in the phylogenetic trees. As expected, the divergences inferred from the MT-CYTB dataset were about twice as high as those from the 16SrRNA dataset because these two genes are functionally different and have different mutation rates. All sequences of N. leucodon derived from recently sampled animals were grouped into five clusters, corresponding to five cryptic species distributed in Serbia. These five clusters were divided into two main groups: an evolutionarily older branch with N. l. montanosyrmiensis and N. l. montanoserbicus in the basal position, and a younger branch with N. l. serbicus, N. l. syrmiensis, and N. l. hungaricus. This topology is consistent with the most recent N. leucodon phylogeny, with the exception of the placement of N. l. syrmiensis, which was marked as a missing taxon and “incertae sedis” [16]. Based on our results from both datasets, N. l. serbicus is the cryptic species closest to N. l. syrmiensis, but they are clearly separated in the phylogenetic trees, with high support. Moreover, the estimated evolutionary divergences between them exceed those between the only four species recognized in the literature, Spalax (=Nannospalax) galili, S. golani, S. carmeli, and S. judaei, which belong to N. ehrenbergi supersp. [9,12,44].
Finally, using a phylogenetic tree derived from both mtDNA markers, we have provided evidence that N. l. syrmiensis is not extinct in Serbia. Old, archived tooth samples (1, 8, 11, and 14 in Figure 1c) clustered with newly collected material, as expected according to karyotyping and known distribution areas [11], with the exception of samples from Mačva district (localities near Bogatić; 15 and 16 in Figure 1c). Mačva was designated as the westernmost part of the distribution range of N. l. syrmiensis, but our results confirmed the occurrence of N. l. montanoserbicus in this region. Thus, the distribution range of N. l. montanoserbicus is larger than previously defined and includes not only localities > 1000 m altitude [45], but also those at lower altitudes (e.g., ~60 m).
Historical overview of N. l. syrmiensis—Historically, the name “syrmiensis” (lat. Syrmium = Srem, a district in the province of Vojvodina between the rivers Sava in the south and Danube in the north) emerged as a result of an early taxonomy based solely on morphological data, when Méhely [46,47] named Spalax monticola syrmiensis as a separate species based on the examination of seven skulls from three localities in the district of Srem (Vojvodina, Serbia): Stara Pazova—locus typicus, Ruma and Sremska Mitrovica, but also a skull from the locality of Lelle (Somogy County, Hungary) in the western part of the Carpathian Basin. Only later, comprehensive karyotyping of BMR specimens [18,19,23] established that the Balkan region was inhabited by different CFs (with 2n = 46–56). Eight different species were proposed, including Spalax leucodon syrmiensis (Méhely 1909), as the monogeneric nomenclature was still in use. After summarizing the differences in chromosome number and morphology, evidence of reproductive isolation, and ecogeographic features, Savić and Soldatović [11] reported that the genus Nannospalax comprises 14 different species with subspecies. They described the species Nannospalax syrmiensis (Méhely 1909) with the karyotypic formula 2n = 54, NF = 90 as a group of seven populations endemic to Serbia, inhabiting the plains of the Srem district and the right bank of the Sava and Danube rivers from the confluence of the Drina, with the Sava in the west and the Great Morava with the Danube in the east [11]. Later, the nomenclature Nannospalax leucodon syrmiensis was accepted and widely used in the literature [6,8,10,14,15,16,21,26].
In contrast to BMRs from Srem District (Vojvodina, Serbia), which were confirmed as N. l. syrmiensis by karyotyping [11], the existence of N. l. syrmiensis in Somogy County (Hungary) has never been proven. Although it was emphasized that the BMRs were not sampled during the extensive cytogenetic mapping in the 1970s and that no karyological information on the populations from the Carpathian Basin is available [10], N. l. syrmiensis was declared extinct in Hungary and possibly extinct in Serbia [15]. Such conclusions may have a negative impact on the conservation of biodiversity, especially the genetic diversity of wildlife.
