Enhancing Immunoglobulin G Goat Colostrum Determination Using Color-Based Techniques and Data Science
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Models
2.2.1. Decision Trees
2.2.2. Neural Network
2.3. Performance Evaluation
2.4. Tools and Development Environment
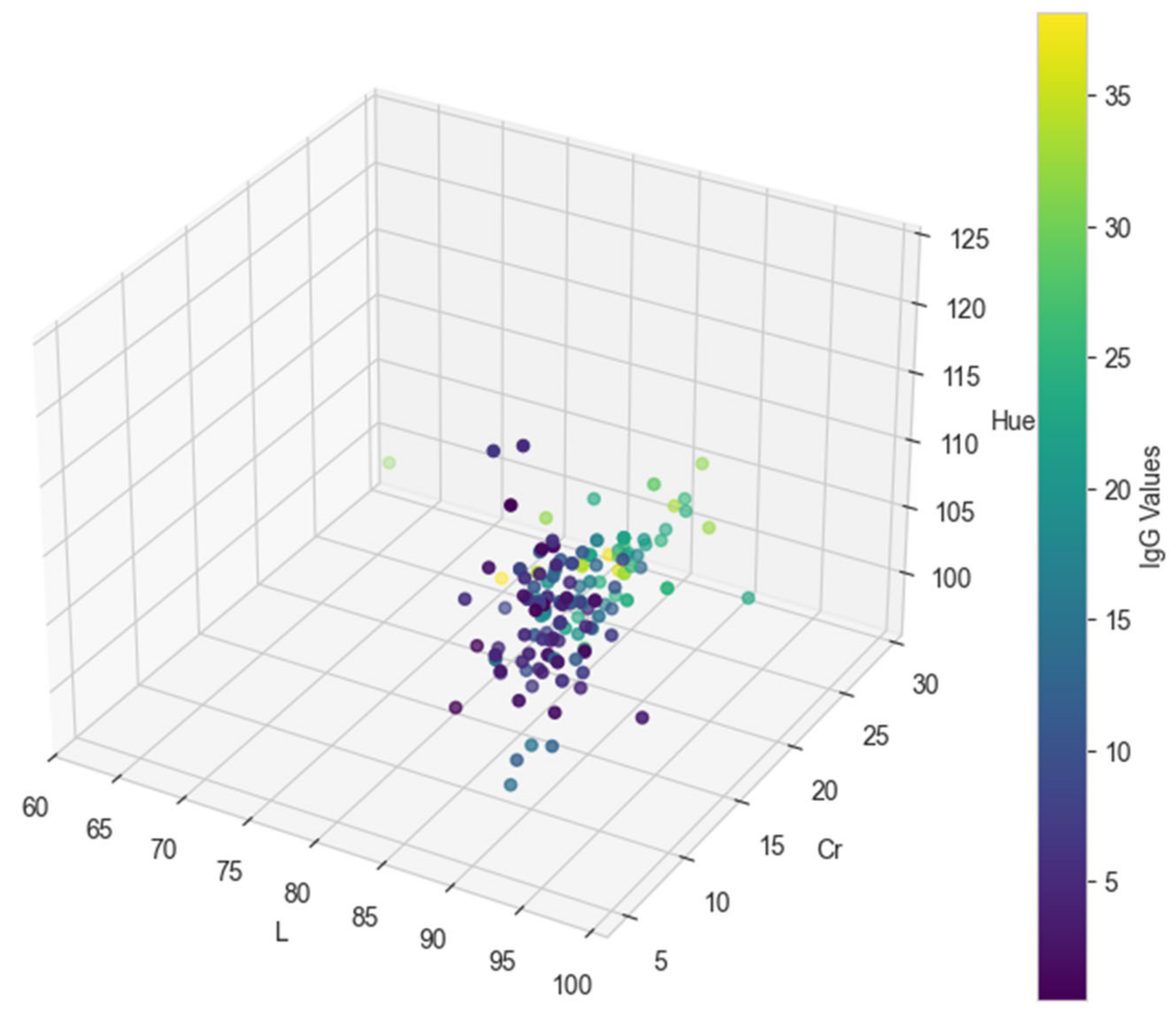
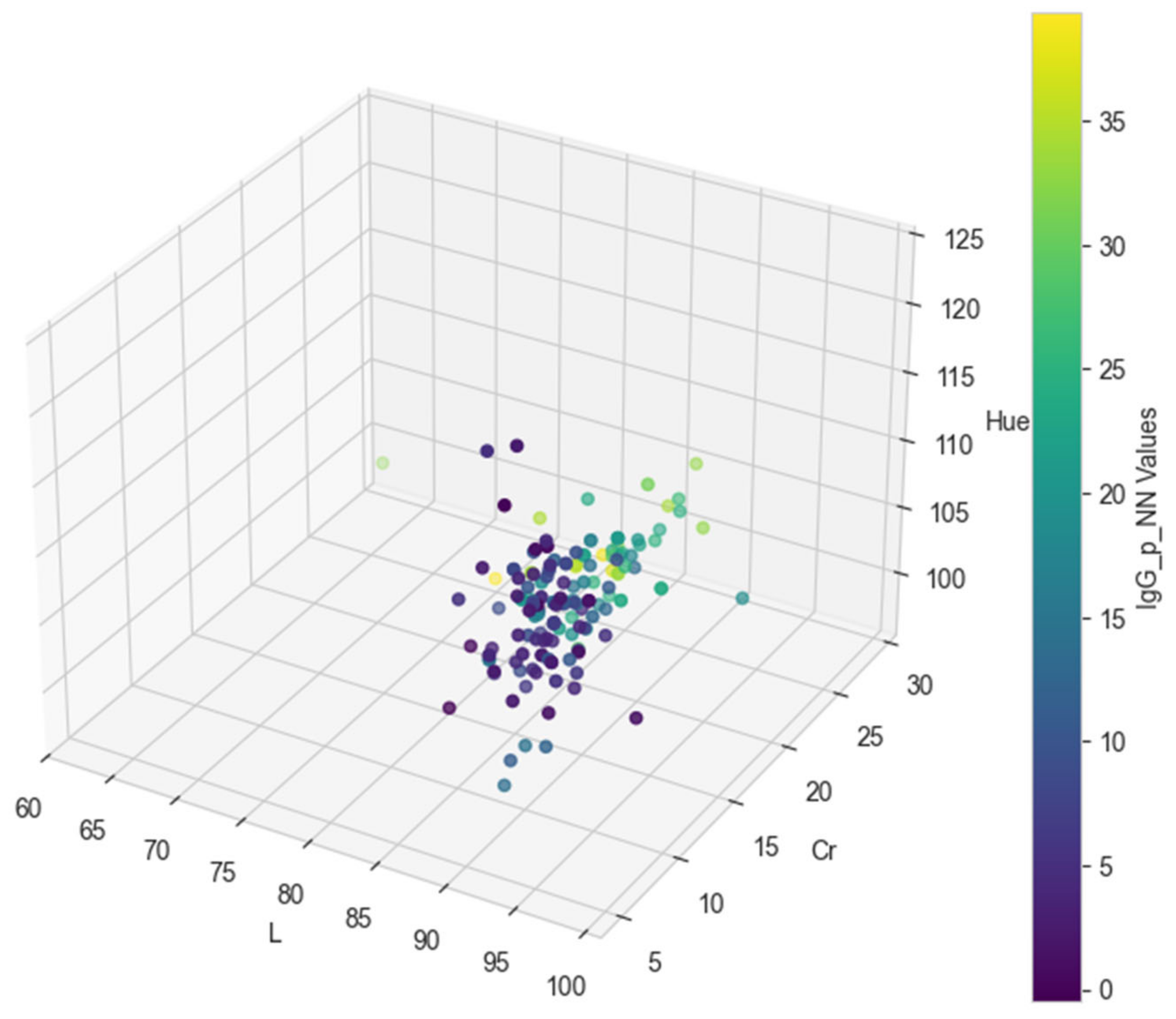
3. Results
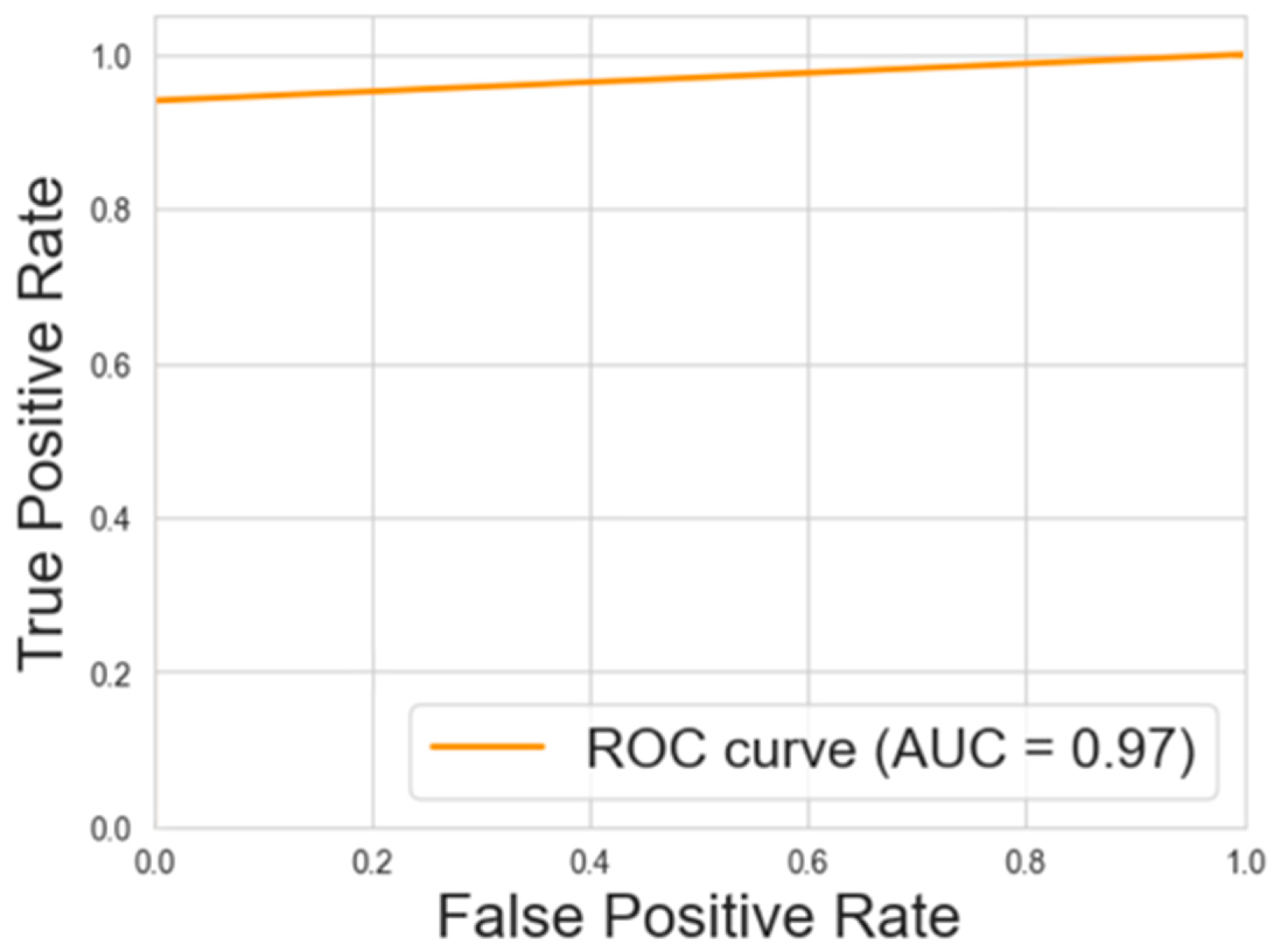
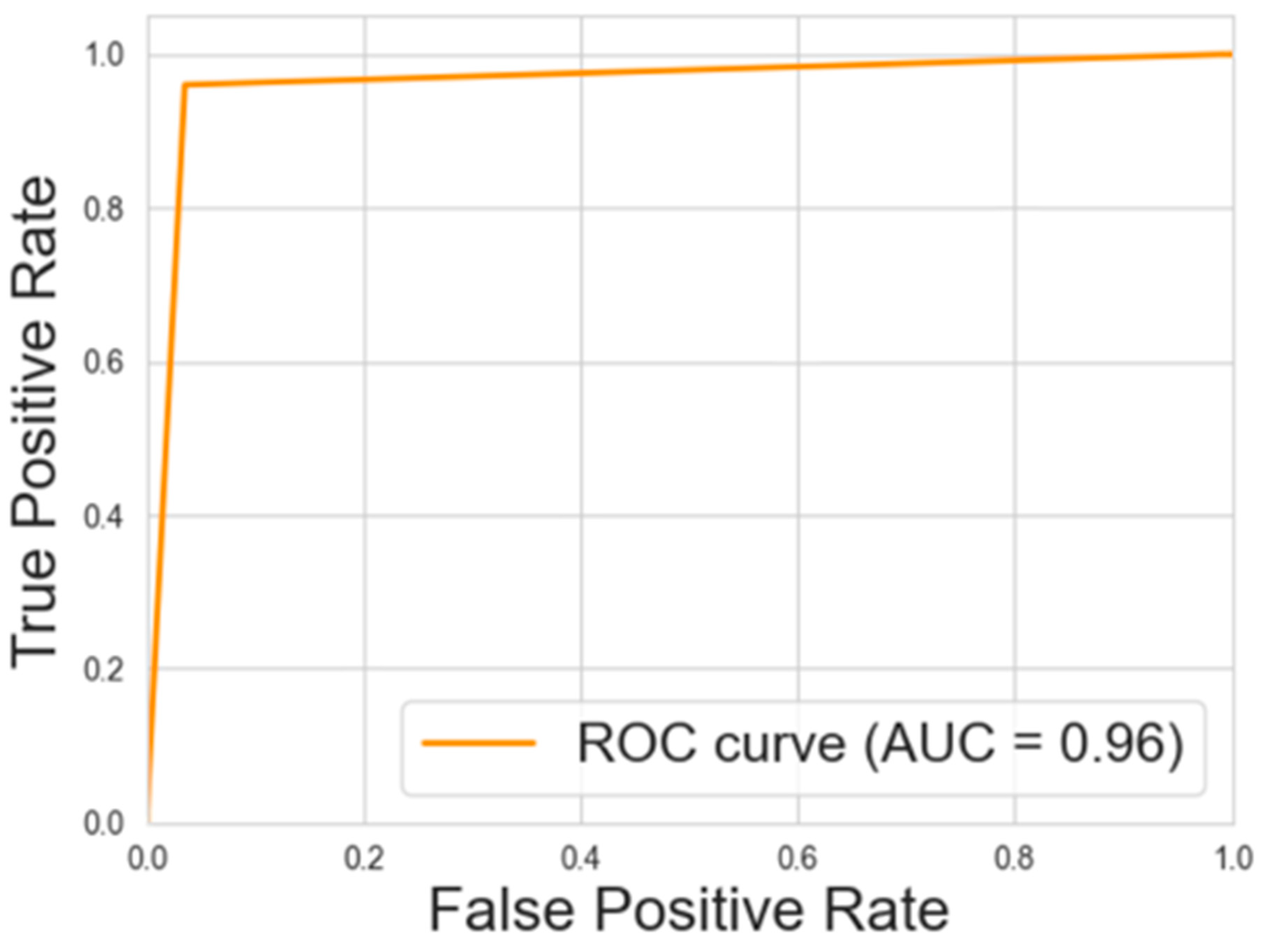
3.1. Predictive Model Performance Evaluation
3.2. Classification Metrics After Factorization
4. Discussion
4.1. Results Overview
4.2. Comparison with the Previous Study
4.3. Advancements Through Machine Learning (ML) and Deep Learning (DL) in Biological Data
4.4. Future Implications
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| L | Cr | Hue | IgG | IgG_p_DT | IgG_p_NN |
|---|---|---|---|---|---|
| 83.86 | 28.40 | 103.56 | 28.71 | 28.71 | 30.98 |
| 94.09 | 7.68 | 113.23 | 3.97 | 3.97 | 3.59 |
| 90.34 | 23.28 | 107.14 | 24.30 | 24.30 | 24.78 |
| 94.62 | 6.48 | 106.67 | 1.78 | 1.78 | 1.45 |
| 77.88 | 21.54 | 99.76 | 38.19 | 38.19 | 39.41 |
| 89.33 | 18.90 | 105.50 | 32.96 | 32.96 | 33.40 |
| 90.17 | 11.12 | 111.03 | 19.69 | 19.69 | 19,00 |
| 90.98 | 10.59 | 114.24 | 1.92 | 1.92 | 0.90 |
| 88.66 | 8.48 | 107.07 | 15.48 | 15.48 | 15.49 |
| 92.59 | 8.70 | 101.80 | 12.80 | 12.80 | 12.45 |
| 91.00 | 8.00 | 108.00 | 5.00 | 5.00 | 4.40 |
| 89.89 | 21.57 | 106.10 | 24.62 | 24.62 | 25.03 |
| 87.97 | 18.06 | 108.19 | 16.87 | 16.87 | 16.82 |
| 87.01 | 20.36 | 105.11 | 37.40 | 37.40 | 38.65 |
| 88.73 | 24.22 | 106.38 | 33.30 | 33.30 | 34.01 |
| 91.10 | 18.47 | 108.83 | 20.71 | 20.71 | 20.39 |
| 88.45 | 15.92 | 105.55 | 16.80 | 16.80 | 16.81 |
| 93.12 | 11.32 | 108.48 | 8.32 | 8.32 | 7.29 |
| 93.50 | 10.03 | 105.26 | 4.60 | 4.60 | 3.93 |
| 84.68 | 16.76 | 105.82 | 33.94 | 33.94 | 33.87 |
| 90.36 | 8.32 | 100.39 | 11.42 | 11.42 | 11.54 |
| 91.08 | 11.01 | 107.17 | 5.08 | 5.08 | 4.59 |
| 90.31 | 6.56 | 109.40 | 4.75 | 4.75 | 4.26 |
| 89.71 | 9.95 | 110.60 | 6.65 | 7.52 | 20.94 |
| 92.00 | 9.00 | 108.00 | 3.00 | 3.00 | 2.89 |
| 94.52 | 9.95 | 111.90 | 2.55 | 2.55 | 1.49 |
| 91.00 | 18.00 | 108.00 | 18.00 | 18.00 | 18.03 |
