Comparative Metagenomic Analysis of the Gut Microbiota of Captive Pangolins: A Case Study of Two Species
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Metagenomic Data
3.2. Gut Microbiota Composition and Structure of Two Pangolin Species
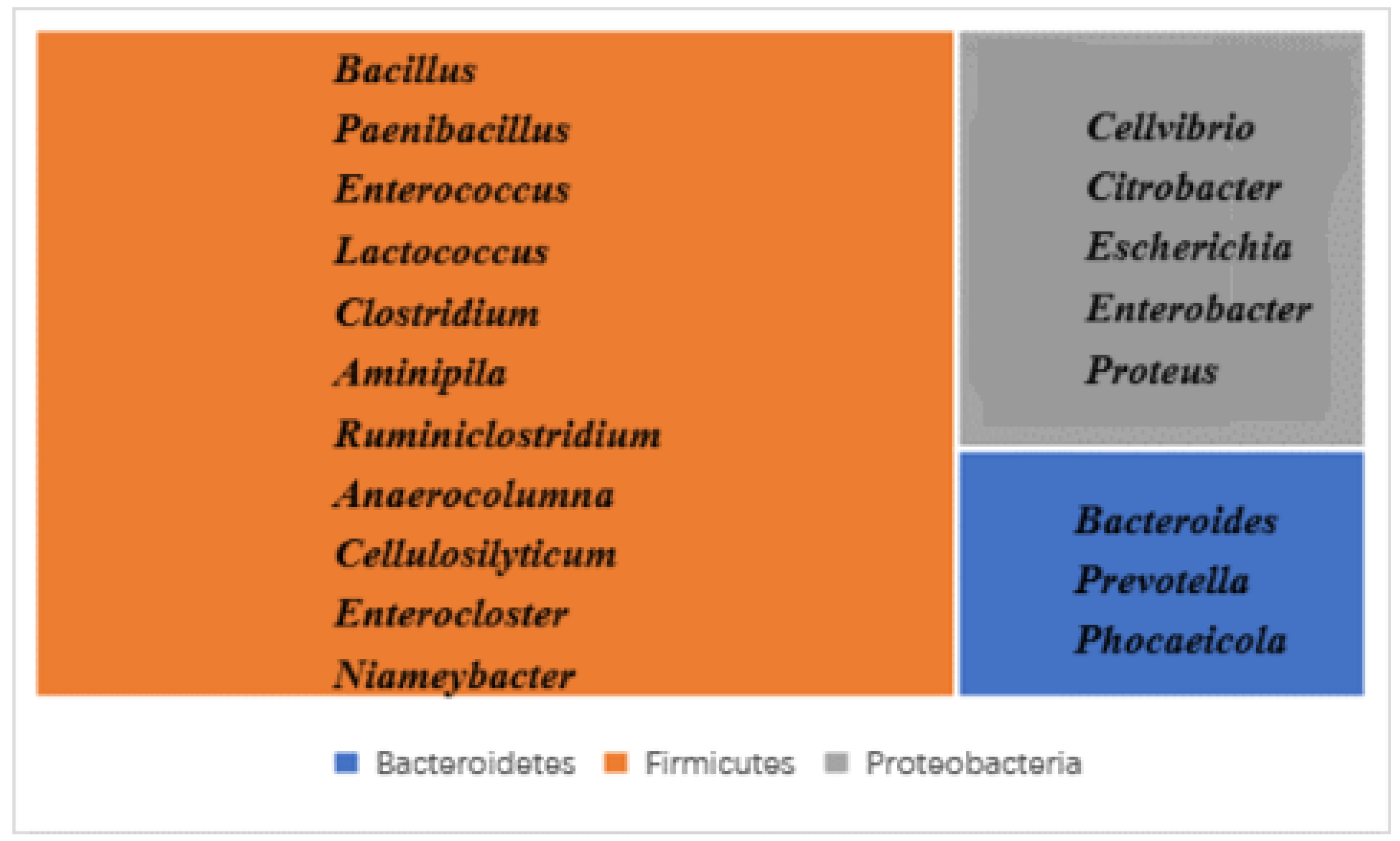
3.2.1. Gut Microbiota Composition
3.2.2. Alpha Diversity
3.2.3. Beta Diversity
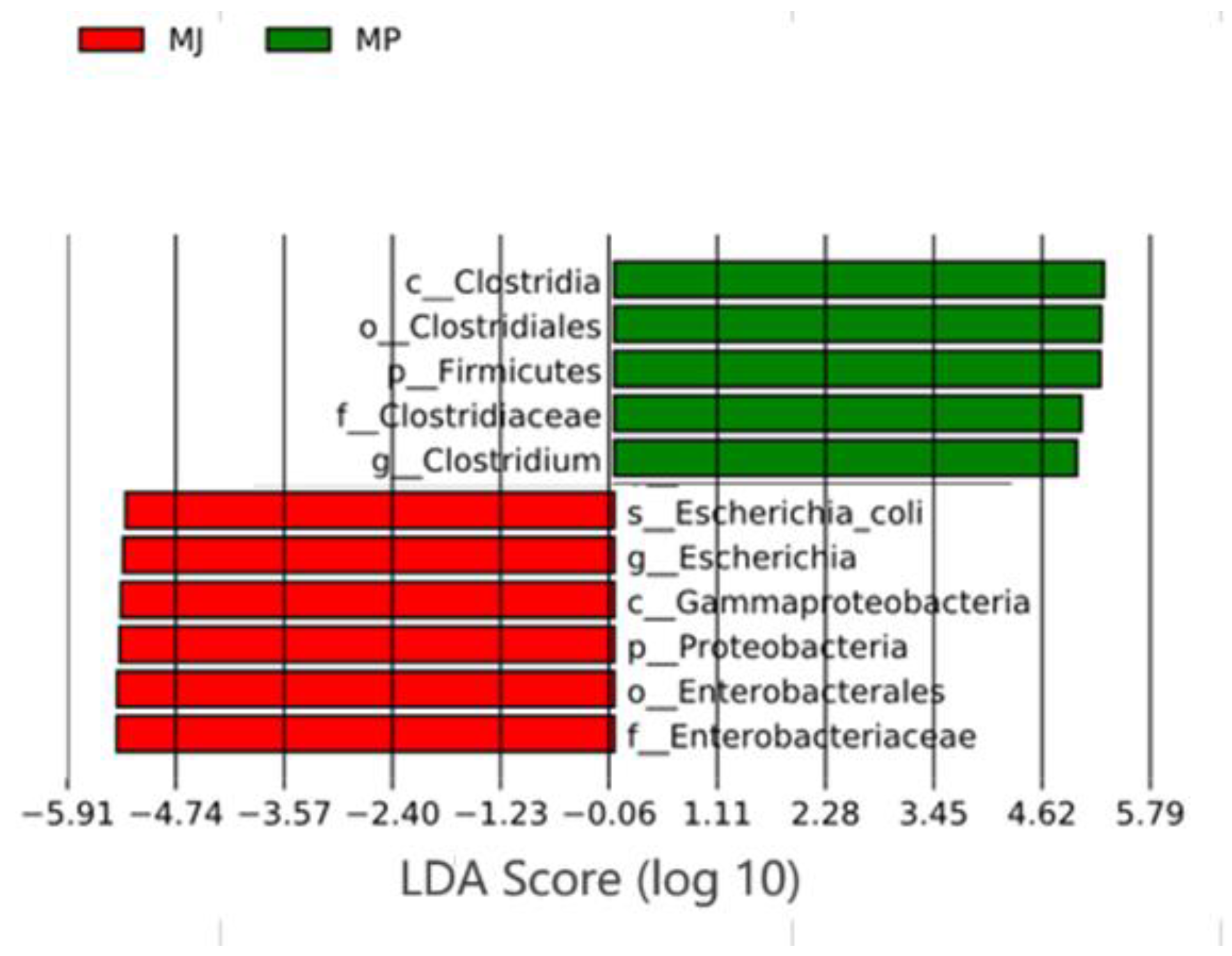
3.3. Analysis of Differences in Gut Microbiota Structure Between Two Pangolin Species
3.4. Functional Differences in Gut Microbiota Between Two Pangolin Species
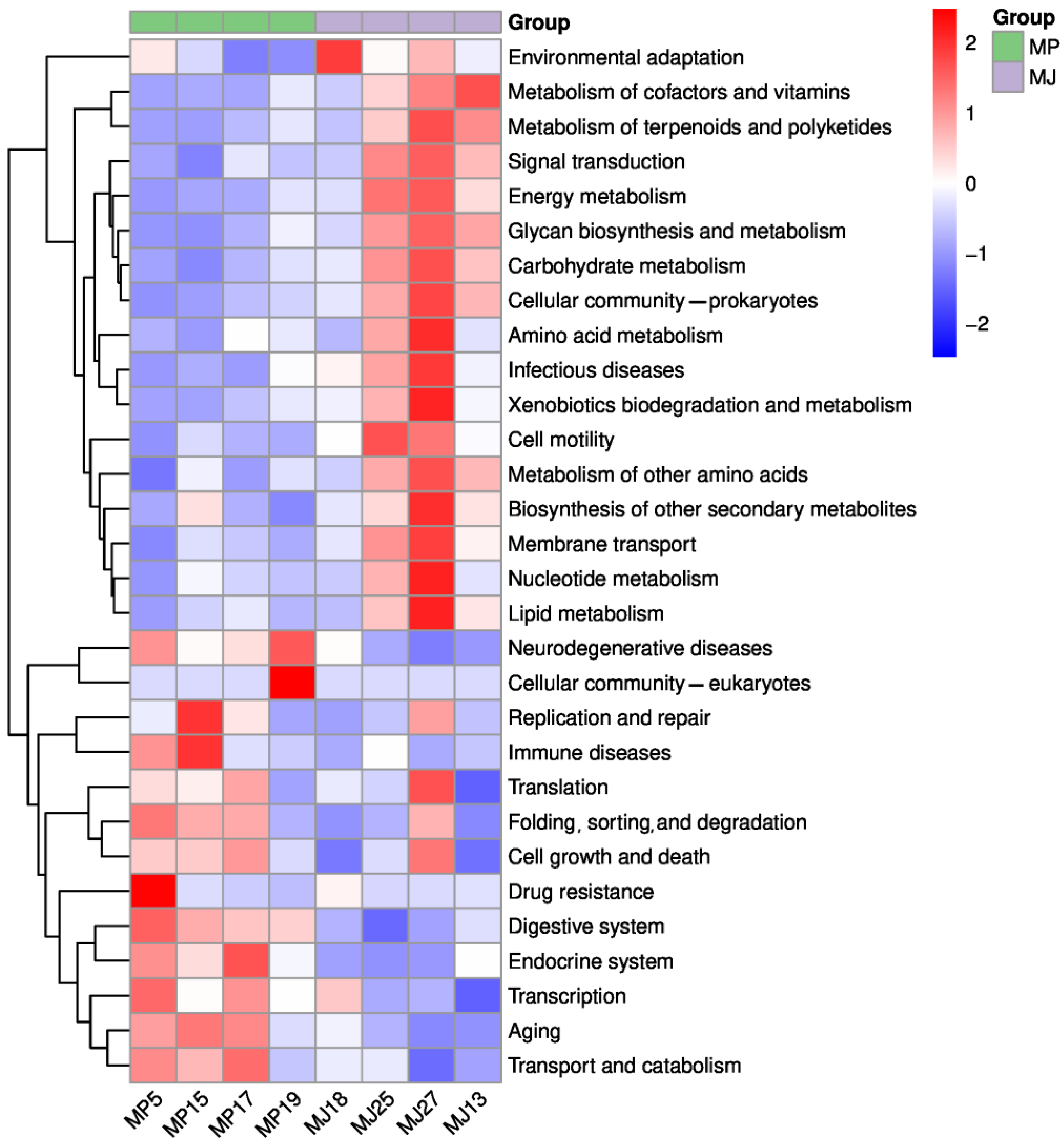
3.4.1. Functional Groups and Metabolic Pathways Statistics and Composition Analysis
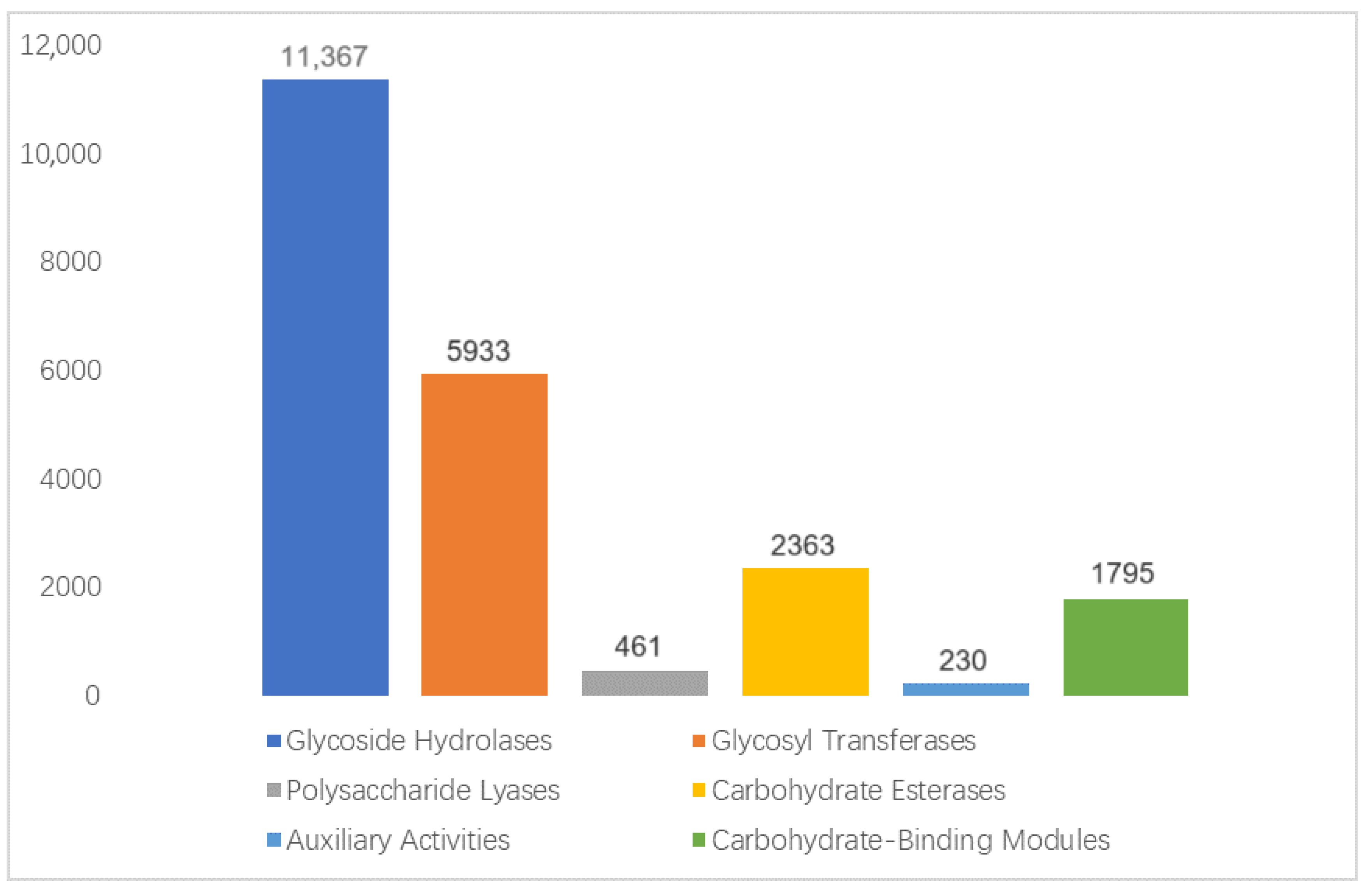
3.4.2. CAZy Functional Annotation
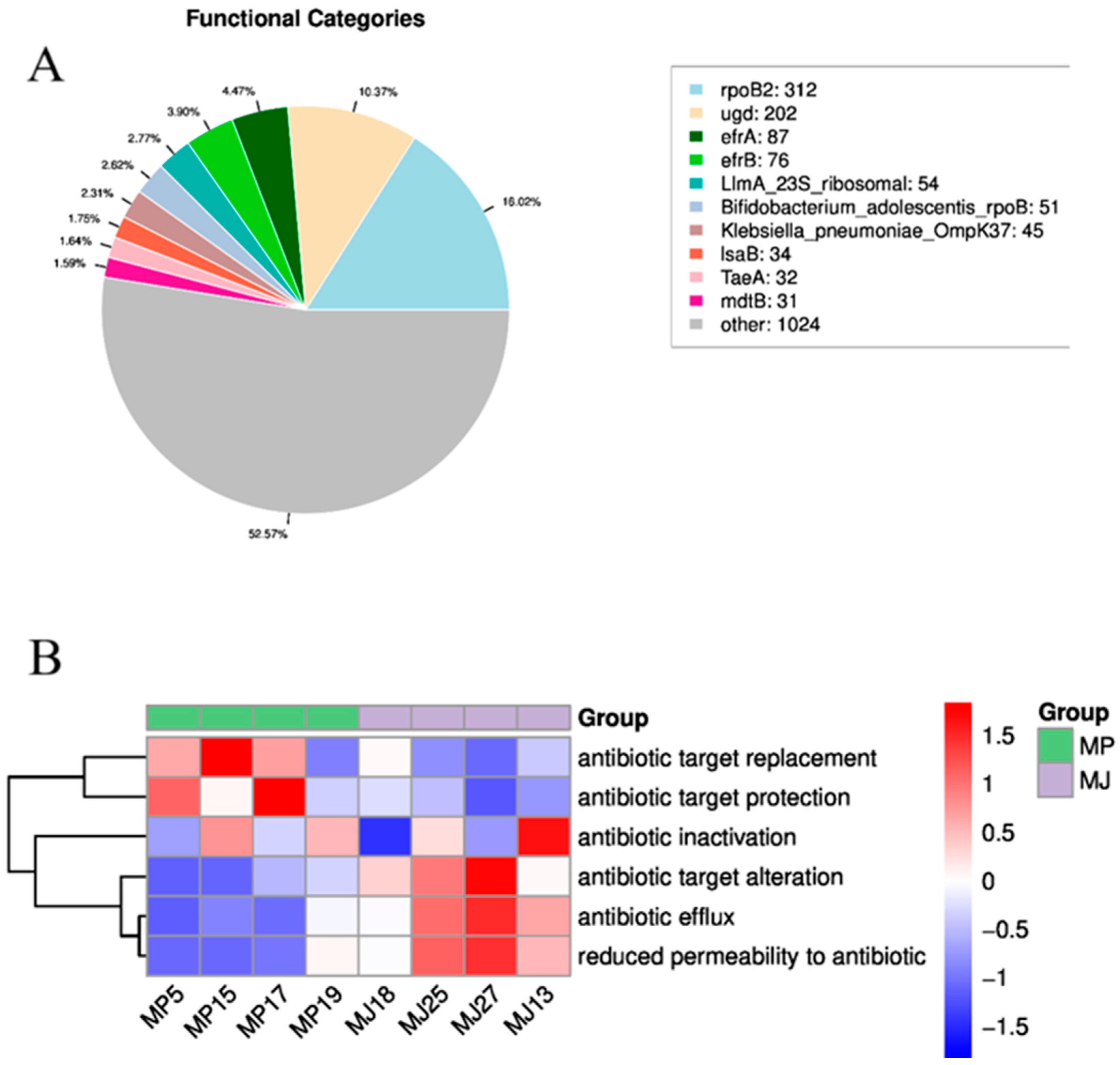
3.4.3. CARD Resistance Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A




References
- Challender, D.W.S.; Harrop, S.R.; MacMillan, D.C. Understanding Markets to Conserve Trade-Threatened Species in CITES. Biol. Conserv. 2015, 187, 249–259. [Google Scholar] [CrossRef]