Chromosomal speciation and cryptic species in the genus Nannospalax—Chromosomal rearrangements are common in rodents, especially in the two most species-rich families, the Cricetidae and Muridae [48], and in the Ctenomyidae [49]. The theoretical basis for speciation mechanism through chromosomal rearrangements is described in detail. These rearrangements have the potential to drive post-zygotic isolation and gradually reduce gene flow, which makes them an important driver of the origination of cryptic species [50,51,52,53]. Chromosomal speciation within the lesser BMRs from the genus Nannospalax represents one of the few examples in nature [53,54,55,56]. It is characterized by extensive chromosomal rearrangements, i.e., karyotype variability. Pre- and post-copulation reproductive isolation between seven cryptic N. leucodon species, including N. l. syrmiensis, was demonstrated by comprehensive experimental crossbreeding and additionally confirmed by artificial insemination performed in similar combinations. Embryos developed only when individuals of the same cryptic species were united [8,11,19]. Reproductive isolation between the cryptic species of N. leucodon, which was caused by rapid chromosomal evolution, led to the deep genetic divergence reported in [14,15,16,21] and in this study. The unclear number of species and uncertain taxonomy have resulted in their inappropriate conservation status. Although many of them are seriously endangered, their status in the IUCN Red List of Threatened Species remains “Data Deficient” (DD) [13]. For the conservation of wildlife genetic diversity, it is important to acknowledge each cryptic species as a group of subpopulations that are genetically and ecologically differentiated [57].
Nannospalax l. syrmiensis—habitat loss and conservation perspectives—Besides cryptic species identification, after a detailed survey of localities formerly attributed to N. l. syrmiensis [11] and nearby areas, we found that the distribution range of N. l. syrmiensis has been highly reduced (Figure 1c). N. l. syrmiensis does not occur at the locality of Stara Pazova (locus typicus) (1 in Figure 1c) or in the wide surroundings in the district of Srem (Vojvodina province) (2–6 in Figure 1c). After detailed field work in cooperation with citizens, we found only moles in the whole area, including in steppe fragments near the villages of Donji Petrovac and Krnješevci (2 and 3 in Figure 1c). The species N. l. syrmiensis is confirmed only in the Belgrade region (7, 8, 12, and 13 in Figure 1c) and Udovice (9 in Figure 1c) near Smederevo, i.e., at the easternmost border of its geographical area—entry of the Great Morava River into the Danube. In the westernmost part of the area, at the entry of the Drina into the Sava in the Mačva district (15 and 16 in Figure 1c), we recorded only the cryptic species N. l. montanoserbicus.
Of all five cryptic species in Serbia, N. l. syrmiensis has the smallest distribution area. Such conditions, in combination with habitat fragmentation, lead to a decline in the number of individuals in populations, to their mutual isolation and frequently to complete extinction. The main risk factors for this cryptic species endemic to Serbia, according to the IUCN—CMP Unified Classification of Direct Threats [58], are as follows: urbanization and infrastructure construction, formation of agroecosystems and transition to intensive agricultural production, road construction, burning of vegetation, systematic transition from small-scale extensive production to large-scale intensive production systems, and habitat pollution, as previously published in [21]. As BMRs are herbivores, they often approach human gardens, farms, and parks, where they are treated as pests. Excessive killing and the use of chemical agents significantly increase the negative pressure on this cryptic species [11,21]. It is therefore necessary to investigate its population structure, to conduct a risk assessment, and to implement the conservation actions proposed by the IUCN—CMP Unified Classification of Conservation Actions Needed [59]. In particular, the following actions are needed: application of protection measures arising from their endemism to preserve their gene pool, informing the local residents about the level of protection of this mammal species and ways to protect their households without killing them, increasing the competence and efficiency of existing protection and monitoring services, creating and funding scientific teams that would increase the amount of research being conducted on insufficiently studied taxa and areas, and creating conditions to increase the diversity of mammal fauna in areas with intensive agricultural production [21].
5. Conclusions
For the conservation of biodiversity, including the genetic diversity of wildlife, declaring a particular cryptic species as probably extinct may have negative consequences. Through five years of field work and molecular genetic studies, we were able to provide evidence that N. l. syrmiensis is not extinct, as was reported in the literature. However, its habitat has been fragmented and reduced in size, primarily due to anthropogenic impact. Consequently, this cryptic species is on the retreat and certain conservation actions are required. For the preservation of wildlife genetic diversity, it is important to acknowledge each cryptic species. Many of these species are seriously endangered, while their status is still classified as Data Deficient (DD) in the IUCN Red List of Threatened Species. It is therefore crucial to clarify their taxonomy in order to assign to them the correct conservation status.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani14050774/s1, Table S1: A list of 22 new N. leucodon and samples imported from GenBank, with IDs (GenBank accession numbers) for mtDNA gene sequences. 2n—diploid chromosomal numbers; NF—fundamental number of chromosomal arms. is—Collection Ivo Savic, IBISS; ISO—ISO country code.