| 90.00 | 15.00 | 110.00 | 22.00 | 22.00 | 22.23 |
| 88.00 | 15.00 | 106.00 | 22.00 | 22.00 | 21.65 |
| 93.03 | 7.30 | 110.77 | 6.00 | 6.00 | 4.64 |
| 92.00 | 11.00 | 113.00 | 9.00 | 9.00 | 8.15 |
| 92.89 | 8.90 | 110.59 | 7.52 | 7.52 | 7.20 |
| 87.33 | 20.62 | 101.95 | 24.44 | 24.44 | 25.02 |
| 89.94 | 11.12 | 108.68 | 17.38 | 17.38 | 16.08 |
| 91.65 | 12.39 | 108.98 | 14.47 | 14.47 | 14,00 |
| 90.98 | 10.59 | 114.24 | 1.92 | 1.92 | 0.90 |
| 88.89 | 13.97 | 101.23 | 6.16 | 6.16 | 5.58 |
| 91.46 | 9.25 | 111.24 | 0.50 | 0.50 | 0.52 |
| 91.68 | 11.21 | 111.39 | 4.91 | 4.91 | 4.02 |
| 91.28 | 9.24 | 110.40 | 18.79 | 18.79 | 17.41 |
| 92.00 | 7.00 | 111.00 | 5.00 | 5.00 | 3.90 |
| 92.00 | 10.00 | 111.00 | 15.00 | 13.33 | 12.65 |
| 87.97 | 18.06 | 108.19 | 16.87 | 16.87 | 16.82 |
| 91.66 | 8.66 | 115.50 | 11.37 | 11.37 | 10.28 |
| 90.00 | 15.00 | 110.00 | 22.00 | 22.00 | 22.23 |
| 90.00 | 20.00 | 107.00 | 22.00 | 22.00 | 22.47 |
| 83.76 | 21.91 | 102.23 | 33.64 | 33.64 | 34.89 |
| 75.00 | 29.00 | 98.00 | 32.00 | 32.00 | 34.56 |
| 90.45 | 9.62 | 111.92 | 10.83 | 10.83 | 10.51 |
| 94.52 | 9.95 | 111.90 | 2.55 | 2.55 | 1.49 |
| 90.87 | 15.93 | 105.62 | 11.51 | 11.51 | 12.84 |
| 92.60 | 8.67 | 114.02 | 11.42 | 11.42 | 8.59 |
| 91.75 | 8.54 | 114.04 | 4.26 | 4.26 | 3.75 |
| 84.62 | 18.87 | 105.15 | 11.21 | 11.21 | 11.86 |
| 94.23 | 7.87 | 113.60 | 2.34 | 2.34 | 2.19 |
| 92.45 | 9.05 | 107.81 | 13.05 | 13.05 | 11.07 |
| 85.08 | 13.08 | 101.34 | 6.67 | 6.67 | 5.92 |
| 89.71 | 9.95 | 110.60 | 6.65 | 7.52 | 20.94 |
| 93.92 | 7.53 | 109.23 | 3.05 | 3.05 | 2.75 |
| 93.36 | 6.30 | 113.68 | 1.03 | 1.03 | 2.37 |
| 91.16 | 8.84 | 109.30 | 10.72 | 10.72 | 9.13 |
| 89.71 | 9.95 | 110.60 | 6.65 | 7.52 | 20.94 |
| 88.34 | 11.48 | 103.81 | 9.89 | 9.89 | 8.96 |
| 88.66 | 8.48 | 107.07 | 15.48 | 15.48 | 15.49 |
| 88.07 | 18.24 | 105.00 | 24.36 | 24.36 | 25.37 |
| 88.58 | 15.82 | 106.68 | 14.85 | 14.85 | 14.86 |
| 92.56 | 7.74 | 116.63 | 1.30 | 1.30 | 0.98 |
| 92.61 | 8.69 | 109.64 | 4.08 | 4.08 | 3.98 |
| 93.04 | 7.00 | 113.49 | 7.26 | 7.26 | 7.29 |
| 90.61 | 14.80 | 109.59 | 13.23 | 13.23 | 14.38 |
| 89.36 | 11.86 | 108.18 | 10.84 | 10.84 | 9.11 |
