Reproductive Performance, Inbreeding, and Genetic Diversity in Montbeliarde Dairy Cattle Obtained by Absorption Crossing
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Genealogical Database
2.3. Absorption Breeding Scheme
2.4. Demographic Structure
2.5. Reproductive Performance-Derived Traits
2.6. Inbreeding and Coancestry
2.7. Gene Origin Probabilities and Ancestral Contributions
2.8. Genetic Diversity
2.9. Data Analysis and Software
3. Results
3.1. Population Structure and Reproductive Performance Evolution in Montbeliarde Cattle Obtained by Absorption Crossing
3.1.1. Demographic Structure and Reproductive Performance
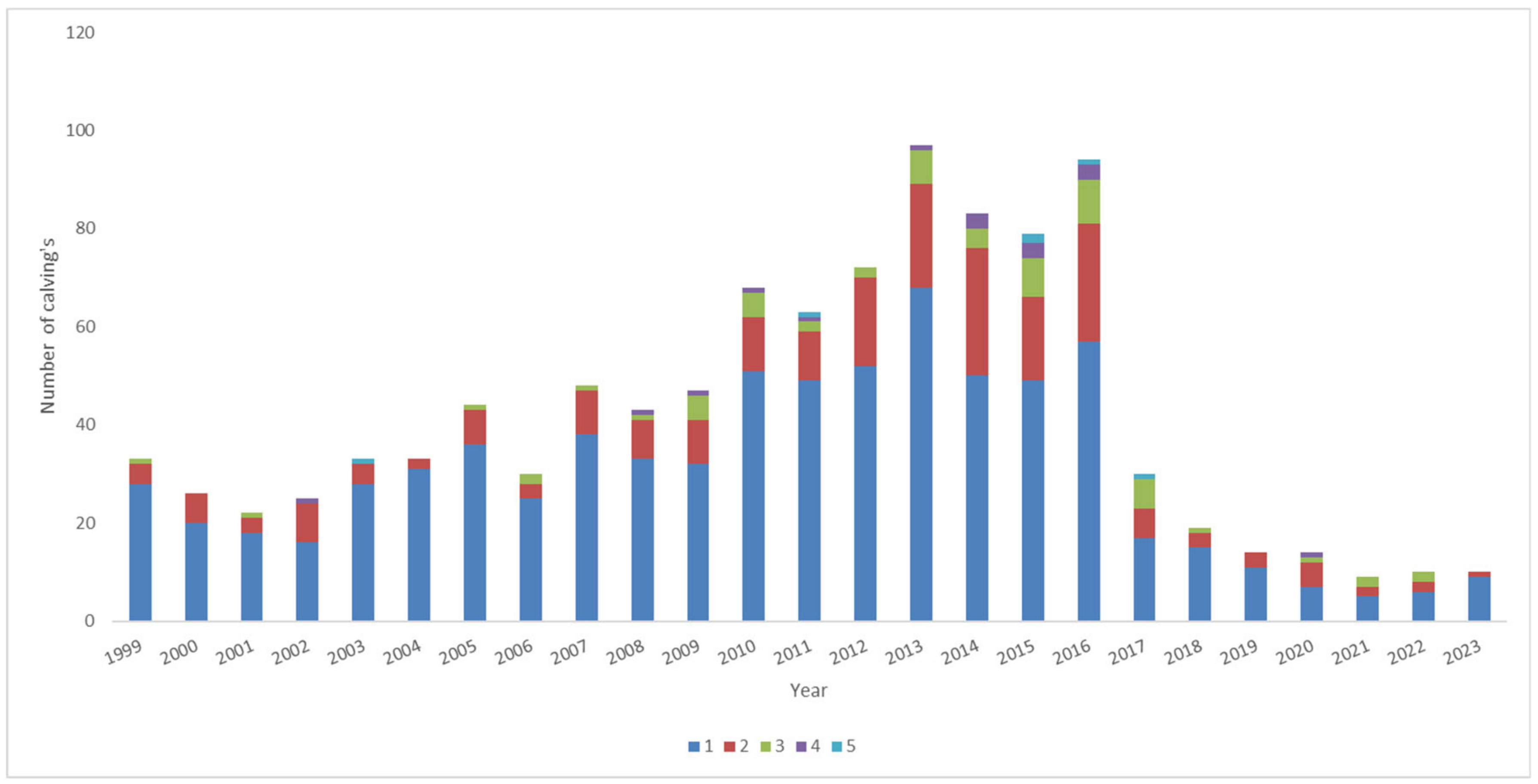
3.1.2. Reproductive Performance per Sire and Dam
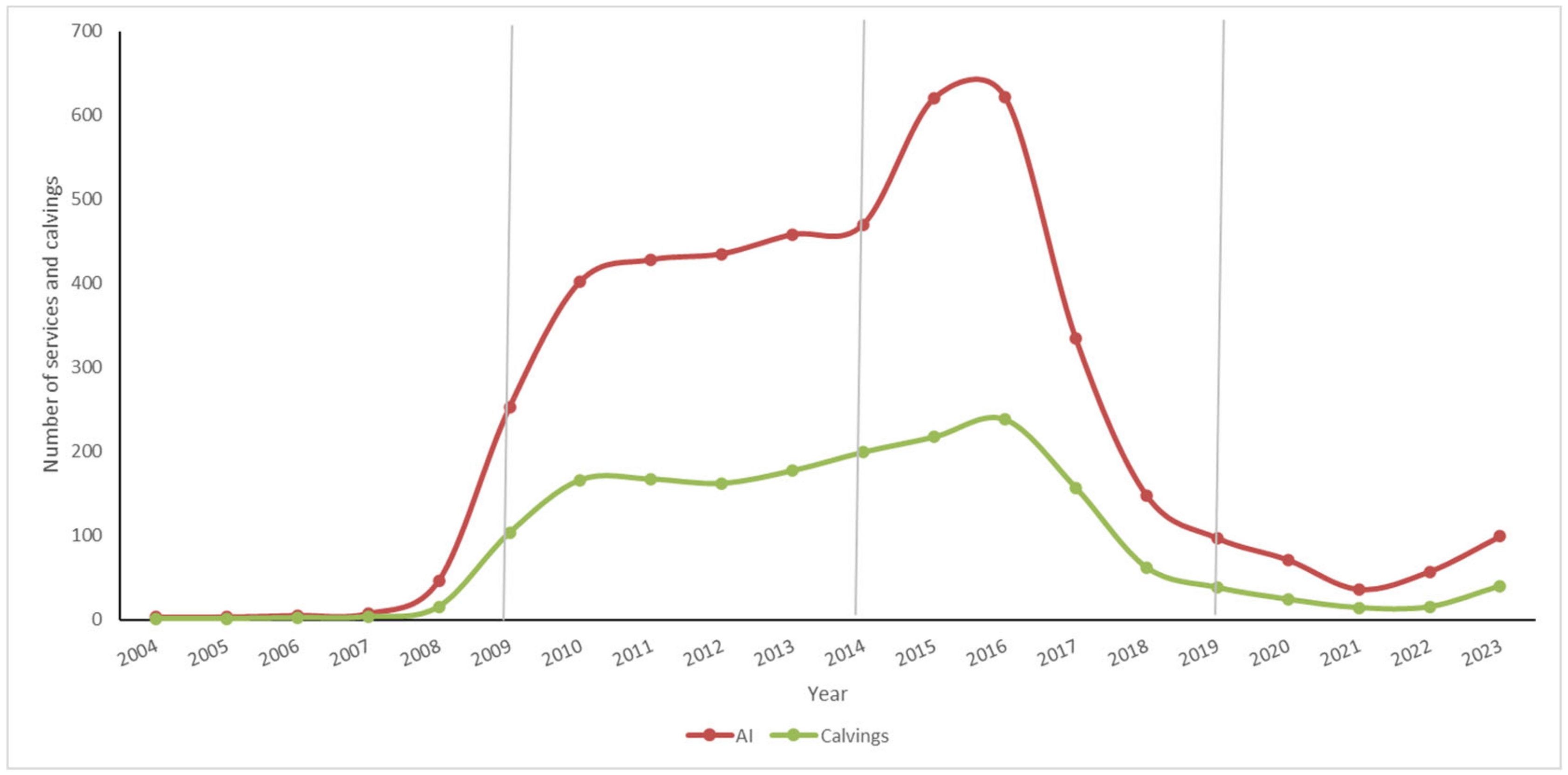
3.1.3. Number of AI Services and Number of Calvings
3.1.4. Evolution of the Number of Calvings
3.2. Pedigree Completeness and Generation Intervals in Ecuadorian Montbeliarde Cattle Obtained by Absorption Crossing
3.2.1. Pedigree Completeness-Derived Parameters
3.2.2. Generation Intervals
3.2.3. Inbreeding, Average Relatedness, Coancestry, and Non-Random Mating
3.2.4. Effective Size (Ne)
3.3. Gene Origin Probability, Ancestral Contributions, and Genetic Diversity in Ecuadorian Montbeliarde Cattle Obtained by Absorption Crossing
