Sex Differences in Cochlear Transcriptomes in Horseshoe Bats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
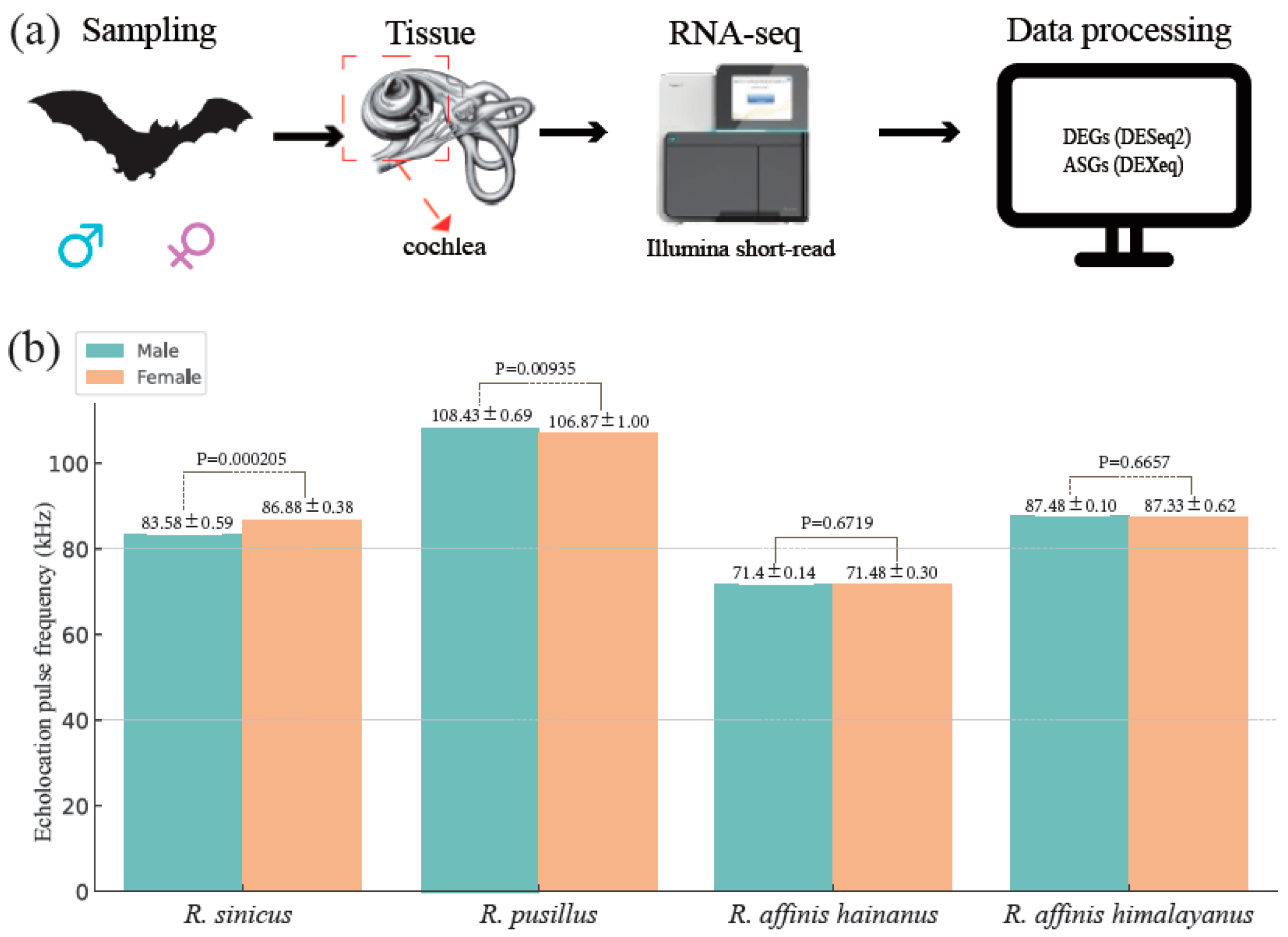
2.1. Study System and Tissue Collection
2.2. RNA Extraction, Library Construction, Sequencing, and Mapping
2.3. Differential Expression Analysis
2.4. Alternative Splicing Analysis
2.5. Comparison of Differential Expression and Alternative Splicing
2.6. Functional Enrichment Analysis
3. Results
3.1. Characterization of Echolocation Pulse Frequency Variation across the Four Rhinolophus Taxa
3.2. Cochlear RNA-Seq Data Collection and Mapping
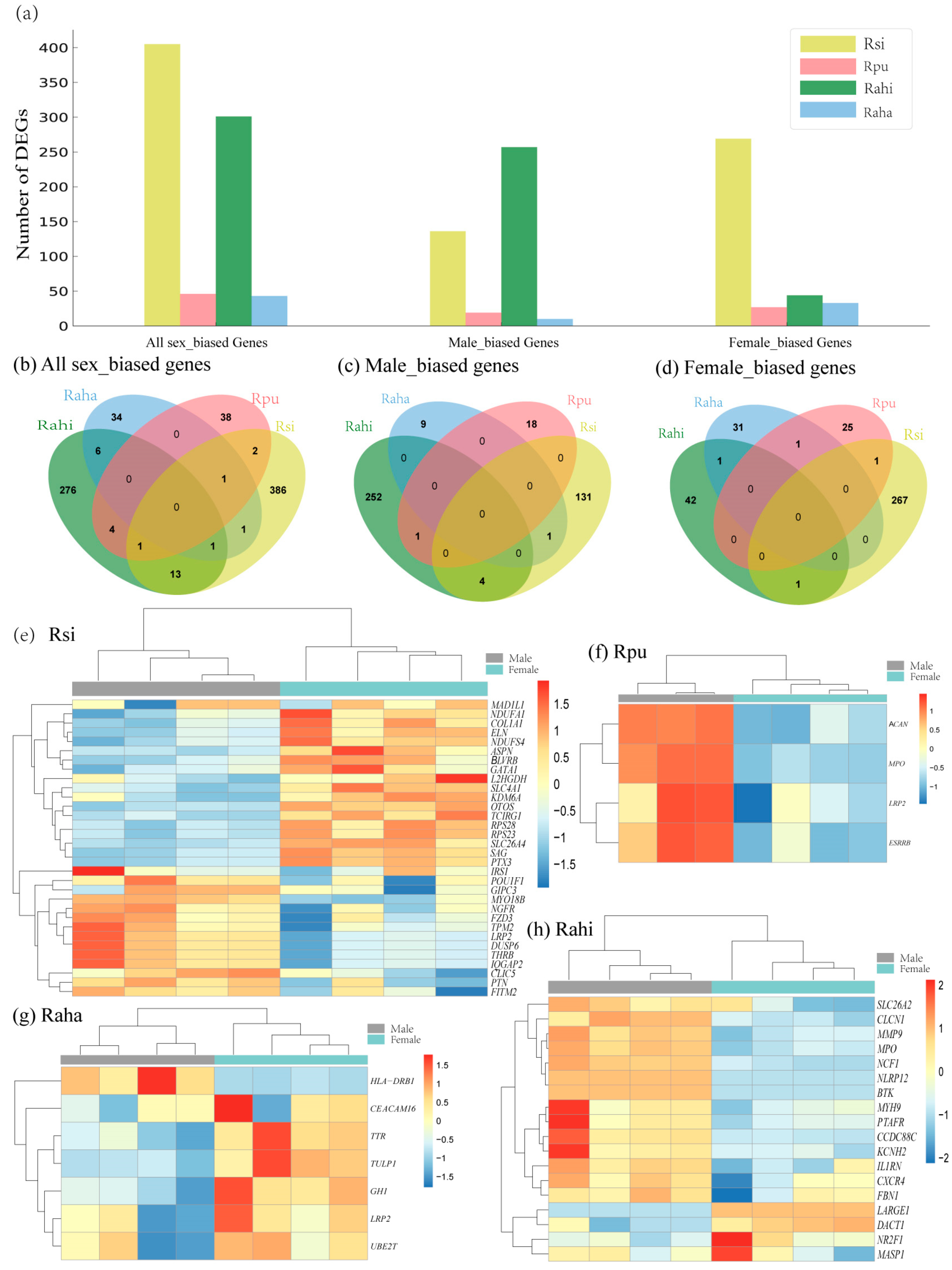
3.3. Characterization of Sex-Biased DEGs in Cochlea of Four Rhinolophus Taxa
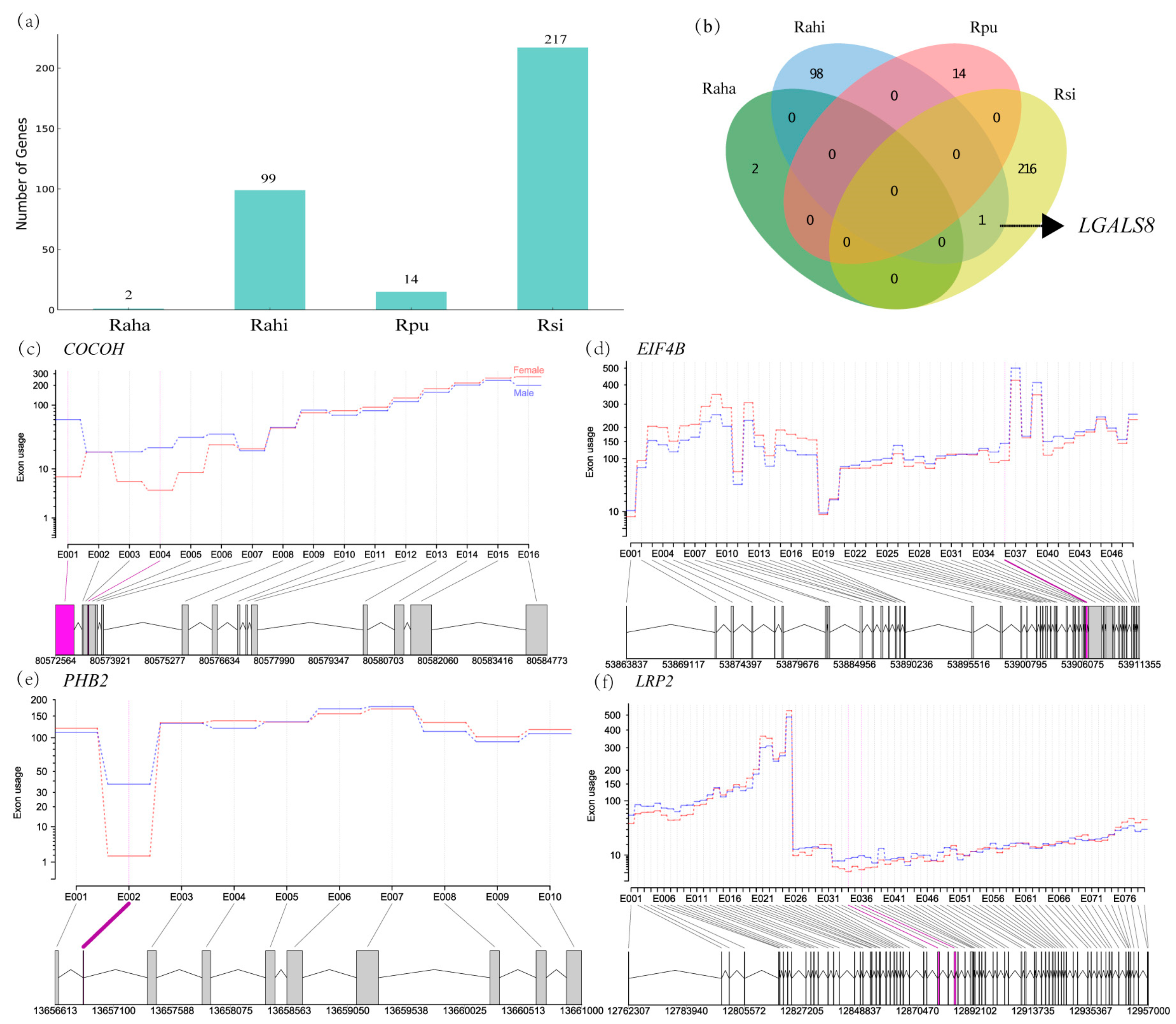
3.4. Characterization of Sex-Biased ASGs in Cochlea of Four Rhinolophus Taxa
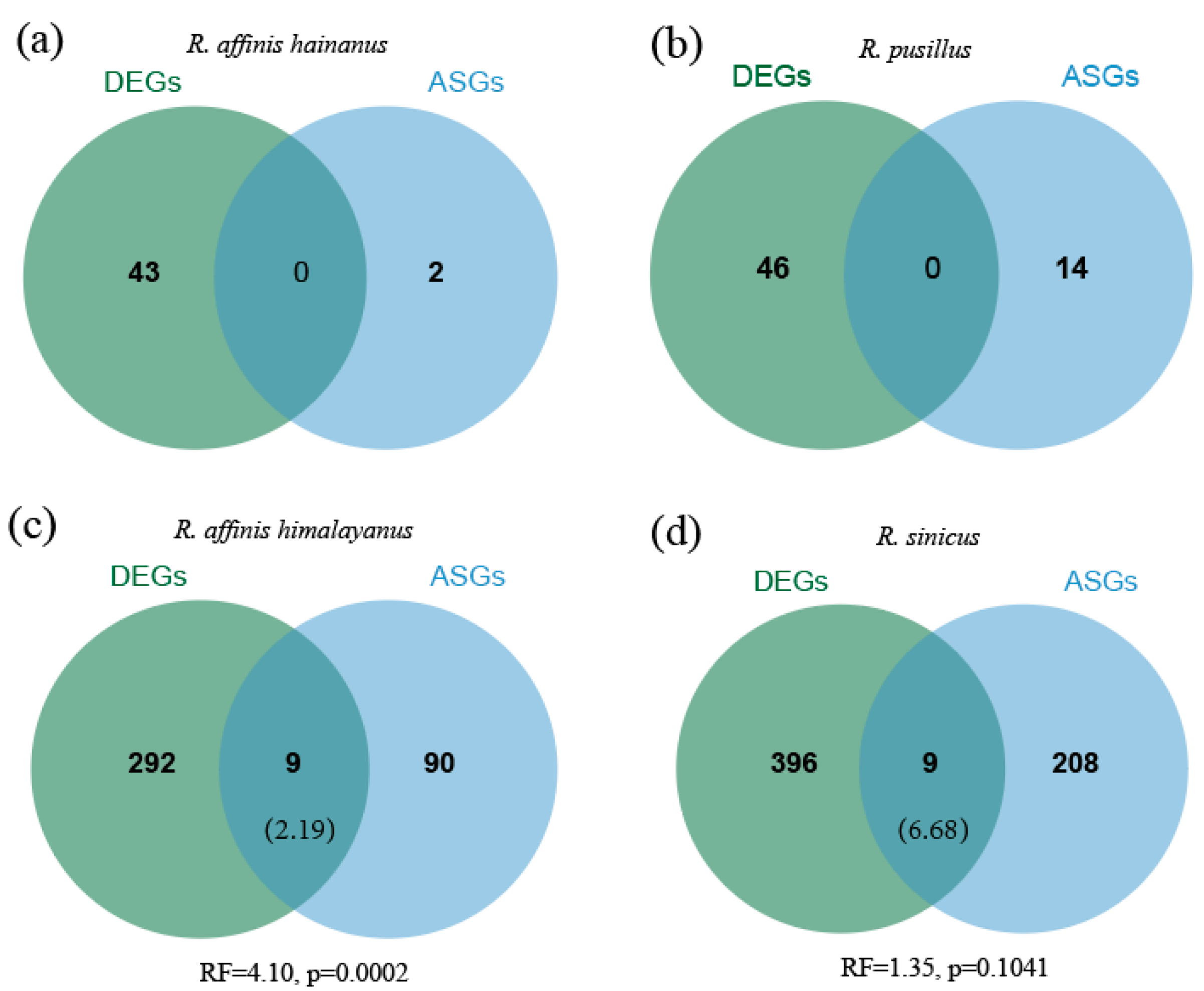
3.5. Comparisons of Differential Expression and Alternative Splicing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tellechea, J.S.; Norbis, W. Sexual dimorphism in sound production and call characteristics in the striped weakfish Cynoscion guatucupa. Zool. Stud. 2012, 51, 946–955. [Google Scholar]
- Emerson, S.B.; Boyd, S.K. Mating vocalizations of female frogs: Control and evolutionary mechanisms. Brain. Behav. Evol. 1999, 53, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Taoka, M.; Sato, T.; Kamada, T.; Okumura, H. Sexual dimorphism of chatter-calls and vocal sex recognition in Leach’s Storm-Petrels (Oceanodroma leucorhoa). The Auk 1989, 106, 498–501. [Google Scholar]
- Yamaguchi, A. A sexually dimorphic learned birdsong in the Northern Cardinal. Condor 1998, 100, 504–511. [Google Scholar] [CrossRef]
- Radford, A. Voice breaking in males results in sexual dimorphism of green woodhoopoe calls. Behaviour 2004, 141, 555–569. [Google Scholar] [CrossRef]
- Efremova, K.O.; Volodin, I.A.; Volodina, E.V.; Frey, R.; Lapshina, E.N.; Soldatova, N.V. Developmental changes of nasal and oral calls in the goitred gazelle Gazella subgutturosa, a nonhuman mammal with a sexually dimorphic and descended larynx. Naturwissenschaften 2011, 98, 919–931. [Google Scholar] [CrossRef] [PubMed]