| 86.66 | 18.03 | 102.13 | 23.45 | 23.45 | 23.87 |
| 92.56 | 9.35 | 110.22 | 4.46 | 4.46 | 5.48 |
| 88.69 | 19.18 | 105.33 | 36.35 | 36.35 | 37.29 |
| 92.97 | 5.41 | 107.87 | 2.60 | 2.60 | 2.43 |
| 88.97 | 7.55 | 114.43 | 2.56 | 2.56 | 2.46 |
| 88.36 | 20.44 | 105.58 | 22.79 | 22.79 | 23.76 |
| 85.01 | 9.32 | 101.77 | 0.82 | 0.82 | -0.45 |
| 87.36 | 14.40 | 103.99 | 8.03 | 8.03 | 7.59 |
| 91.12 | 7.63 | 114.89 | 8.46 | 8.46 | 8.47 |
| 88.00 | 20.00 | 106.00 | 22.00 | 22.00 | 23.04 |
| 88.94 | 9.46 | 97.16 | 14.31 | 14.31 | 13.85 |
| 84.68 | 16.76 | 105.82 | 33.94 | 33.94 | 33.87 |
| 90.83 | 13.84 | 107.23 | 5.44 | 5.44 | 5.15 |
| 92.00 | 7.00 | 115.00 | 6.00 | 6.00 | 3.92 |
| 86.07 | 20.07 | 109.20 | 23.44 | 23.44 | 23.58 |
| 87.26 | 21.4 | 105.06 | 28.10 | 28.10 | 31.99 |
| 93.00 | 8.11 | 117.07 | 6.14 | 6.14 | 4.22 |
| 86.24 | 11.53 | 104.77 | 4.72 | 4.72 | 3.38 |
| 92.75 | 6.49 | 110.28 | 4.48 | 4.48 | 3.80 |
| 91.62 | 23.87 | 105.83 | 32.37 | 32.37 | 32.73 |
| 84.62 | 18.87 | 105.15 | 11.21 | 11.21 | 11.86 |
| 88.55 | 13.03 | 108.02 | 5.51 | 5.51 | 4.65 |
| 93.31 | 10.57 | 107.51 | 7.04 | 7.04 | 6.75 |
| 89.83 | 6.93 | 123.23 | 6.92 | 6.92 | 5.15 |
| 91.71 | 15.88 | 111.07 | 22.54 | 8.77 | 21.10 |
| 93.92 | 7.53 | 109.23 | 3.05 | 3.05 | 2.75 |
| 93.50 | 9.58 | 107.68 | 11.40 | 11.40 | 10.91 |
| 88.55 | 13.03 | 108.02 | 5.51 | 5.51 | 4.65 |
| 88.68 | 23.52 | 105.11 | 22.35 | 22.35 | 23.11 |
| 92.00 | 9.00 | 108.00 | 3.00 | 3.00 | 2.89 |
| 92.56 | 9.35 | 110.22 | 4.46 | 4.46 | 5.48 |
| 96.75 | 21.11 | 104.17 | 23.06 | 23.06 | 18.93 |
| 92.72 | 13.89 | 109.30 | 10.60 | 10.60 | 10.76 |
| 83.76 | 21.91 | 102.23 | 33.64 | 33.64 | 34.89 |
| 89.50 | 18.99 | 103.49 | 23.98 | 23.98 | 23.67 |
| 94.68 | 11.24 | 108.47 | 7.27 | 7.27 | 5.22 |
| 93.24 | 8.08 | 115.11 | 13.80 | 13.80 | 8.19 |
| 87.22 | 7.59 | 111.66 | 5.87 | 5.87 | 5.60 |
| 91.32 | 11.58 | 109.58 | 20.27 | 20.27 | 19.79 |
| 91.08 | 9.32 | 106.45 | 5.09 | 5.09 | 5.44 |
| 93.82 | 17.25 | 106.99 | 25.06 | 25.06 | 23.91 |
| 89.80 | 18.5 | 107.26 | 22.38 | 22.38 | 22.54 |
| 98.53 | 9.27 | 105.27 | 3.51 | 3.51 | 1.40 |
| 90.99 | 17.46 | 107.60 | 25.35 | 25.35 | 25.33 |
| 88.37 | 18.39 | 103.30 | 17.51 | 17.51 | 18.40 |