Author Contributions
Conceptualization, V.B.-S., V.J. and I.S.; methodology, V.B.-S., M.Đ., G.S. and O.S.; formal analysis, V.B.-S., M.Đ. and G.S.; investigation, V.B.-S., M.Đ., N.B.K. and V.J.; writing—original draft preparation, V.B.-S. and V.J.; writing—review and editing, M.Đ., I.S. and N.B.K.; visualization, V.B.-S.; project administration, V.B.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Ministry of Education, Science, and Technological Development of the Republic of Serbia (Grant No. 451-03-66/2024-03/200007).
Institutional Review Board Statement
As the BMR is officially strictly protected in the Republic of Serbia, this research was conducted under permits issued annually by the Ministry of Environmental Protection of the Republic of Serbia (numbers: 353-01-2892/2018-04; 353-01-2699/2020-04; 353-01-26/2022-04; 353-01-175/2023-04). All animal procedures were approved by the Veterinary Directorate of the Ministry of Agriculture, Forestry, and Water Economy of the Republic of Serbia (Licence No. 323-07-11307).
Informed Consent Statement
Not applicable.
Data Availability Statement
DNA nucleotide sequences were deposited in the NCBI GenBank, and can be assessed at https://www.ncbi.nlm.nih.gov/genbank, accessed on 26 February 2024.
Acknowledgments
We are most grateful to Nada Ćosić, Pavle Lukić, and Đurđe Đuković for assistance with fieldwork and Mihajlo Jeličić for his expertise in technical support. We thank Dragan Kataranovski, the curator of the Collection Ivo Savić, IBISS. The authors would also like to thank two anonymous reviewers for their valuable suggestions that improved our manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gorbunova, V.; Hine, C.; Tian, X.; Ablaeva, J.; Gudkov, A.V.; Nevo, E.; Seluanov, A. Cancer resistance in the blind mole rat is mediated by concerted necrotic cell death mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 19392–19396. [Google Scholar] [CrossRef]
- Azpurua, J.; Seluanov, A. Long-lived cancer resistant rodents as new model species for cancer research. Front. Genet. 2013, 3, 319. [Google Scholar] [CrossRef] [PubMed]
- Manov, I.; Hirsh, M.; Iancu, T.; Malik, A.; Sotnichenko, N.; Band, M.; Avivi, A.; Shams, I. Pronounced cancer resistence in a subterranean rodent, the blind mole-rat, Spalax: In Vivo and in vitro evidence. BMC Biol. 2013, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Shams, I.; Malik, A.; Manov, I.; Joel, A.; Band, M.; Avivi, A. Transcription Pattern of p53-targeted DNA repair genes in the hypoxiatolerant subterranean mole rat Spalax. J. Mol. Biol. 2013, 425, 1111–1118. [Google Scholar] [CrossRef]
- Fang, X.D.; Nevo, E.; Han, L.J.; Levanon, E.Y.; Zhao, J.; Avivi, A.; Larkin, D.; Jiang, X.; Feranchuk, S.; Zhu, Y.; et al. Genome-wide adaptive complexes to undergound stresses in blind mole rats. Nat. Commun. 2014, 5, 3966. [Google Scholar] [CrossRef] [PubMed]
- Arslan, A.; Kryštufek, B.; Matur, F.; Zima, J. Review of chromosome races in blind mole rats (Spalax and Nannospalax). Folia Zool. 2016, 65, 249–301. [Google Scholar] [CrossRef]
- Kankılıç, T.; Arslan, S.; ¸Seker, P.S.; Kankılıç, T.; Toyran, K.; Zima, J. A new chromosomal race (2n = 44) of Nannospalax xanthodon from Turkey (Mammalia: Rodentia). Zool. Middle East 2017, 63, 181–188. [Google Scholar] [CrossRef]