- Challender, D.W.S.; Helen, C.N.; Carly, W. Pangolins: Science, Society and Conservation; Academic Press: New York, NY, USA, 2019. [Google Scholar]
- Gu, T.; Hu, J.; Yu, L. Evolution and Conservation Genetics of Pangolins. Integr. Zool. 2024, 19, 426–441. [Google Scholar] [CrossRef]
- Wang, X.M.; Janssens, G.P.J.; Xie, C.-G.; Xie, B.-W.; Xie, Z.-G.; He, H.-J.; Wang, Y.-N.; Xu, J. To Save Pangolins: A Nutritional Perspective. Animals 2022, 12, 3137. [Google Scholar] [CrossRef]
- Liu, P.; Chen, W.; Chen, J.P. Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica). Viruses 2019, 11, 979. [Google Scholar] [CrossRef]
- Yan, D.; Zeng, X.; Jia, M.; Guo, X.; Deng, S.; Tao, L.; Huang, X.; Li, B.; Huang, C.; Que, T.; et al. Successful captive breeding of a Malayan pangolin population to the third filial generation. Commun. Biol. 2021, 4, 1212. [Google Scholar] [CrossRef]
- Morgan, K.N.; Tromborg, C.T. Sources of stress in captivity. Appl. Anim. Behav. Sci. 2007, 102, 262–302. [Google Scholar] [CrossRef]