| 88.53 | 11.77 | 102.47 | 6.51 | 6.51 | 6.10 |
| 93.87 | 9.90 | 108.07 | 1.03 | 1.03 | 0.82 |
| 91.32 | 13.65 | 106.07 | 18.38 | 18.38 | 17.65 |
| 83.28 | 15.55 | 103.72 | 8.43 | 8.43 | 8.94 |
| 93.58 | 11.45 | 109.34 | 14.51 | 14.51 | 12.83 |
| 91.28 | 11.33 | 107.89 | 22.45 | 22.45 | 21.75 |
| 89.50 | 18.99 | 103.49 | 23.98 | 23.98 | 23.67 |
| 82.19 | 23.73 | 100.27 | 34.00 | 34.00 | 36.05 |
| 89.33 | 18.90 | 105.50 | 32.96 | 32.96 | 33.40 |
| 92.00 | 10.00 | 111.00 | 10.00 | 13.33 | 12.65 |
| 92.00 | 10.88 | 110.22 | 9.53 | 9.53 | 9.08 |
| 92.00 | 11.00 | 113.00 | 9.00 | 9.00 | 8.15 |
| 91.71 | 15.88 | 111.07 | 22.54 | 8.77 | 21.10 |
| 91.17 | 7.89 | 112.83 | 2.38 | 2.38 | 2.53 |
| 92.00 | 5.00 | 110.00 | 3.00 | 3.00 | 2.30 |
| 91.39 | 7.54 | 123.58 | 5.41 | 5.41 | 2.51 |
| 88.00 | 17.00 | 101.00 | 28.00 | 28.00 | 29.07 |
| 90.09 | 23.46 | 107.85 | 25.97 | 25.97 | 26.55 |
| 88.89 | 13.97 | 101.23 | 6.16 | 6.16 | 5.584 |
| 92.03 | 15.42 | 109.95 | 8.77 | 8.77 | 8.59 |
| 91.80 | 8.75 | 106.88 | 3.83 | 3.83 | 3.77 |
| 89.25 | 18.46 | 107.11 | 28.49 | 28.49 | 28.13 |
| 91.02 | 18.30 | 106.90 | 13.07 | 13.07 | 13.49 |
| 92.80 | 8.24 | 109.86 | 4.07 | 4.07 | 3.65 |
| 91.12 | 7.63 | 114.89 | 8.46 | 8.46 | 8.47 |
| 92.31 | 7.85 | 112.78 | 8.98 | 8.98 | 9.34 |
| 79.99 | 22.05 | 98.56 | 22.85 | 22.85 | 23.76 |
| 90.24 | 10.20 | 110.42 | 19.73 | 19.73 | 19.05 |
| 93.46 | 10.48 | 109.33 | 4.68 | 4.68 | 3.93 |
| 88.13 | 27.58 | 106.92 | 31.66 | 31.66 | 32.91 |
| 85.24 | 10.87 | 105.15 | 0.90 | 0.90 | 0.33 |
| 90.90 | 12.23 | 109.00 | 15.46 | 15.46 | 15.13 |
| 94.69 | 8.77 | 107.73 | 7.43 | 7.43 | 6.35 |
| 93.38 | 7.99 | 115.57 | 4.67 | 4.67 | 5.12 |
| 93.82 | 17.25 | 106.99 | 25.06 | 25.06 | 23.91 |
| 91.74 | 12.05 | 109.34 | 7.22 | 7.22 | 7.64 |
| 90.31 | 6.56 | 109.40 | 4.75 | 4.75 | 4.26 |
| 91.52 | 6.39 | 120.38 | 0.70 | 0.70 | -0.11 |
| 90.00 | 14.00 | 105.00 | 20.00 | 20.00 | 19.71 |
| 91.70 | 12.93 | 110.13 | 17.20 | 17.20 | 17.07 |
| 75.00 | 29.00 | 98.00 | 32.00 | 32.00 | 34.56 |
| 62.54 | 28.82 | 98.94 | 31.39 | 31.39 | 32.34 |
| 92.95 | 10.73 | 114.27 | 11.27 | 11.27 | 9.60 |
| 90.00 | 10.00 | 100.00 | 15.00 | 15.00 | 14.56 |
