Characterization of the Effects of a Novel Probiotic on Salmonella Colonization of a Piglet-Derived Intestinal Microbiota Using Improved Bioreactor
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Probiotic Description
2.2. Disk Diffusion Assay of PRO-P2702 against Salmonella Typhimurium, Escherichia coli, Staphylococcus aureus and Enterococcus faecalis
2.3. PRO-P2702 Broth Inhibition Test against Salmonella Typhimurium
2.4. Recovery of Intestinal Contents for the Bioreactor’s Inoculum
2.4.1. Animal Experimentation
2.4.2. Inoculum’s Recovery
2.5. Bioreactor Experiments
2.5.1. Bioreactor Description
2.5.2. Bioreactor Experiments
2.6. SCFAs Analysis, Total Gross Energy, Total Crude Protein and Dry Matter Analysis
2.7. DNA Extraction and PCR for DNA Amplification of V4 Region
2.8. Sequence Analysis, Microbiota Diversity and Composition Determination
2.9. Statistical Analysis
3. Results
3.1. Disk Diffusion Assay of PRO-P2702 against Salmonella Typhimurium, E. coli, Staphylococcus aureus and Enterococcus faecalis
3.2. PRO-P2702 Broth Inhibition Test against Salmonella Typhimurium-R Obtained by Cultural Approach in Liquid