- Savić, I.; Ćirović, D.; Bugarski-Stanojević, V. Exceptional Chromosomal Evolution and Cryptic Speciation of Blind Mole Rats Nannospalax leucodon (Spalacinae, Rodentia) from South-Eastern Europe. Genes 2017, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Ivanitskaya, E.; Beiles, A. Adaptive Radiation of Blind Subterranean Mole Rats: Naming and Revisiting the Four Sibling Species of the Spalax Ehrenbergi Superspecies in Israel: Spalax galili (2n = 52), S. golani (2n = 54), S. carmeli (2n = 58) and S. judaei (2n = 60); Bachkhuys Publishers: Leiden, The Netherlands, 2001. [Google Scholar]
- Csorba, G.; Krivek, G.; Sendula, T.; Homonnay, Z.G.; Hegyeli, Z.; Sugár, S.; Farkas, J.; Stojnić, N.; Németh, A. How can scientific researches change conservation priorities? A review of decade-long research on blind mole-rats (Rodentia: Spalacinae) in the Carpathian Basin. Therya 2015, 6, 103–121. [Google Scholar] [CrossRef]
- Savić, I.; Soldatović, B. Karyotype Evolution and Taxonomy of the Genus Nannospalax Palmer, 1903, Mammalia, in Europe; Separate Editions; Serbian Academy of Sciences and Arts: Belgrade, Serbia, 1984; Volume 59, pp. 1–104. [Google Scholar]
- Musser, G.G.; Carleton, M.D. Order Rodentia. In Mammal Species of the World: A Taxonomic and Geographic Reference; Wilson, D.E., Reeder, D.M., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 745–1601. [Google Scholar]
- Kryštufek, B.; Amori, G. Nannospalax leucodon (amended version of 2008 assessment). In The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2017. [Google Scholar]
- Bugarski-Stanojević, V.; Stamenković, G.; Ćirović, D.; Ćirić, D.; Stojković, O.; Veličković, J.; Kataranovski, D.; Savić, I.R. 16S rRNA gene polymorphism supports cryptic speciation within the lesser blind mole rat Nannospalax leucodon superspecies (Rodentia: Spalacidae). Mamm. Biol. 2020, 100, 315–324. [Google Scholar] [CrossRef]
- Németh, A.; Csorba, G.; Laczkó, L.; Mizsei, E.; Bereczki, J.; Pásztor, J.A.; Petró, P.; Sramkó, G. Multi-locus genetic identification of a newly discovered population reveals a deep genetic divergence in European blind mole rats (Rodentia: Spalacidae: Nannospalax). Ann. Zool. Fenn. 2020, 57, 89–98. [Google Scholar] [CrossRef]
- Németh, A.; Mizsei, E.; Laczkó, L.; Czabán, D.; Hegyeli, Z.; Lengyel, S.; Csorba, G.; Sramkó, G. Evolutionary history and systematics of European blind mole rats (Rodentia: Spalacidae: Nannospalax): Multilocus phylogeny and species delimitation in a puzzling group. Mol. Phylogenet. Evol. 2024, 190, 107958. [Google Scholar] [CrossRef]
- Németh, A.; Krnács, G.; Krizsik, V.; Révay, T.; Czabán, D.; Stojnić, N.; Farkas, J.; Csorba, G. European rodent on the edge: Status and distribution of the Vojvodina blind mole rat. Springerplus 2013, 2, 2. [Google Scholar] [CrossRef]
- Savić, I.; Soldatović, B. Contribution to the study of ecogeographic distribution and evolution of chromosomal forms of the Spalacidae from the Balkan Peninsula. Arch. Biol. Sci. 1977, 29, 141–156, (In Serbian with English Summary). [Google Scholar]
- Soldatović, B. Karyotype analysis and cytogenetic aspects of speciation in the genus Spalax. Zb. Prir. Nauk. Mat. Srp. 1977, 52, 5–58, (In Serbian with English Summary). [Google Scholar]
- Hadid, Y.; Németh, A.; Snir, S.; Pavlíček, T.; Csorba, G.; Kázmér, M.; Major, A.; Mezhzherin, S.; Rusin, M.; Coşkun, Y.; et al. Is evolution of blind mole rats determined by climate oscillations? PLoS ONE 2012, 7, e30043. [Google Scholar] [CrossRef] [PubMed]
- Bugarski-Stanojević, V.; Stamenković, G.; Jojić, V.; Ćosić, N.; Ćirović, D.; Stojković, O.; Veličković, J.; Savić, I.R. Cryptic Diversity of the European Blind Mole Rat Nannospalax leucodon Species Complex: Implications for Conservation. Animals 2022, 12, 1097. [Google Scholar] [CrossRef] [PubMed]