| 91.71 | 15.88 | 111.07 | 22.54 | 8.77 | 21.10 |
| 83.86 | 28.40 | 103.56 | 28.71 | 28.71 | 30.98 |
| 92.00 | 10.00 | 111.00 | 10.00 | 13.33 | 12.65 |
References
- Castro, N.; Capote, J.; Bruckmaier, R.M.; Argüello, A. Management Effects on Colostrogenesis in Small Ruminants: A Review. J. Appl. Anim. Res. 2011, 39, 85–93. [Google Scholar] [CrossRef]
- Constant, S.B.; LeBlanc, M.M.; Klapstein, E.F.; Beebe, D.E.; Leneau, H.M.; Nunier, C.J. Serum Immunoglobulin G Concentration in Goat Kids Fed Colostrum or a Colostrum Substitute. J. Am. Vet. Med. Assoc. 1994, 205, 1759–1762. [Google Scholar] [CrossRef] [PubMed]
- Argüello, A.; Castro, N.; Capote, J.; Tyler, J.W.; Holloway, N.M. Effect of Colostrum Administration Practices on Serum IgG in Goat Kids. Livest. Prod. Sci. 2004, 90, 235–239. [Google Scholar] [CrossRef]
- Castro, N.; Capote, J.; Morales-Delanuez, A.; Rodríguez, C.; Argüello, A. Effects of Newborn Characteristics and Length of Colostrum Feeding Period on Passive Immune Transfer in Goat Kids. J. Dairy Sci. 2009, 92, 1616–1619. [Google Scholar] [CrossRef]
- Hernández-Castellano, L.E.; Almeida, A.M.; Renaut, J.; Argüello, A.; Castro, N. A Proteomics Study of Colostrum and Milk from the Two Major Small Ruminant Dairy Breeds from the Canary Islands: A Bovine Milk Comparison Perspective. J. Dairy Res. 2016, 83, 366–374. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Sánchez-Macías, D.; Castro, N.; Morales-delaNuez, A.; Hernández-Castellano, L.E.; Capote, J.; Argüello, A. Chemical Composition and Immune Status of Dairy Goat Colostrum Fractions during the First 10h after Partum. Small Rumin. Res. 2012, 103, 220–224. [Google Scholar] [CrossRef]
- Mancini, G.; Carbonara, A.O.; Heremans, J.F. Immunochemical Quantitation of Antigens by Single Radial Immunodiffusion. Immunochemistry 1965, 2, 235–486. [Google Scholar] [CrossRef]
- Bartier, A.; Windeyer, M.; Doepel, L. Evaluation of On-Farm Tools for Colostrum Quality Measurement. J. Dairy Sci. 2015, 98, 1878–1884. [Google Scholar] [CrossRef]
- Buranakarl, C.; Thammacharoen, S.; Nuntapaitoon, M.; Semsirmboon, S.; Katoh, K. Validation of Brix Refractometer to Estimate Immunoglobulin G Concentration in Goat Colostrum. Vet. World 2021, 14, 3194–31999. [Google Scholar] [CrossRef]