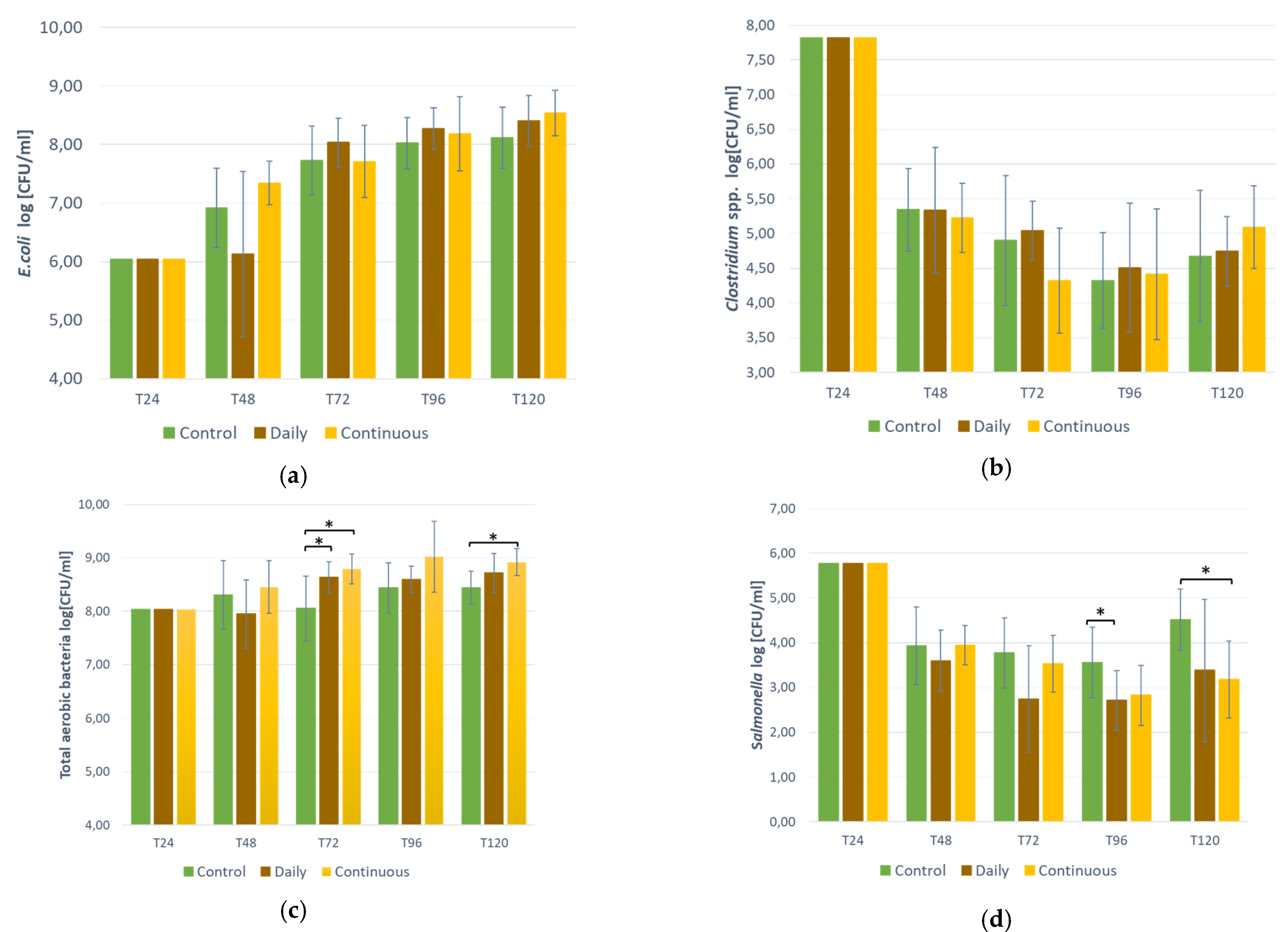
3.3. Bacteria Counts in the Bioreactor
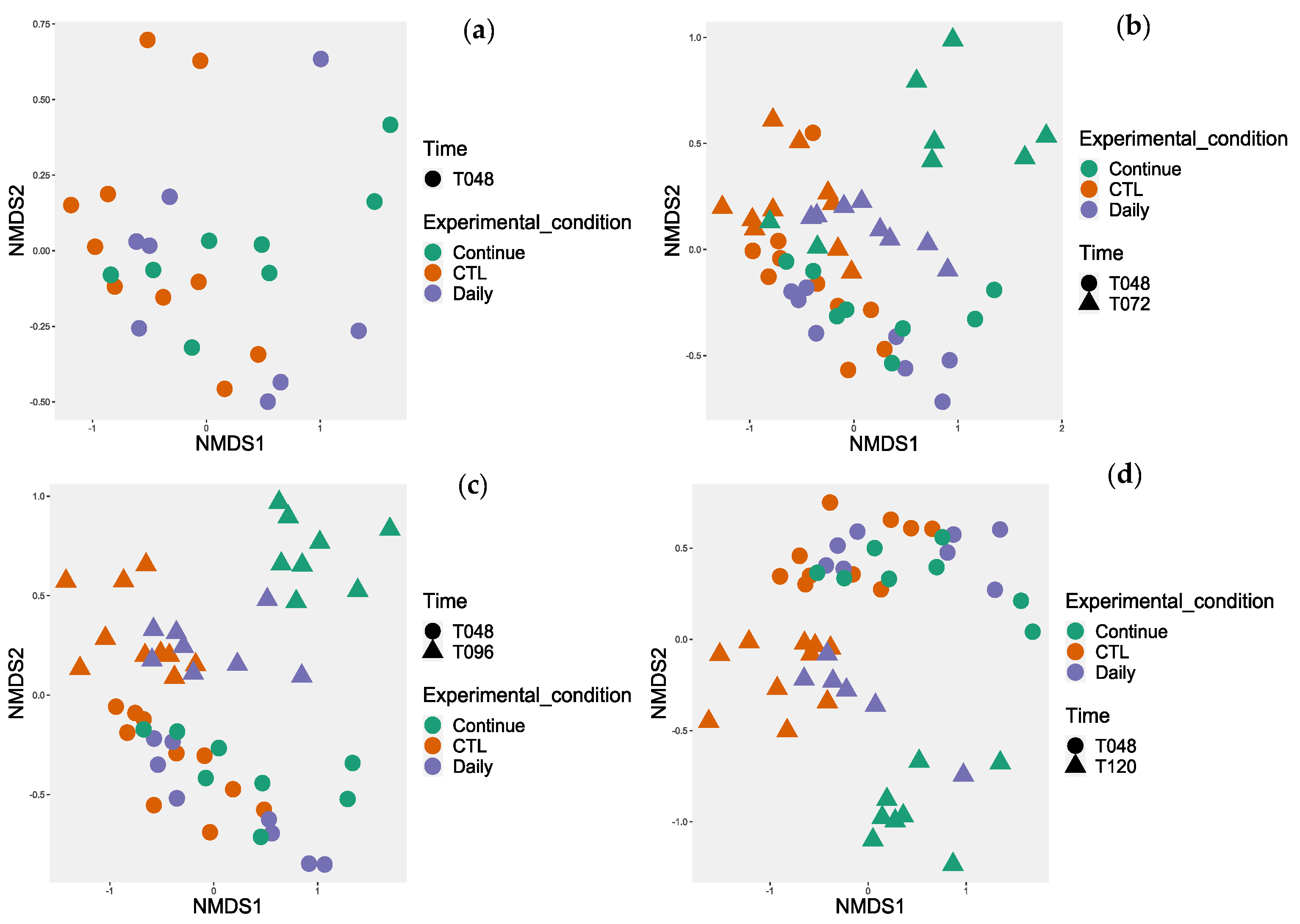
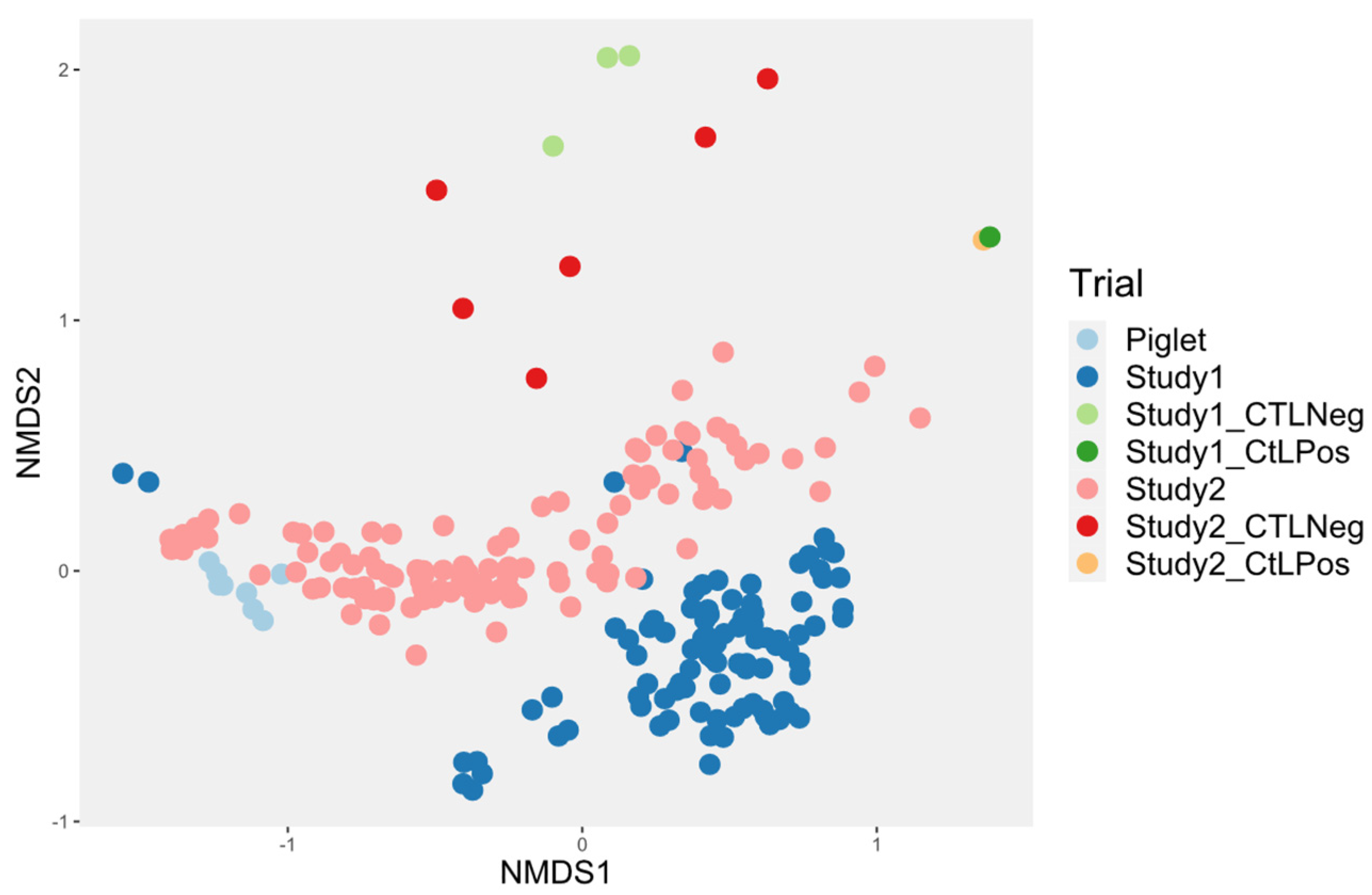
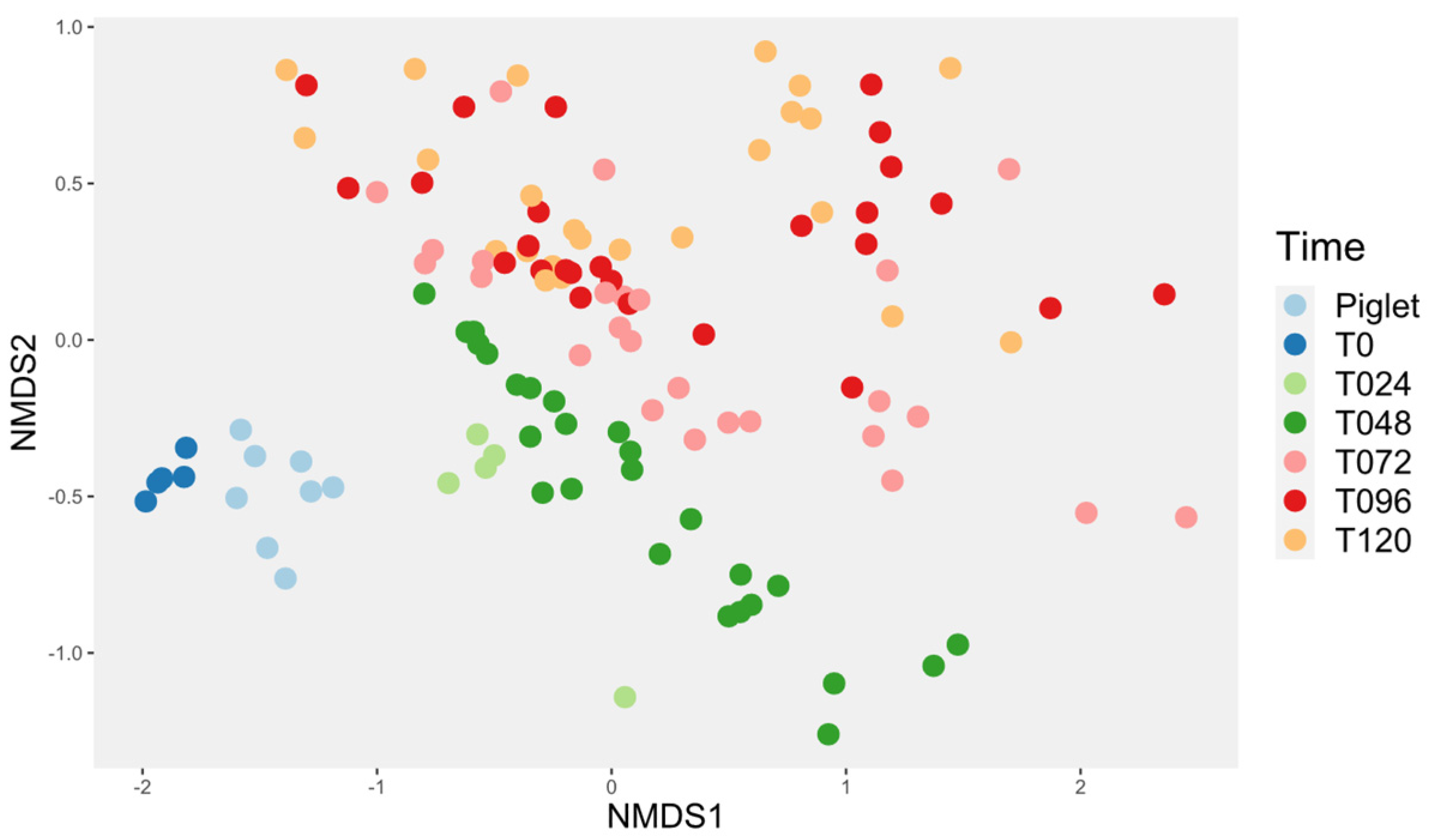
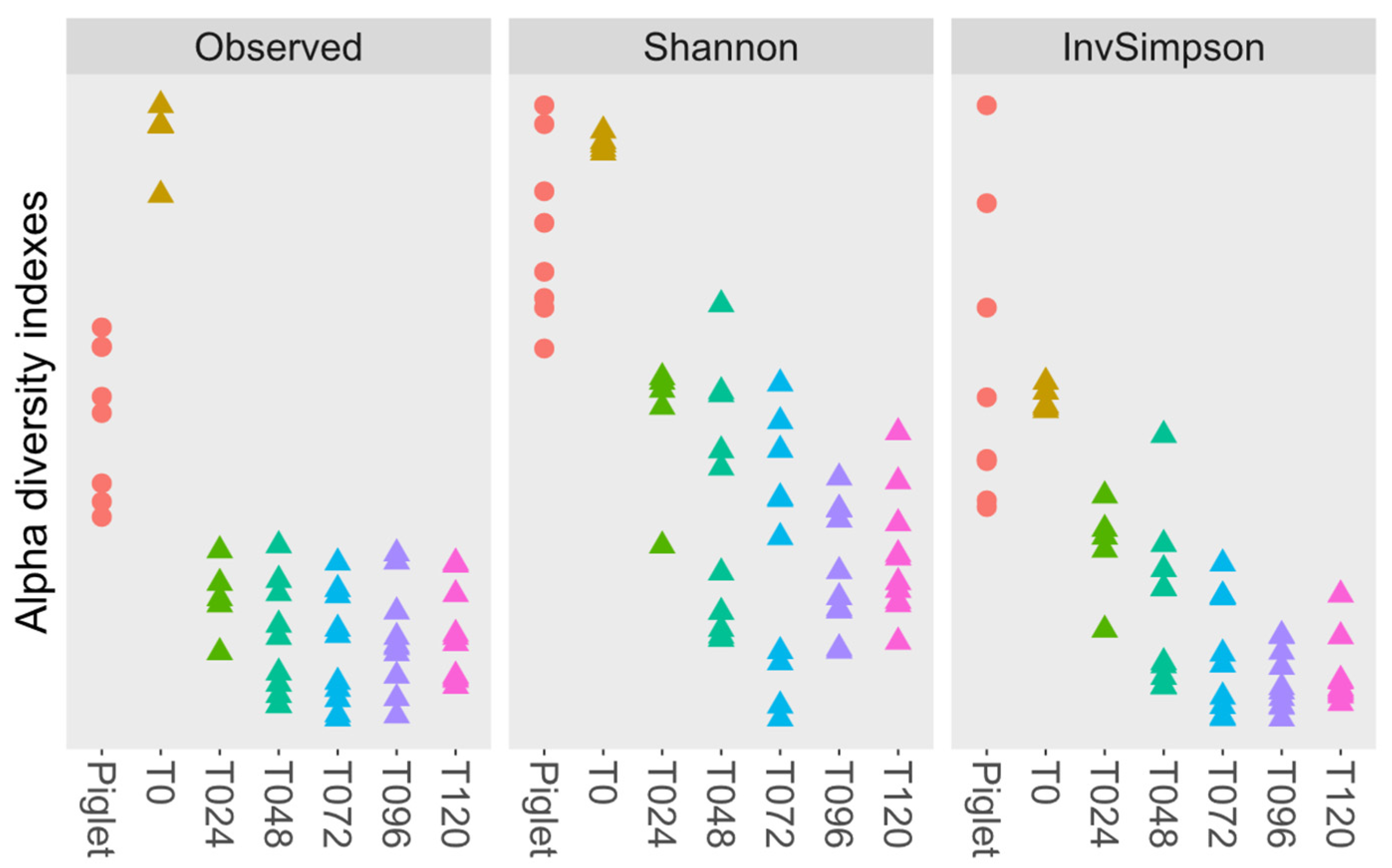
3.4. Microbiota Diversity
3.4.1. Probiotic Effect on Microbiota Diversity
3.4.2. Probiotic Effect on the Microbiota Composition
3.4.3. Investigation of the Alteration in Microbial Composition in the Bioreactor under Different Conditions
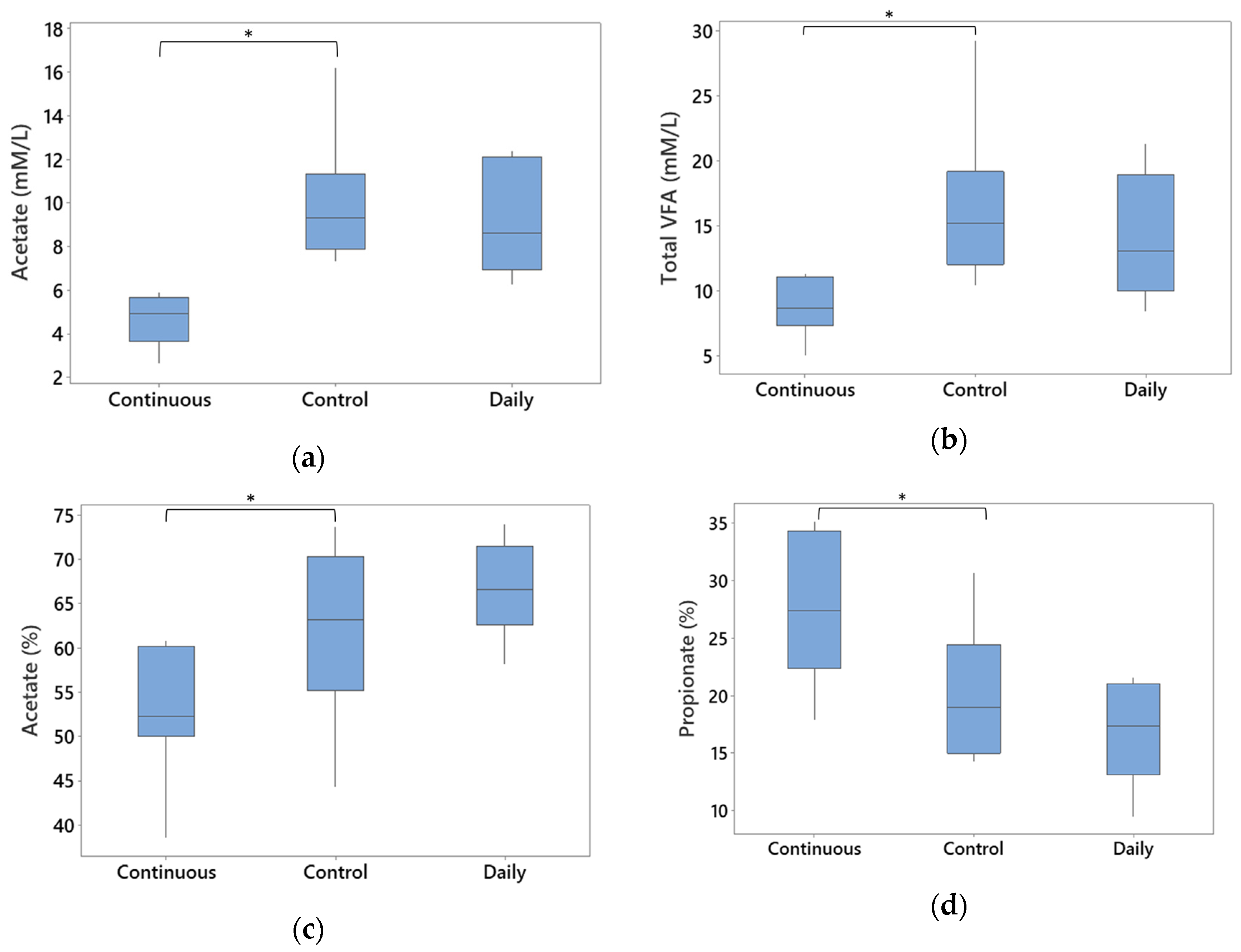
3.5. Total Gross Energy and SCFAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thomas, M.; Murray, R. Estimating the burden of food-borne illness in Canada. Can. Commun. Dis. Rep. 2014, 40, 299–302. [Google Scholar] [CrossRef]
- U.S. Food And Drug Administration. Get the Facts about Salmonella. 2023. Available online: https://www.fda.gov/animal-veterinary/animal-health-literacy/get-facts-about-salmonella#:~:text=sick%20since%201885.-,The%20Statistics%20at%20a%20Glance,for%20most%20of%20these%20cases (accessed on 15 December 2023).