- Fast, M.D.; Hosoya, S.; Johnson, S.C.; Afonso, L.O.B. Cortisol Response and Immune-Related Effects of Atlantic Salmon (Salmo salar Linnaeus) Subjected to Short- and Long-Term Stress. Fish. Shellfish. Immunol. 2008, 24, 194–204. [Google Scholar] [CrossRef]
- Bravo, J.A.; Julio-Pieper, M.; Forsythe, P.; Kunze, W.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Communication between Gastrointestinal Bacteria and the Nervous System. Curr. Opin. Pharmacol. 2012, 12, 667–672. [Google Scholar] [CrossRef]
- Trevelline, B.K.; Kohl, K.D. The Gut Microbiome Influences Host Diet Selection Behavior. Proc. Natl. Acad. Sci. USA 2022, 119, e2117537119. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Frankel, J.S.; Mallott, E.K.; Hopper, L.M.; Ross, S.R.; Amato, K.R. The Effect of Captivity on the Primate Gut Microbiome Varies with Host Dietary Niche. Am. J. Primatol. 2019, 81, e23061. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Mishra, S.; Wang, C.; Zhang, H.; Ning, R.; Kong, F.; Zeng, B.; Zhao, J.; Li, Y. Comparative Study of Gut Microbiota in Wild and Captive Giant Pandas (Ailuropoda melanoleuca). Genes 2019, 10, 827. [Google Scholar] [CrossRef] [PubMed]