- Charlton, B.; Reby, D. The evolution of acoustic size exaggeration in terrestrial mammals. Nat. Commun. 2016, 7, 12739. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, J.W.; Vehrencamp, S.L. Principles of Animal Communication; Sinauer Associates: Sunderland, MA, USA, 1998; Volume 132. [Google Scholar]
- Puts, D.A.; Hill, A.K.; Bailey, D.H.; Walker, R.S.; Rendall, D.; Wheatley, J.R.; Welling, L.L.; Dawood, K.; Cárdenas, R.; Burriss, R.P. Sexual selection on male vocal fundamental frequency in humans and other anthropoids. Proc. R. Soc. B-Biol. Sci. 2016, 283, 20152830. [Google Scholar] [CrossRef]
- Schnitzler, H.-U.; Moss, C.F.; Denzinger, A. From spatial orientation to food acquisition in echolocating bats. Trends Ecol. Evol. 2003, 18, 386–394. [Google Scholar] [CrossRef]
- Henson, O.W., Jr.; Bishop, A.; Keating, A.; Kobler, J.; Henson, M.; Wilson, B.; Hansen, R. Biosonar imaging ofinsects by Pteronotus p. parnellii, the mustached bat. Natl. Geogr. Res. 1987, 3, 82–101. [Google Scholar]
- Grinnell, A.D.; Gould, E.; Fenton, M.B. A history of the study of echolocation. Bat Bioacoustics 2016, 54, 1–24. [Google Scholar] [CrossRef]
- Jones, G.; Siemers, B.M. The communicative potential of bat echolocation pulses. J. Comp. Physiol. A 2011, 197, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Siemers, B.M.; Beedholm, K.; Dietz, C.; Dietz, I.; Ivanova, T. Is species identity, sex, age or individual quality conveyed by echolocation call frequency in European horseshoe bats? Acta Chiropterol. 2005, 7, 259–274. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, H.; Huang, X.; Sun, K.; Feng, J. Comparative cochlear transcriptomics of echolocating bats provides new insights into different nervous activities of CF bat species. Sci. Rep. 2018, 8, 15934. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, H.; Liu, T.; Liu, S.; Jin, L.; Huang, X.; Dai, W.; Sun, K.; Feng, J. Gene expression vs. sequence divergence: Comparative transcriptome sequencing among natural Rhinolophus ferrumequinum populations with different acoustic phenotypes. Front. Zool. 2019, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Chen, W.; Wang, J.; Zhang, L.; Rossiter, S.J.; Mao, X. Echolocation call frequency variation in horseshoe bats: Molecular basis revealed by comparative transcriptomics. Proc. Roy. Soc. B-Biol. Sci. 2020, 287, 20200875. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, W.; Mao, X. Characterization of microRNA and gene expression in the cochlea of an echolocating bat (Rhinolophus affinis). Ecol. Evol. 2022, 12, e9025. [Google Scholar] [CrossRef]
- Hutson, A.; Rossiter, S.; Csorba, G.; Burgin, C. Family Rhinolophidae (horseshoe bats). In Handbook of the Mammals of the World: Bats; Lynx Edicions: Barcelona, Spain, 2019; pp. 260–332. [Google Scholar]
- Schuller, G.; Pollak, G. Disproportionate frequency representation in the inferior colliculus of Doppler-compensating greater horseshoe bats: Evidence for an acoustic fovea. J. Comp. Physiol. 1979, 132, 47–54. [Google Scholar] [CrossRef]
- Jones, G.; Gordon, T.; Nightingale, J. Sex and age differences in the echolocation calls of the lesser horseshoe bat, Rhinolophus hipposideros. Mammalia 1992, 56, 189–194. [Google Scholar] [CrossRef]
- Yoshino, H.; Matsumura, S.; Kinjo, K.; Tamura, H.; Ota, H.; Izawa, M. Geographical variation in echolocation call and body size of the Okinawan least horseshoe bat, Rhinolophus pumilus (Mammalia: Rhinolophidae), on Okinawa-jima Island, Ryukyu Archipelago, Japan. Zool. Sci. 2006, 23, 661–667. [Google Scholar] [CrossRef]
- Chen, S.-F.; Jones, G.; Rossiter, S.J. Determinants of echolocation call frequency variation in the Formosan lesser horseshoe bat (Rhinolophus monoceros). Proc. Biol. Sci. 2009, 276, 3901–3909. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Metzner, W.; You, Y.; Liu, S.; Lu, G.; Li, S.; Wang, L.; Feng, J. Variation in the resting frequency of Rhinolophus pusillus in Mainland China: Effect of climate and implications for conservation. J. Acoust. Soc. Am. 2010, 128, 2204–2211. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Luo, L.; Kimball, R.T.; Wei, X.; Jin, L.; Jiang, T.; Li, G.; Feng, J. Geographic variation in the acoustic traits of greater horseshoe bats: Testing the importance of drift and ecological selection in evolutionary processes. PLoS ONE 2013, 8, e70368. [Google Scholar] [CrossRef] [PubMed]