- Gong, B.; Li, H.; Feng, Y.; Zeng, S.; Zhuo, Z.; Luo, J.; Chen, X.; Li, X. Prevalence, Serotype Distribution and Antimicrobial Resistance of Non-Typhoidal Salmonella in Hospitalized Patients in Conghua District of Guangzhou, China. Front. Cell. Infect. Microbiol. 2022, 12, 805384. [Google Scholar] [CrossRef]
- Gouvernement du Québec. Salmonellose. Available online: https://www.quebec.ca/sante/problemes-de-sante/a-z/salmonellose (accessed on 15 December 2023).
- Soliani, L.; Rugna, G.; Prosperi, A.; Chiapponi, C.; Luppi, A. Salmonella Infection in Pigs: Disease, Prevalence, and a Link between Swine and Human Health. Pathogens 2023, 12, 1267. [Google Scholar] [CrossRef]
- Duggan, S.J.; Mannion, C.; Prendergast, D.M.; Leonard, N.; Fanning, S.; Gonzales-Barron, U.; Egan, J.; Butler, F.; Duffy, G. Tracking the Salmonella Status of Pigs and Pork from Lairage through the Slaughter Process in the Republic of Ireland. J. Food Prot. 2010, 73, 2148–2160. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S. Salmonella in the pork production chain and its impact on human health in the European Union. Epidemiol. Infect. 2017, 145, 1513–1526. [Google Scholar] [CrossRef]
- Kim, H.B.; Isaacson, R.E. Salmonella in Swine: Microbiota Interactions. Annu. Rev. Anim. Biosci. 2017, 5, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Griffith, R.W.; Carlson, S.A.; Krull, A.C. Salmonellosis. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2019; pp. 912–925. [Google Scholar]
- Bearson, S.M.D. Salmonella in Swine: Prevalence, Multidrug Resistance, and Vaccination Strategies. Annu. Rev. Anim. Biosci. 2022, 10, 373–393. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Soufi, L.; Cenatus, S.; Archambault, M.; Butaye, P. Current Insights Regarding the Role of Farm Animals in the Spread of Antimicrobial Resistance from a One Health Perspective. Vet. Sci. 2022, 9, 480. [Google Scholar] [CrossRef]
- Ojha, S.; Kostrzynska, M.; Delhalle, L.; Saegerman, C.; Messens, W.; Farnir, F.; Korsak, N.; van der Stede, Y.; Daube, G. Approaches for Reducing Salmonella in Pork Production. J. Food Prot. 2007, 70, 2676–2694. [Google Scholar] [CrossRef]
- Boyen, F.; Haesebrouck, F.; Vanparys, A.; Volf, J.; Mahu, M.; Van Immerseel, F.; Rychlik, I.; Dewulf, J.; Ducatelle, R.; Pasmans, F. Coated fatty acids alter virulence properties of Salmonella Typhimurium and decrease intestinal colonization of pigs. Vet. Microbiol. 2008, 132, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Nazzaro, F.; Fratianni, F.; De Martino, L.; Coppola, R.; De Feo, V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals 2013, 6, 1451–1474. [Google Scholar] [CrossRef]
- FAO; WHO. Probiotics in Food—Health and Nutritional Properties and Guidelines for Evaluation. 2006. p. 56. Available online: https://www.fao.org/documents/card/en?details=7c102d95-2fd5-5b22-8faf-f0b2e68dfbb6/ (accessed on 15 December 2023).