- Khairulmunir, M.; Gani, M.; Mohd-Ridwan, A.R.; Karuppannan, K.V.; Abdul-Latiff, M.A.B.; Md-Zain, B.M. Alteration of the gut microbial composition of critically endangered Malayan tigers (Panthera tigris jacksoni) in captivity during enrichment phase. Mol. Biol. Rep. 2024, 51, 742. [Google Scholar] [CrossRef]
- Delsuc, F.; Metcalf, J.L.; Wegener Parfrey, L.; Song, S.J.; González, A.; Knight, R. Convergence of Gut Microbiomes in Myrmecophagous Mammals. Mol. Ecol. 2014, 23, 1301–1317. [Google Scholar] [CrossRef]
- Ma, J.; Jiang, H.; Li, L.; Zhang, X.; Li, G.; Li, H.; Jin, X.; Chen, J. The Fecal Metagenomics of Malayan Pangolins Identifies an Extensive Adaptation to Myrmecophagy. Front. Microbiol. 2018, 9, 2793. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Bunæs, A.C.; Mathiesen, G.; Eijsink, V.G.H. The Chitinolytic System of Lactococcus Lactis Ssp. Lactis Comprises a Nonprocessive Chitinase and a Chitin-Binding Protein That Promotes the Degradation of α- and β-Chitin. FEBS J. 2009, 276, 2402–2415. [Google Scholar] [CrossRef]