- Castro, N.; Gómez-González, L.A.; Earley, B.; Argüello, A. Use of Clinic Refractometer at Farm as a Tool to Estimate the Igg Content in Goat Colostrum. J. Appl. Anim. Res. 2018, 46, 1505–1508. [Google Scholar] [CrossRef]
- Zobel, G.; Rodriguez-Sanchez, R.; Hea, S.; Weatherall, A.; Sargent, R. Validation of Brix Refractometers and a Hydrometer for Measuring the Quality of Caprine Colostrum. J. Dairy Sci. 2020, 103, 9277–9289. [Google Scholar] [CrossRef] [PubMed]
- Mechor, G.D.; Gröhn, Y.T.; McDowell, L.R.; Van Saun, R.J. Specific Gravity of Bovine Colostrum Immunoglobulins as Affected by Temperature and Colostrum Components. J. Dairy Sci. 1992, 75, 3131–3135. [Google Scholar] [CrossRef] [PubMed]
- Argüello, A.; Castro, N.; Capote, J. Short Communication: Evaluation of a Color Method for Testing Immunoglobulin g Concentration in Goat Colostrum. J. Dairy Sci. 2005, 88, 1752–1754. [Google Scholar] [CrossRef] [PubMed]
- Chui, M.; McCarthy, B.; McKinsey. An Executive’s Guide to AI. Available online: https://www.mckinsey.com/capabilities/quantumblack/our-insights/an-executives-guide-to-ai (accessed on 23 December 2024).
- Ali AlZubi, A. Artificial Intelligence and Its Application in the Prediction and Diagnosis of Animal Diseases: A Review. Indian J. Anim. Res. 2023, 57, 1265–1271. [Google Scholar] [CrossRef]
- Denholm, S.J.; Brand, W.; Mitchell, A.P.; Wells, A.T.; Krzyzelewski, T.; Smith, S.L.; Wall, E.; Coffey, M.P. Predicting Bovine Tuberculosis Status of Dairy Cows from Mid-Infrared Spectral Data of Milk Using Deep Learning. J. Dairy Sci. 2020, 103, 9355–9367. [Google Scholar] [CrossRef]
- Neto, H.A.; Tavares, W.L.F.; Ribeiro, D.C.S.Z.; Alves, R.C.O.; Fonseca, L.M.; Campos, S.V.A. On the Utilization of Deep and Ensemble Learning to Detect Milk Adulteration. BioData Min. 2019, 12, 13. [Google Scholar] [CrossRef]
- Schanda, J. Colorimetry: Understanding the CIE System; Schanda, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2007; ISBN 9780470049044. [Google Scholar]
- Berns, R.S. Billmeyer and Saltzman’s: Principles of Color Technology, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2019; ISBN 9781119367314. [Google Scholar]