- Neuweiler, G.; Metzner, W.; Heilmann, U.; Rübsamen, R.; Eckrich, M.; Costa, H. Foraging behaviour and echolocation in the rufous horseshoe bat (Rhinolophus rouxi) of Sri Lanka. Behav. Ecol. Sociobiol. 1987, 20, 53–67. [Google Scholar] [CrossRef]
- Xie, L.; Sun, K.; Jiang, T.; Liu, S.; Lu, G.; Jin, L.; Feng, J. The effects of cultural drift on geographic variation in echolocation calls of the Chinese rufous horseshoe bat (Rhinolophus sinicus). Ethology 2017, 123, 532–541. [Google Scholar] [CrossRef]
- Mao, X.; He, G.; Zhang, J.; Rossiter, S.J.; Zhang, S. Lineage divergence and historical gene flow in the Chinese horseshoe bat (Rhinolophus sinicus). PLoS ONE 2013, 8, e56786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Russo, D.; Jones, G.; Mucedda, M. Influence of age, sex and body size on echolocation calls of Mediterranean and Mehely’s horseshoe bats, Rhinolophus euryale and R. mehelyi (Chiroptera: Rhinolophidae). Mammalia 2001, 65, 429–436. [Google Scholar] [CrossRef]
- Mao, X.; Zhu, G.; Zhang, L.; Zhang, S.; Rossiter, S.J. Differential introgression among loci across a hybrid zone of the intermediate horseshoe bat (Rhinolophus affinis). BMC Evol. Biol. 2014, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H.; Parsch, J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007, 8, 689–698. [Google Scholar] [CrossRef]
- Mank, J.E. The transcriptional architecture of phenotypic dimorphism. Nat. Ecol. Evol. 2017, 1, 0006. [Google Scholar] [CrossRef]
- Pointer, M.A.; Harrison, P.W.; Wright, A.E.; Mank, J.E. Masculinization of gene expression is associated with exaggeration of male sexual dimorphism. PLoS Genet. 2013, 9, e1003697. [Google Scholar] [CrossRef] [PubMed]
- Dick, C.; Reznick, D.N.; Hayashi, C.Y. Sex-biased expression between guppies varying in the presence of ornamental coloration. PeerJ 2018, 6, e5782. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, H.; Wang, W.; Fu, C.; Zhang, W.; Han, F.; Wu, H. Genomic and transcriptomic insights into molecular basis of sexually dimorphic nuptial spines in Leptobrachium leishanense. Nat. Commun. 2019, 10, 5551. [Google Scholar] [CrossRef] [PubMed]