- Gooday, G.W. The ecology of chitin degradation. In Advances in Microbial Ecology; Springer: Boston, MA, USA, 1990; pp. 387–430. [Google Scholar] [CrossRef]
- Liu, C.; Hu, J.; Wu, Y.; Irwin, D.M.; Chen, W.; Zhang, Z.; Yu, L. Comparative Study of Gut Microbiota from Captive and Confiscated-Rescued Wild Pangolins. J. Genet. Genom. 2021, 48, 825–835. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiol. Mol. Biol. Rev. 2005, 69, 195. [Google Scholar] [CrossRef]
- Dinsdale, E.A.; Edwards, R.A.; Hall, D.; Angly, F.; Breitbart, M.; Brulc, J.M.; Furlan, M.; Desnues, C.; Haynes, M.; Li, L.; et al. Functional metagenomic profiling of nine biomes. Nature 2008, 452, 629–632. [Google Scholar] [CrossRef]
- Cheng, S.; Liu, C.; Yao, X.; Hu, J.; Yin, T.; Lim, B.; Chen, W.; Wang, G.; Zhang, C.; Irwin, D.M.; et al. Hologenomic insights into mammalian adaptations to myrmecophagy. Natl. Sci. Rev. 2023, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Supple, M.A.; Shapiro, B. Conservation of biodiversity in the genomics era. Genome Biol. 2018, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- de Flamingh, A.; Ishida, Y.; Pečnerová, P.; Vilchis, S.; Siegismund, H.R.; van Aarde, R.J.; Malhi, R.S.; Roca, A.L. Combining Methods for Non-Invasive Fecal DNA Enables Whole Genome and Metagenomic Analyses in Wildlife Biology. Front. Genet. 2023, 13, 1021004. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Zeng, X.; Jia, M.; Guo, X.; Que, C.; Tao, L.; Li, M.; Li, B.; Chen, J.; Xu, S.; et al. Weaning period and growth patterns of captive Sunda pangolin (Manis javanica) cubs. PLoS ONE 2022, 17, e0272020. [Google Scholar] [CrossRef]
- Nelson, K.E. An Update on the Status of Current Research on the Mammalian Microbiome. ILAR J. 2015, 56, 163–168. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Baselga, A.; Orme, C.D.L. betapart: An R package for the study of beta diversity. Methods Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. Available online: https://www.jstor.org/stable/4615964 (accessed on 15 September 2022).