- Konica Minolta Sensing. Color Spaces. Available online: https://sensing.konicaminolta.us/us/learning-center/color-measurement/color-spaces/ (accessed on 12 June 2024).
- Ziegler, A. An Introduction to Statistical Learning with Applications; James, R.G., Witten, D., Hastie, T., Tibshirani, R., Eds.; Springer: Berlin, Germany, 2013; p. 440. ISBN 978-1-4614-7138-7. [Google Scholar]
- McClarren, R.G. Decision Trees and Random Forests for Regression and Classification. In Machine Learning for Engineers: Using Data to Solve Problems for Physical Systems; McClarren, R.G., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 55–82. ISBN 978-3-030-70388-2. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT: Cambridge, MA, USA, 2016. [Google Scholar]
- Molnar, C. Interpretable Machine Learning, 2nd ed.; Leanpub: Victoria, BC, Canada, 2022; ISBN 978-1-09-812017-0. [Google Scholar]
- Quinlan, J.R. Pruning Decision Trees. In C4.5; Elsevier: Amsterdam, The Netherlands, 1993; pp. 35–43. ISBN 978-1-55860-238-0. [Google Scholar]
- Haykin, S.S. Neural Networks and Learning Machines; Prentice Hall/Pearson: London, UK, 2009; ISBN 9780131471399. [Google Scholar]
- Bebis, G.; Georgiopoulos, M. Feed-Forward Neural Networks. IEEE Potentials 1994, 13, 27–31. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: Berlin/Heidelberg, Germany, 2009; ISBN 9780387848570. [Google Scholar]
- Agarap, A.F. Deep Learning Using Rectified Linear Units (ReLU). arXiv 2018, arXiv:1803.08375. [Google Scholar]
- Cortes, C.; Research, G.; York, N.; Mohri, M.; Rostamizadeh, A. L 2 Regularization for Learning Kernels. arXiv 2012, arXiv:1205.2653. [Google Scholar]
- Hodson, T.O. Root-Mean-Square Error (RMSE) or Mean Absolute Error (MAE): When to Use Them or Not. Geosci. Model. Dev. 2022, 15, 5481–5487. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A Systematic Analysis of Performance Measures for Classification Tasks. Inf. Process Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Python 3.9.6 Documentation. Available online: https://docs.python.org/release/3.9.6/ (accessed on 23 December 2024).