- Gibilisco, L.; Zhou, Q.; Mahajan, S.; Bachtrog, D. Alternative splicing within and between Drosophila species, sexes, tissues, and developmental stages. PLoS Genet. 2016, 12, e1006464. [Google Scholar] [CrossRef] [PubMed]
- Naftaly, A.S.; Pau, S.; White, M.A. Long-read RNA sequencing reveals widespread sex-specific alternative splicing in threespine stickleback fish. Genome Res. 2021, 31, 1486–1497. [Google Scholar] [CrossRef]
- Rogers, T.F.; Palmer, D.H.; Wright, A.E. Sex-specific selection drives the evolution of alternative splicing in birds. Mol. Biol. Evol. 2021, 38, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Agrawal, A.F. Two forms of sexual dimorphism in gene expression in Drosophila melanogaster: Their coincidence and evolutionary genetics. Mol. Biol. Evol. 2023, 40, msad091. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, W.; Li, Q.; Mao, X. Sex differences in gene expression and alternative splicing in the Chinese horseshoe bat. PeerJ 2023, 11, e15231. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Liang, R.; Cui, C.; Zhu, Y.; Zhang, F.; Zhang, J.; Chen, X. Comparative transcriptome analysis provides insights into the molecular mechanisms of high-frequency hearing differences between the sexes of Odorrana tormota. BMC Genom. 2022, 23, 296. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Ren, L.; Wu, C.; Guo, L.; Yao, J.; Wang, C.; Xiao, Y.; Pisco, A.O.; Wu, Z.; Lei, X.; Liu, Y. Single-cell transcriptional atlas of the Chinese horseshoe bat (Rhinolophus sinicus) provides insight into the cellular mechanisms which enable bats to be viral reservoirs. biorxiv 2020, 175778. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Bodner, M.; Castriilo, J.-L.; Theill, L.E.; Deerinck, T.; Ellisman, M.; Karin, M. The pituitary-specific transcription factor GHF-1 is a homeobox-containing protein. Cell 1988, 55, 505–518. [Google Scholar] [CrossRef]
- Delprat, B.; Boulanger, A.; Wang, J.; Beaudoin, V.; Guitton, M.J.; Venteo, S.; Dechesne, C.J.; Pujol, R.; Lavigne-Rebillard, M.; Puel, J.-L. Downregulation of otospiralin, a novel inner ear protein, causes hair cell degeneration and deafness. J. Neurosci. 2002, 22, 1718–1725. [Google Scholar] [CrossRef] [PubMed]