- Dickman, M. Some Indices of Diversity. Ecology 1968, 49, 1191–1193. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Ma, J.; Li, L.M.; Jiang, H.; Zhang, X.; Li, J.; Li, G. Acidic mammalian chitinase gene is highly expressed in the special oxyntic glands of Manis javanica. FEBS Open Bio 2018, 8, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.D.; Wang, K.; An, F.Y.; Xu, X.L.; Yan, H.M.; Kuang, Y.J.; Zou, C.S.; Hua, Y. Dietary Supplementation of Termitarium Soil Ameliorated the Gut Microbiota in Captive Chinese Pangolins. Available online: https://www.researchsquare.com/article/rs-2032613/v1 (accessed on 15 September 2022).
- Zhang, F.; Xu, N.; Wang, W.; Yu, Y.; Wu, S. The Gut Microbiome of the Sunda Pangolin (Manis javanica) Reveals Its Adaptation to Specialized Myrmecophagy. PeerJ 2021, 9, e11490. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Moore, R.J.; Stanley, D.; Chousalkar, K.K. The Gut Microbiota of Laying Hens and Its Manipulation with Prebiotics and Probiotics To Enhance Gut Health and Food Safety. Appl. Environ. Microbiol. 2020, 86, e00600-20. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Li, X.; Yu, Y.; Li, C.; Yan, Q. Comparative Study on the Gut Microbiotas of Four Economically Important Asian Carp Species. Sci. China Life Sci. 2018, 61, 696–705. [Google Scholar] [CrossRef]
- Scott, K.P.; Gratz, S.W.; Sheridan, P.O.; Flint, H.J.; Duncan, S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013, 69, 52–60. [Google Scholar] [CrossRef]
- Li, Y.; Hu, X.; Yang, S.; Zhou, J.; Zhang, T.; Qi, L.; Sun, X.; Fan, M.; Xu, S.; Cha, M.; et al. Comparative analysis of the gut microbiota composition between captive and wild forest musk deer. Front. Microbiol. 2017, 8, 1705. [Google Scholar] [CrossRef]
- Guan, Y.; Yang, H.; Han, S.; Feng, L.; Wang, T.; Ge, J. Comparison of the Gut Microbiota Composition between Wild and Captive Sika Deer (Cervus nippon hortulorum) from Feces by High-Throughput Sequencing. AMB Express 2017, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.A.; Hesta, M.; Hollants, J.; Janssens, G.P.J.; Huys, G. Phylogenetic analysis of faecal microbiota from captive cheetahs reveals under representation of Bacteroidetes and Bifidobacteriaceae. BMC Microbiol. 2014, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- Cabana, F.; Plowman, A.; Van Nguyen, T.; Chin, S.; Wu, S.; Lo, H.; Watabe, H.; Yamamoto, F. Feeding Asian Pangolins: An Assessment of Current Diets Fed in Institutions Worldwide. Zoo. Biol. 2017, 36, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Hu, D.; Li, K.; Li, B.; Zeng, X.; Chen, J.; Li, Y.; Wronski, T. Effects of Chronic Stress on the Fecal Microbiome of Malayan Pangolins (Manis javanica) Rescued from the Illegal Wildlife Trade. Curr. Microbiol. 2021, 78, 1017–1025. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, L.; Liu, X.; Lan, Y.; Wu, L.; Wang, J.; Wu, K.; Yang, C.; Lv, R.; Yi, D.; et al. Red pandas with different diets and environments exhibit different gut microbial functional composition and capacity. Integr. Zool. 2024, 19, 662–682. [Google Scholar] [CrossRef]