- Vasić, V.; Džukić, G.; Janković, D.; Simonov, N.; Petrov, B.; Savić, I. Preliminarni spisak vrsta za crvenu listu kičmenjaka Srbije. Zaštita Prir. Prot. Nat. 1991, 43–44, 121–132, (In Serbian and Latin). [Google Scholar]
- Soldatović, B. Cytogenetic Study of the Speciation of the Genus Spalax in Yugoslavia. Ph.D. Thesis, University of Beograd, Belgrade, Serbia, 1971. (In Serbian). [Google Scholar]
- Savić, I.; Soldatović, B. Die Verbreitung der Karyotypen der Blindmaus Spalax (Mesospalax) in Jugoslawien. Arch. Biol. Nauka 1974, 26, 115–122. [Google Scholar]
- Nižetić, D.; Stevanović, M.; Soldatović, B.; Savić, I.; Crkvenjakov, R. Limited polymorphism of both classes of MHC genes in four different species of the Balkan mole rat. Immunogenetics 1988, 28, 91–98. [Google Scholar] [CrossRef]
- Németh, A.; Révay, T.; Hegyeli, Z.; Farkas, J.; Czabán, D.; Rózsás, A.; Csorba, G. Chromosomal forms and risk assessment of Nannospalax (superspecies leucodon) (Mammalia: Rodentia) in the Carpathian Basin. Folia Zool. 2009, 58, 349–361. [Google Scholar]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P., Jr. How Close Is Close: 16s RNA Sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 1992, 42, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G. Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Res. 2002, 30, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tan, Z.; Wang, D.; Xue, L.; Guan, M.; Huang, T.; Li, R. Species identifcation through mitochondrial rRNA genetic analysis. Sci. Rep. 2014, 4, 4089. [Google Scholar] [CrossRef] [PubMed]
- Arslan, E.; Gülbahçe, E.; Arıkoğlu, H.; Arslan, A.; Bužan, E.V.; Kryštufek, B. Mitochondrial divergence between three cytotypes of the Anatolian Mole Rat, Nannospalax xanthodon (Nordmann, 1840). Zool. Middle East 2010, 50, 27–34. [Google Scholar] [CrossRef]
- Kryštufek, B.; Ivanitskaya, E.; Arslan, A.; Arslan, E.; Buzan, E. Evolutionary history of mole rats (genus Nannospalax) inferred from mitochondrial cytochrome b sequences. Biol. J. Linn. Soc. 2012, 105, 446–455. [Google Scholar] [CrossRef]
- Matur, F.; Yanchukov, A.; Çolak, F.; Sözen, M. Two major clades of blind mole rats (Nannospalax sp.) revealed by mtDNA and microsatellite genotyping in Western and Central Turkey. Mamm. Biol. 2018, 94, 38–47. [Google Scholar] [CrossRef]
- Savić, I.R. Ecology of the mole rat Spalax leucodon Nordm, in Yugoslavia. Proc. Nat. Sci. Matica Srp. Novi Sad 1973, 44, 5–70, (In Serbian with English Summary). [Google Scholar]
- Veith, M.; Kosuch, J.; Vences, M. Climatic oscillations triggered post-Messinian speciation of Western Palearctic brown frogs (Amphibia, Ranidae). Mol. Phylogenet. Evol. 2003, 26, 310–327. [Google Scholar] [CrossRef]
- Irwin, D.M.; Kocher, T.D.; Wilson, A.C. Evolution of the Cytochrome b Gene of Mammals. J. Mol. Evol. 1991, 32, 128–144. [Google Scholar] [CrossRef]
- Schlegel, M.; Ali, H.S.; Stieger, N.; Groschup, M.H.; Wolf, R.; Ulrich, R.G. Molecular identification of small mammal species using novel cytochrome B gene-derived degenerated primers. Biochem. Genet. 2012, 50, 440–447. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computingplatforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Tsai, C.-L. Regression and time series model selection in small samples. Biometrika 1989, 76, 297–307. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Karanth, K.P.; Avivi, A.; Beharav, A.; Nevo, E. Microsatellite diversity in populations of blind subterranean mole rats (Spalax ehrenbergi superspecies) in Israel: Speciation and adaptation. Biol. J. Linn. Soc. 2004, 83, 229–241. [Google Scholar] [CrossRef]