- Delprat, B.; Ruel, J.; Guitton, M.J.; Hamard, G.; Lenoir, M.; Pujol, R.; Puel, J.-L.; Brabet, P.; Hamel, C.P. Deafness and cochlear fibrocyte alterations in mice deficient for the inner ear protein otospiralin. Mol. Cell. Biol. 2005, 25, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, Y.; Zhang, P.; Liu, C.; Wang, J.; Gao, H.; Hoelzel, A.R.; Seim, I.; Lv, M.; Lin, M. Comparative genomics provides insights into the aquatic adaptations of mammals. Proc. Natl. Acad. Sci. USA 2021, 118, e2106080118. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Lei, M.; Liu, Y.; Zhang, S. Comparative inner ear transcriptome analysis between the Rickett’s big-footed bats (Myotis ricketti) and the greater short-nosed fruit bats (Cynopterus sphinx). BMC Genom. 2013, 14, 916. [Google Scholar] [CrossRef]
- Wei, Y.; Chiang, W.-C.; Sumpter, R.; Mishra, P.; Levine, B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 2017, 168, 224–238e210. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Guan, M.; Shang, H.; Teng, Y.; Gao, Y.; Wang, B.; Ma, Z.; Cao, X.; Li, Y. The expression of PHB2 in the cochlea: Possible relation to age-related hearing loss. Cell Biol. Int. 2021, 45, 2490–2498. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-Y.; Liang, L.; Li, G.-S.; Murphy, R.W.; Zhang, Y.-P. Parallel evolution of auditory genes for echolocation in bats and toothed whales. PLoS Genet. 2012, 8, e1002788. [Google Scholar] [CrossRef] [PubMed]
- Jebb, D.; Huang, Z.; Pippel, M.; Hughes, G.M.; Lavrichenko, K.; Devanna, P.; Winkler, S.; Jermiin, L.S.; Skirmuntt, E.C.; Katzourakis, A. Six reference-quality genomes reveal evolution of bat adaptations. Nature 2020, 583, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Bidon-Wagner, N.; Le Pennec, J.-P. Human galectin-8 isoforms and cancer. Glycoconj. J. 2002, 19, 557–563. [Google Scholar] [CrossRef]
- Schuchmann, M.; Puechmaille, S.J.; Siemers, B.M. Horseshoe bats recognise the sex of conspecifics from their echolocation calls. Acta Chiropterol. 2012, 14, 161–166. [Google Scholar] [CrossRef]