- Liu, W.C.; Ye, M.; Liao, J.H.; Zhao, Z.H.; Kim, I.H.; An, L.L. Application of Complex Probiotics in Swine Nutrition—A Review. Ann. Anim. Sci. 2018, 18, 335–350. [Google Scholar] [CrossRef]
- Park, S.-O.; Seo, K.-H. Digital livestock systems and probiotic mixtures can improve the growth performance of swine by enhancing immune function, cecal bacteria, short-chain fatty acid, and nutrient digestibility. Front. Vet. Sci. 2023, 10, 1126064. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, D.; Wang, J.-H.; Li, H.-H.; Liu, J.-L.; Liu, A.-H.; Wang, J.-M.; Guan, G.-Q.; Luo, J.-X.; Yin, H.; et al. Assessment of probiotic adhesion and inhibitory effect on Escherichia coli and Salmonella adhesion. Arch. Microbiol. 2021, 203, 6267–6274. [Google Scholar] [CrossRef]
- Pahumunto, N.; Dahlen, G.; Teanpaisan, R. Evaluation of Potential Probiotic Properties of Lactobacillus and Bacillus Strains Derived from Various Sources for Their Potential Use in Swine Feeding. Probiotics Antimicrob. Proteins 2023, 15, 479–490. [Google Scholar] [CrossRef]
- Barba-Vidal, E.; Martín-Orúe, S.M.; Castillejos, L. Review: Are we using probiotics correctly in post-weaning piglets? Animal 2018, 12, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Thorsen, L.; Kpikpi, E.N.; Stuer-Lauridsen, B.; Cantor, M.D.; Nielsen, B.; Brockmann, E.; Derkx, P.M.F.; Jespersen, L. Characterization of Bacillus spp. strains for use as probiotic additives in pig feed. Appl. Microbiol. Biotechnol. 2014, 98, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.T.; Hoon, J.; Mun, H.-S.; Yang, C.-J. Evaluation of Lactobacillus and Bacillus-based probiotics as alternatives to antibiotics in enteric microbial challenged weaned piglets. Afr. J. Microbiol. Res. 2014, 8, 96–104. [Google Scholar]
- Wang, Y.; Cho, J.; Chen, Y.; Yoo, J.; Huang, Y.; Kim, H.; Kim, I. The effect of probiotic BioPlus 2B® on growth performance, dry matter and nitrogen digestibility and slurry noxious gas emission in growing pigs. Livest. Sci. 2009, 120, 35–42. [Google Scholar] [CrossRef]
- Danilova, I.; Sharipova, M. The Practical Potential of Bacilli and Their Enzymes for Industrial Production. Front. Microbiol. 2020, 11, 1782. [Google Scholar] [CrossRef] [PubMed]
- Magar, S.T. Bioreactor-Definition, Design, Principle, Parts, Types, Applications, Limitations. 2023. Available online: https://microbenotes.com/bioreactor/ (accessed on 15 December 2023).
- Bellerose, M.; Fravalo, P.; Mainville, I.; Arcand, Y.; Thibodeau, A. A short-term bioreactor assay to assess the effect of essential oils on a microbiota derived from piglet’s intestinal content. Acta Vet. Scand. 2023, 65, 17. [Google Scholar] [CrossRef]
- Cordonnier, C.; Thévenot, J.; Etienne-Mesmin, L.; Denis, S.; Alric, M.; Livrelli, V.; Blanquet-Diot, S. Dynamic In Vitro Models of the Human Gastrointestinal Tract as Relevant Tools to Assess the Survival of Probiotic Strains and Their Interactions with Gut Microbiota. Microorganisms 2015, 3, 725–745. [Google Scholar] [CrossRef] [PubMed]
- Westfall, S.; Carracci, F.; Estill, M.; Zhao, D.; Wu, Q.-L.; Shen, L.; Simon, J.; Pasinetti, G.M. Optimization of probiotic therapeutics using machine learning in an artificial human gastrointestinal tract. Sci. Rep. 2021, 11, 1067. [Google Scholar] [CrossRef] [PubMed]
- Fleury, M.A.; Le Goff, O.; Denis, S.; Chaucheyras-Durand, F.; Jouy, E.; Kempf, I.; Alric, M.; Blanquet-Diot, S. Development and validation of a new dynamic in vitro model of the piglet colon (PigutIVM): Application to the study of probiotics. Appl. Microbiol. Biotechnol. 2017, 101, 2533–2547. [Google Scholar] [CrossRef]
- Tanner, S.A.; Chassard, C.; Berner, A.Z.; Lacroix, C. Synergistic effects of Bifidobacterium thermophilum RBL67 and selected prebiotics on inhibition of Salmonella colonization in the swine proximal colon PolyFermS model. Gut Pathog. 2014, 6, 44. [Google Scholar] [CrossRef]
- Azaroual, S.E.; Kasmi, Y.; Aasfar, A.; El Arroussi, H.; Zeroual, Y.; El Kadiri, Y.; Zrhidri, A.; Elfahime, E.; Sefiani, A.; Kadmiri, I.M. Investigation of bacterial diversity using 16S rRNA sequencing and prediction of its functionalities in Moroccan phosphate mine ecosystem. Sci. Rep. 2022, 12, 3741. [Google Scholar] [CrossRef]
- Peterson, C.T.; Santiago, J.P.; Iablokov, S.N.; Chopra, D.; Rodionov, D.A.; Peterson, S.N. Short-Chain Fatty Acids Modulate Healthy Gut Microbiota Composition and Functional Potential. Curr. Microbiol. 2022, 79, 128. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Zhan, Z.; Tang, H.; Zhang, Y.; Huang, X.; Xu, M. Potential of gut-derived short-chain fatty acids to control enteric pathogens. Front. Microbiol. 2022, 13, 976406. [Google Scholar] [CrossRef]
- Gao, X.; Yu, J.; Chang, L.; Wang, Y.; Sun, X.; Mu, G.; Qian, F. In vitro antibacterial activity of Bacillus coagulans T242 on Caco-2 cells infected with Salmonella Typhimurium. Food Biosci. 2023, 52, 102512. [Google Scholar] [CrossRef]
- Sudan, S.; Flick, R.; Nong, L.; Li, J. Potential Probiotic Bacillus subtilis Isolated from a Novel Niche Exhibits Broad Range Antibacterial Activity and Causes Virulence and Metabolic Dysregulation in Enterotoxic E. coli. Microorganisms 2021, 9, 1483. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, I.; Choi, Y.; Cho, J. Bacillus strains as feed additives: In vitro evaluation of its potential probiotic properties. Rev. Colomb. Cienc. Pecu. 2012, 25, 577–585. [Google Scholar]