- API Reference—Scikit-Learn 1.4.2 Documentation. Available online: https://scikit-learn.org/1.4/modules/classes.html (accessed on 13 June 2024).
- API Documentation. Available online: https://www.tensorflow.org/api_docs (accessed on 23 December 2024).
- Keras 3 API Documentation. Available online: https://keras.io/api/ (accessed on 23 December 2024).
- Zhang, L.; Han, G.; Qiao, Y.; Xu, L.; Chen, L.; Tang, J. Interactive Dairy Goat Image Segmentation for Precision Livestock Farming. Animals 2023, 13, 3250. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, P.; Marques, M.d.R.; Belo, A.T.; Monteiro, A.; Morais, J.; Riegel, I.; Braz, F. Exploring the Potential of Machine Learning Algorithms Associated with the Use of Inertial Sensors for Goat Kidding Detection. Animals 2024, 14, 938. [Google Scholar] [CrossRef]
- Solis, I.L.; de Oliveira-Boreli, F.P.; de Sousa, R.V.; Martello, L.S.; Pereira, D.F. Using Thermal Signature to Evaluate Heat Stress Levels in Laying Hens with a Machine-Learning-Based Classifier. Animals 2024, 14, 1996. [Google Scholar] [CrossRef]
- Pedrosa, V.B.; Chen, S.-Y.; Gloria, L.S.; Doucette, J.S.; Boerman, J.P.; Rosa, G.J.M.; Brito, L.F. Machine Learning Methods for Genomic Prediction of Cow Behavioral Traits Measured by Automatic Milking Systems in North American Holstein Cattle. J. Dairy Sci. 2024, 107, 4758–4771. [Google Scholar] [CrossRef]
- García-Infante, M.; Castro-Valdecantos, P.; Delgado-Pertíñez, M.; Teixeira, A.; Guzmán, J.L.; Horcada, A. Effectiveness of Machine Learning Algorithms as a Tool to Meat Traceability System. A Case Study to Classify Spanish Mediterranean Lamb Carcasses. Food Control 2024, 164, 110604. [Google Scholar] [CrossRef]
- Hu, H.; Zhou, H.; Cao, K.; Lou, W.; Zhang, G.; Gu, Q.; Wang, J. Biomass Estimation of Milk Vetch Using UAV Hyperspectral Imagery and Machine Learning. Remote Sens. 2024, 16, 2183. [Google Scholar] [CrossRef]
- Chen, X.; Zheng, H.; Wang, H.; Yan, T. Can Machine Learning Algorithms Perform Better than Multiple Linear Regression in Predicting Nitrogen Excretion from Lactating Dairy Cows. Sci. Rep. 2022, 12, 12478. [Google Scholar] [CrossRef]
- Hansen, B.G.; Li, Y.; Sun, R.; Schei, I. Forecasting Milk Delivery to Dairy—How Modern Statistical and Machine Learning Methods Can Contribute. Expert Syst. Appl. 2024, 248, 123475. [Google Scholar] [CrossRef]


| HIGH | LOW | ||
| HIGH | 47 | 3 | 50 |
| LOW | 0 | 113 | 113 |
| 47 | 116 |
| HIGH | LOW | ||
| HIGH | 47 | 3 | 50 |
| LOW | 3 | 110 | 113 |
| 50 | 113 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betancor-Sánchez, M.; González-Cabrera, M.; Morales-delaNuez, A.; Hernández-Castellano, L.E.; Argüello, A.; Castro, N. Enhancing Immunoglobulin G Goat Colostrum Determination Using Color-Based Techniques and Data Science. Animals 2025, 15, 31. https://doi.org/10.3390/ani15010031
Betancor-Sánchez M, González-Cabrera M, Morales-delaNuez A, Hernández-Castellano LE, Argüello A, Castro N. Enhancing Immunoglobulin G Goat Colostrum Determination Using Color-Based Techniques and Data Science. Animals. 2025; 15(1):31. https://doi.org/10.3390/ani15010031
Chicago/Turabian StyleBetancor-Sánchez, Manuel, Marta González-Cabrera, Antonio Morales-delaNuez, Lorenzo E. Hernández-Castellano, Anastasio Argüello, and Noemí Castro. 2025. "Enhancing Immunoglobulin G Goat Colostrum Determination Using Color-Based Techniques and Data Science" Animals 15, no. 1: 31. https://doi.org/10.3390/ani15010031
APA StyleBetancor-Sánchez, M., González-Cabrera, M., Morales-delaNuez, A., Hernández-Castellano, L. E., Argüello, A., & Castro, N. (2025). Enhancing Immunoglobulin G Goat Colostrum Determination Using Color-Based Techniques and Data Science. Animals, 15(1), 31. https://doi.org/10.3390/ani15010031