- Fu, Z.-Y.; Dai, X.-Y.; Xu, N.; Shi, Q.; Li, G.-J.; Li, B.; Li, J.; Li, J.; Tang, J.; Jen, P.H.-S. Sexual dimorphism in echolocation pulse parameters of the CF-FM bat, Hipposideros pratti. Zool. Stud. 2015, 54, 44. [Google Scholar] [CrossRef]
- Yin, W.; Mendoza, L.; Monzon-Sandoval, J.; Urrutia, A.O.; Gutierrez, H. Emergence of co-expression in gene regulatory networks. PLoS ONE 2021, 16, e0247671. [Google Scholar] [CrossRef] [PubMed]
- Biederman, J.; Kim, J.W.; Doyle, A.E.; Mick, E.; Fagerness, J.; Smoller, J.W.; Faraone, S.V. Sexually dimorphic effects of four genes (COMT, SLC6A2, MAOA, SLC6A4) in genetic associations of ADHD: A preliminary study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008, 147, 1511–1518. [Google Scholar] [CrossRef]
- Kantarci, S.; Al-Gazali, L.; Hill, R.S.; Donnai, D.; Black, G.C.; Bieth, E.; Chassaing, N.; Lacombe, D.; Devriendt, K.; Teebi, A. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat. Genet. 2007, 39, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Faridi, R.; Yousaf, R.; Gu, S.; Inagaki, S.; Turriff, A.E.; Pelstring, K.; Guan, B.; Naik, A.; Griffith, A.J.; Adadey, S.M. Variants of LRP2, encoding a multifunctional cell-surface endocytic receptor, associated with hearing loss and retinal dystrophy. Clin. Genet. 2023, 103, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, K.C.; Sabir, S.; Shum, M.; Meng, Y.; Acín-Pérez, R.; Lang, J.M.; Floyd, R.R.; Vergnes, L.; Seldin, M.M.; Fuqua, B.K. Sex-specific metabolic functions of adipose Lipocalin-2. Mol. Metab. 2019, 30, 30–47. [Google Scholar] [CrossRef] [PubMed]
- Catalán, A.; Macias-Munoz, A.; Briscoe, A.D. Evolution of sex-biased gene expression and dosage compensation in the eye and brain of Heliconius butterflies. Mol. Biol. Evol. 2018, 35, 2120–2134. [Google Scholar] [CrossRef]
- Price, P.D.; Palmer Droguett, D.H.; Taylor, J.A.; Kim, D.W.; Place, E.S.; Rogers, T.F.; Mank, J.E.; Cooney, C.R.; Wright, A.E. Detecting signatures of selection on gene expression. Nat. Ecol. Evol. 2022, 6, 1035–1045. [Google Scholar] [CrossRef]