- Wang, X.; Howe, S.; Wei, X.; Deng, F.; Tsai, T.; Chai, J.; Xiao, Y.; Yang, H.; Maxwell, C.V.; Li, Y.; et al. Comprehensive Cultivation of the Swine Gut Microbiome Reveals High Bacterial Diversity and Guides Bacterial Isolation in Pigs. mSystems 2021, 6, e0047721. [Google Scholar] [CrossRef]
- Guo, X.; Li, D.; Lu, W.; Piao, X.; Chen, X. Screening of Bacillus strains as potential probiotics and subsequent confirmation of the in vivo effectiveness of Bacillus subtilis MA139 in pigs. Antonie Van Leeuwenhoek 2006, 90, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, J.; Liu, Y.; Cao, L.; Qiu, W.; Qin, M. Probiotic Effects of Bacillus subtilis on Growth Performance and Intestinal Microecological Balance of Growing-to-Finishing Pigs. J. Food Biochem. 2023, 2023, 7150917. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Lawlor, P.G.; Ranjitkar, S.; Cormican, P.; Villodre, C.; Bouwhuis, M.A.; Marsh, A.; Crispie, F.; Rattigan, R.; Gardiner, G.E. Intestinal microbiota modulation and improved growth in pigs with post-weaning antibiotic and ZnO supplementation but only subtle microbiota effects with Bacillus altitudinis. Sci. Rep. 2021, 11, 23304. [Google Scholar] [CrossRef] [PubMed]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Saladrigas-García, M.; Solà-Oriol, D.; López-Vergé, S.; D’Angelo, M.; Collado, M.C.; Nielsen, B.; Faldyna, M.; Pérez, J.F.; Martín-Orúe, S.M. Potential effect of two Bacillus probiotic strains on performance and fecal microbiota of breeding sows and their piglets. J. Anim. Sci. 2022, 100, skac163. [Google Scholar] [CrossRef]
- Wang, X.; Tian, Z.; Azad, M.; Zhang, W.; Blachier, F.; Wang, Z.; Kong, X. Dietary supplementation with Bacillus mixture modifies the intestinal ecosystem of weaned piglets in an overall beneficial way. J. Appl. Microbiol. 2021, 130, 233–246. [Google Scholar] [CrossRef]
- Jo, H.E.; Kwon, M.-S.; Whon, T.W.; Kim, D.W.; Yun, M.; Lee, J.; Shin, M.-Y.; Kim, S.-H.; Choi, H.-J. Alteration of Gut Microbiota After Antibiotic Exposure in Finishing Swine. Front. Microbiol. 2021, 12, 596002. [Google Scholar] [CrossRef]
- Li, J.; Wang, H.; Li, N.; Zhang, Y.; Lü, X.; Liu, B. Antibiotic susceptibility and biofilm-forming ability of Veillonella strains. Anaerobe 2022, 78, 102667. [Google Scholar] [CrossRef]
- Kolenbrander, P. The Genus Veillonella. In The Prokaryotes: Volume 4: Bacteria: Firmicutes, Cyanobacteria; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 1022–1040. [Google Scholar]
- Bescucci, D.M.; Moote, P.E.; Polo, R.O.; Uwiera, R.R.E.; Inglis, G.D. Salmonella enterica Serovar Typhimurium Temporally Modulates the Enteric Microbiota and Host Responses To Overcome Colonization Resistance in Swine. Appl. Environ. Microbiol. 2020, 86, e01569-20. [Google Scholar] [CrossRef]
- Langlais, M.; Thibodeau, A.; Fravalo, P. A metagenomic analysis of the pre-enrichment step for the isolation of Salmonella spp. from pig feces. J. Microbiol. Methods 2019, 157, 43–46. [Google Scholar] [CrossRef]
- Argüello, H.; Estellé, J.; Leonard, F.C.; Crispie, F.; Cotter, P.D.; O’sullivan, O.; Lynch, H.; Walia, K.; Duffy, G.; Lawlor, P.G.; et al. Influence of the Intestinal Microbiota on Colonization Resistance to Salmonella and the Shedding Pattern of Naturally Exposed Pigs. mSystems 2019, 4, e00021-19. [Google Scholar] [CrossRef] [PubMed]
- Larivière-Gauthier, G.; Thibodeau, A.; Letellier, A.; Yergeau, E.; Fravalo, P. Reduction of Salmonella Shedding by Sows during Gestation in Relation to Its Fecal Microbiome. Front. Microbiol. 2017, 8, 2219. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Park, W. Gut microbiomes and their metabolites shape human and animal health. J. Microbiol. 2018, 56, 151–153. [Google Scholar] [CrossRef] [PubMed]
- La Reau, A.J.; Suen, G. The Ruminococci: Key symbionts of the gut ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Precup, G.; Vodnar, D.-C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: A comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia-a new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef]
- Vasquez, R.; Oh, J.K.; Song, J.H.; Kang, D.-K. Gut microbiome-produced metabolites in pigs: A review on their biological functions and the influence of probiotics. J. Anim. Sci. Technol. 2022, 64, 671–695. [Google Scholar] [CrossRef] [PubMed]
- Ilinskaya, O.N.; Ulyanova, V.V.; Yarullina, D.R.; Gataullin, I.G. Secretome of Intestinal Bacilli: A Natural Guard against Pathologies. Front. Microbiol. 2017, 8, 1666. [Google Scholar] [CrossRef] [PubMed]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef]
- Berge, A.C.; Wierup, M. Nutritional strategies to combat Salmonella in mono-gastric food animal production. Animal 2012, 6, 557–564. [Google Scholar] [CrossRef]
- Kurian, S.J.; Baral, T.; M, S.S.; Rao, M. Chapter 28—Role of probiotics and prebiotics in digestion, metabolism, and immunity. In Nutrition and Functional Foods in Boosting Digestion, Metabolism and Immune Health; Bagchi, D., Ohia, S.E., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 501–522. [Google Scholar]