| MP5 | MP15 | MP17 | MP19 | MJ13 | MJ18 | MJ25 | MJ27 | |
|---|---|---|---|---|---|---|---|---|
| Gender | Male | Male | Male | Female | Male | Male | Male | Male |
| Species | M. pentadactyla | M. pentadactyla | M. pentadactyla | M. pentadactyla | M. javanica | M. javanica | M. javanica | M. javanica |
| Captivity duration (year) | 2.5 | 2.5 | 2 | 2.5 | 5 | 5 | 5 | 5 |
| Maturity | Adult | Adult | Adult | Adult | Adult | Adult | Adult | Adult |
| Body weight (kg) | 6.68 | 7.46 | 9.16 | 5.6 | 9.96 | 9.14 | 6.54 | 6.35 |
| Body length (cm) | 47 | 53 | 55 | 36 | 65 | 64 | 57 | 53 |
| Tail length (cm) | 32 | 35 | 34 | 23 | 53 | 47 | 43 | 37 |
| Rectal temperature (°C) | 31.43 ± 1.54 | 31.58 ± 1.25 | 31.20 ± 1.09 | 32.01 ± 1.85 | 31.44 ± 1.51 | 31.89 ± 1.48 | 32.45 ± 1.78 | 30.68 ± 1.66 |
| Heart rate (beats/min) | 57.50 ± 7.48 | 59.50 ± 10.48 | 56.60 ± 2.22 | 57.90 ± 9.19 | 58.80 ± 5.89 | 55.30 ± 4.13 | 56.90 ± 6.24 | 58.70 ± 6.30 |
| Respiratory rate (breaths/min) | 21.70 ± 1.16 | 21.80 ± 1.14 | 22.10 ± 1.52 | 22.40 ± 1.78 | 21.00 ± 0.47 | 21.10 ± 0.89 | 21.90 ± 2.22 | 21.90 ± 1.55 |
| Health status | Healthy | Healthy | Healthy | Healthy | Healthy | Healthy | Healthy | Healthy |
| Samples | Chao1 | Shannon | Simpson | ACE |
|---|---|---|---|---|
| MP5 | 7100.904 | 5.197 | 0.983 | 7038.848 |
| MP15 | 6964.906 | 4.701 | 0.971 | 6898.871 |
| MP17 | 7102.242 | 5.206 | 0.981 | 7054.912 |
| MP19 | 6164.520 | 3.353 | 0.853 | 6104.062 |
| MJ13 | 6026.771 | 3.167 | 0.805 | 5930.006 |
| MJ18 | 6845.826 | 3.808 | 0.872 | 6733.518 |
| MJ25 | 6909.893 | 2.479 | 0.636 | 6824.279 |
| MJ27 | 3509.821 | 2.135 | 0.619 | 3451.216 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Z.; Xie, B.; Xie, C.; Xiang, J.; Wang, X.; Li, J.; Zheng, R.; Wang, Y. Comparative Metagenomic Analysis of the Gut Microbiota of Captive Pangolins: A Case Study of Two Species. Animals 2025, 15, 57. https://doi.org/10.3390/ani15010057
Dai Z, Xie B, Xie C, Xiang J, Wang X, Li J, Zheng R, Wang Y. Comparative Metagenomic Analysis of the Gut Microbiota of Captive Pangolins: A Case Study of Two Species. Animals. 2025; 15(1):57. https://doi.org/10.3390/ani15010057
Chicago/Turabian StyleDai, Zhengyu, Bowen Xie, Chungang Xie, Jinsuo Xiang, Xinmei Wang, Jing Li, Rongquan Zheng, and Yanni Wang. 2025. "Comparative Metagenomic Analysis of the Gut Microbiota of Captive Pangolins: A Case Study of Two Species" Animals 15, no. 1: 57. https://doi.org/10.3390/ani15010057
APA StyleDai, Z., Xie, B., Xie, C., Xiang, J., Wang, X., Li, J., Zheng, R., & Wang, Y. (2025). Comparative Metagenomic Analysis of the Gut Microbiota of Captive Pangolins: A Case Study of Two Species. Animals, 15(1), 57. https://doi.org/10.3390/ani15010057