- Meyfour, A.; Pahlavan, S.; Ansari, H.; Baharvand, H.; Salekdeh, G.H. Down-regulation of a male-specific H3K4 demethylase, KDM5D, impairs cardiomyocyte differentiation. J. Proteome Res. 2019, 18, 4277–4282. [Google Scholar] [CrossRef]
- Godfrey, A.K.; Naqvi, S.; Chmátal, L.; Chick, J.M.; Mitchell, R.N.; Gygi, S.P.; Skaletsky, H.; Page, D.C. Quantitative analysis of Y-Chromosome gene expression across 36 human tissues. Genome Res. 2020, 30, 860–873. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Mao, X. The roles of different gene expression regulators in acoustic variation in the intermediate horseshoe bat revealed by long-read and short-read RNA sequencing data. Curr. Zool. 2023, zoad045. [Google Scholar] [CrossRef]
- Blekhman, R.; Marioni, J.C.; Zumbo, P.; Stephens, M.; Gilad, Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010, 20, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Karlebach, G.; Veiga, D.F.; Mays, A.D.; Chatzipantsiou, C.; Barja, P.P.; Chatzou, M.; Kesarwani, A.K.; Danis, D.; Kararigas, G.; Zhang, X.A. The impact of biological sex on alternative splicing. BioRxiv 2018, 490904. [Google Scholar] [CrossRef]
- Teeling, E.C.; Jones, G.; Rossiter, S.J. Phylogeny, genes, and hearing: Implications for the evolution of echolocation in bats. Bat Bioacoustics 2016, 54, 25–54. [Google Scholar] [CrossRef]
- Mantica, F.; Irimia, M. The 3D-Evo space: Evolution of gene expression and alternative splicing regulation. Annu. Rev. Genet. 2022, 56, 315–337. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Duan, C.; Lan, L.; Chen, W.; Mao, X. Sex Differences in Cochlear Transcriptomes in Horseshoe Bats. Animals 2024, 14, 1177. https://doi.org/10.3390/ani14081177
Wu J, Duan C, Lan L, Chen W, Mao X. Sex Differences in Cochlear Transcriptomes in Horseshoe Bats. Animals. 2024; 14(8):1177. https://doi.org/10.3390/ani14081177
Chicago/Turabian StyleWu, Jianyu, Can Duan, Linjing Lan, Wenli Chen, and Xiuguang Mao. 2024. "Sex Differences in Cochlear Transcriptomes in Horseshoe Bats" Animals 14, no. 8: 1177. https://doi.org/10.3390/ani14081177
APA StyleWu, J., Duan, C., Lan, L., Chen, W., & Mao, X. (2024). Sex Differences in Cochlear Transcriptomes in Horseshoe Bats. Animals, 14(8), 1177. https://doi.org/10.3390/ani14081177