- Savić, I.; Soldatović, B. Distribution range and evolution of chromosomal forms in the Spalacidae of the Balkan Peninsula and bordering regions. J. Biogeogr. 1979, 6, 363–374. [Google Scholar] [CrossRef]
- Méhely, L. Species generis Spalax. In A Földi Kutyák Fajai Származás- És Rendszertani Tekintetben; Magyar Tudományos Akadémia: Budapest, Hungary, 1909. (In Hungarian) [Google Scholar]
- Méhely, L. Species generis Spalax. In Die Arten der Blindmäuse in Systematischer und Phylogenetischer Beziehung; Teubner, B.G., Ed.; Berichte aus Ungarn: Leipzig, Germany, 1913; pp. 1–390. [Google Scholar]
- Castiglia, R. Sympatric sister species in rodents are more chromosomally differentiated than allopatric ones: Implications for the role of chromosomal rearrangements in speciation. Mammal Rev. 2013, 44, 1–4. [Google Scholar] [CrossRef]
- Oliveira, T.D.; Freitas, T.R.O. Investigating the evolutionary dynamics of diploid number variation in Ctenomys (Ctenomyidae, Rodentia). Genet. Mol. Biol. 2024, 46, e20230180. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Barton, N.H. Chromosomal speciation and molecular divergence—Accelerated evolution in rearranged chromosomes. Science 2003, 300, 321–324. [Google Scholar] [CrossRef]
- Ayala, F.J.; Coluzzi, M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. USA 2005, 102, 6535–6542. [Google Scholar] [CrossRef]
- Faria, R.; Navarro, A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 2010, 25, 660–669. [Google Scholar] [CrossRef]
- Mills, P.J.; Cook, L.G. Rapid chromosomal evolution in a morphologically cryptic radiation. Mol. Phylogenet. Evol. 2014, 77, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Pansonato-Alves, J.C.; Costa-Silva, G.J.; Mendonça, F.F.; Scacchetti, P.C.; Oliveira, C.; Foresti, F. Molecular and cytogenetic analyses of cryptic species within the Synbranchus marmoratus Bloch, 1795 (Synbranchiformes: Synbranchidae) grouping: Species delimitations, karyotypic evolution and intraspecific diversification. Neotrop. Ichthyol. 2014, 12, 903–911. [Google Scholar] [CrossRef]
- Baskevich, M.I.; Potapov, S.G.; Mironova, T.A. Caucasian Cryptic Species of Rodents as Models in Research on the Problems of Species and Speciation. Biol. Bull. Rev. 2016, 6, 245–259. [Google Scholar] [CrossRef]
- Malcher, S.M.; Pieczarka, J.C.; Geise, L.; Rossi, R.V.; Pereira, A.L.; O’Brien, P.C.M.; Asfora, P.H.; da Silva, V.F.; Sampaio, M.I.; Ferguson-Smith, M.A.; et al. Oecomys catherinae (Sigmodontinae, Cricetidae): Evidence for chromosomal speciation? PLoS ONE 2017, 12, e0181434. [Google Scholar] [CrossRef]
- Angeler, D.G.; Fried-Petersen, H.B. Parallels of quantum superposition in ecological models: From counterintuitive patterns to eco-evolutionary interpretations of cryptic species. BMC Ecol. Evol. 2024, 24, 15. [Google Scholar] [CrossRef]
- IUCN. Threats Classification Scheme. Version 3.3. 2022. Available online: https://www.iucnredlist.org/resources/threat-classification-scheme (accessed on 8 February 2024).
- IUCN. Conservation Actions Classification Scheme. Version 2.0. 2012. Available online: www.iucnredlist.org/resources/conservation-actions-classification-scheme (accessed on 8 February 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).