- Kaewtapee, C.; Burbach, K.; Tomforde, G.; Hartinger, T.; Camarinha-Silva, A.; Heinritz, S.; Seifert, J.; Wiltafsky, M.; Mosenthin, R.; Rosenfelder-Kuon, P. Effect of Bacillus subtilis and Bacillus licheniformis supplementation in diets with low- and high-protein content on ileal crude protein and amino acid digestibility and intestinal microbiota composition of growing pigs. J. Anim. Sci. Biotechnol. 2017, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Mun, D.; Kyoung, H.; Kong, M.; Ryu, S.; Jang, K.B.; Baek, J.; Park, K.I.; Song, M.; Kim, Y. Effects of Bacillus-based probiotics on growth performance, nutrient digestibility, and intestinal health of weaned pigs. J. Anim. Sci. Technol. 2021, 63, 1314–1327. [Google Scholar] [CrossRef]
- Mahmoud, K.; Obeidat, B.; Al-Sadi, M.; Hatahet, S. Effect of Bacillus subtilis supplementation and dietary crude protein level on growth performance and intestinal morphological changes of meat type chicken. Livest. Sci. 2017, 195, 99–104. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Tang, W.; Diao, H.; Zhang, H.; Yan, H.; Liu, J. Dietary Complex Probiotic Supplementation Changed the Composition of Intestinal Short-Chain Fatty Acids and Improved the Average Daily Gain of Growing Pigs. Vet. Sci. 2023, 10, 79. [Google Scholar] [CrossRef]
- Hu, J.; Kim, I.H. Effect of Bacillus subtilis C-3102 Spores as a Probiotic Feed Supplement on Growth Performance, Nutrient Digestibility, Diarrhea Score, Intestinal Microbiota, and Excreta Odor Contents in Weanling Piglets. Animals 2022, 12, 316. [Google Scholar] [CrossRef]
- Azizi, A.F.N.; Uemura, R.; Omori, M.; Sueyoshi, M.; Yasuda, M. Effects of Probiotics on Growth and Immunity of Piglets. Animals 2022, 12, 1786. [Google Scholar] [CrossRef]

| Dilution of the Supernatant | ||||||
|---|---|---|---|---|---|---|
| Inhibition diameter (mm) | Strains | Not diluted | 1/2 | 1/5 | 1/10 | 1/100 |
| Salmonella Typhimurium | 11 | 10 | 9 | 0 | 0 | |
| Staphylococcus aureus ATCC 25923 | 25 | 21 | 15 | 10 | 0 | |
| Enterococcus faecalis ATCC 29212 | 21 | 17 | 10 | 0 | 0 | |
| Escherichia coli ATCC 25922 | 13 | 11 | 8 | 7 | 0 | |
| Dilution of the Pellet | ||||||
|---|---|---|---|---|---|---|
| Inhibition diameter (mm) | Strains | Not diluted | 1/2 | 1/5 | 1/10 | 1/100 |
| Salmonella Typhimurium | 0 | 0 | 0 | 0 | 0 | |
| Staphylococcus aureus ATCC 25923 | 26 | 20 | 13 | 0 | 0 | |
| Enterococcus faecalis ATCC 29212 | 21 | 17 | 0 | 0 | 0 | |
| Escherichia coli ATCC 25922 | 17 | 13 | 11 | 11 | 0 | |
| Conditions (in CFU) | T0 | T10 min | T6, 24 and 120 h |
|---|---|---|---|
| PRO-P2702 whole + 106 Salmonella Typhimurium | Present | Present | Absent |
| PRO-P2702 whole + 103 Salmonella Typhimurium | Present | Absent | Absent |
| PRO-P2702 supernatant + 106 Salmonella Typhimurium | Present | Present | Absent |
| PRO-P2702 supernatant + 103 Salmonella Typhimurium | Present | Absent | Absent |
| 106 Salmonella Typhimurium control | Present | Present | Present |
| 103 Salmonella Typhimurium control | Present | Present | Present |
| PRO-P2702 control | Absent | Absent | Absent |
| Negative control | Absent | Absent | Absent |
| Control | Daily | Continuous | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Time | Obsv | Sh | Isimp | Obsv | Sh | Isimp | Obsv | Sh | Isimp |
| T048 | 232 | 2.8 | 10.3 | 200 | 2.5 | 10.3 | 198 | 2.4 | 7.4 |
| T072 | 240 | 2.6 | 7.7 | 170 | 2.3 | 6.6 | 172 | 2.0 | 4.8 |
| T096 | 242 a | 2.6 d | 6.4 f | 184 a | 2.4 d | 5.7 f | 138 b | 2.0 e | 5.0 g |
| T120 | 238 a | 2.6 d | 6.7 f | 175 b | 2.4 | 5.6 g | 124 c | 2.1 e | 4.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grandmont, A.; Rhouma, M.; Létourneau-Montminy, M.-P.; Thériault, W.; Mainville, I.; Arcand, Y.; Leduc, R.; Demers, B.; Thibodeau, A. Characterization of the Effects of a Novel Probiotic on Salmonella Colonization of a Piglet-Derived Intestinal Microbiota Using Improved Bioreactor. Animals 2024, 14, 787. https://doi.org/10.3390/ani14050787
Grandmont A, Rhouma M, Létourneau-Montminy M-P, Thériault W, Mainville I, Arcand Y, Leduc R, Demers B, Thibodeau A. Characterization of the Effects of a Novel Probiotic on Salmonella Colonization of a Piglet-Derived Intestinal Microbiota Using Improved Bioreactor. Animals. 2024; 14(5):787. https://doi.org/10.3390/ani14050787
Chicago/Turabian StyleGrandmont, Amely, Mohamed Rhouma, Marie-Pierre Létourneau-Montminy, William Thériault, Isabelle Mainville, Yves Arcand, Roland Leduc, Bruno Demers, and Alexandre Thibodeau. 2024. "Characterization of the Effects of a Novel Probiotic on Salmonella Colonization of a Piglet-Derived Intestinal Microbiota Using Improved Bioreactor" Animals 14, no. 5: 787. https://doi.org/10.3390/ani14050787
APA StyleGrandmont, A., Rhouma, M., Létourneau-Montminy, M.-P., Thériault, W., Mainville, I., Arcand, Y., Leduc, R., Demers, B., & Thibodeau, A. (2024). Characterization of the Effects of a Novel Probiotic on Salmonella Colonization of a Piglet-Derived Intestinal Microbiota Using Improved Bioreactor. Animals, 14(5), 787. https://doi.org/10.3390/ani14050787