3.3.1. Gene Origin Probability and Ancestral Contributions
3.3.2. Genetic Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thibier, M.; Wagner, H.G. World Statistics for Artificial Insemination in Cattle. Livest. Prod. Sci. 2002, 74, 203–212. [Google Scholar] [CrossRef]
- NAAB National Association of Animal Breeders. Available online: https://www.naab-css.org/ (accessed on 15 December 2024).
- Ministry of Agriculture and Livestock. MAG Ministerial Agreement 037: Authorize the Brown Swiss Association of Ecuador to Manage the Genealogical Records of the Brown Swiss Bovine Breed. Available online: https://www.agricultura.gob.ec/asociaciones-ganaderas-reciben-autorizaciones-para-hacer-registros-genealogicos-y-mejorar-la-genetica/ (accessed on 14 December 2024).
- Ministry of Agriculture and Livestock. MAG Ministerial Agreement 039: Authorize the Gyr-Girolando Breeders Association of Ecuador to Manage the Genealogical Records of the Gyr-Girolando Bovine Breed. Available online: https://app.vlex.com/vid/873876734 (accessed on 14 December 2024).
- Ministry of Agriculture and Livestock. MAG Ministerial Agreement 040: Authorize the Holstein Friesian Association of Ecuador to Manage the Genealogical Records of the Holstein Bovine Breed. Available online: https://app.vlex.com/vid/874235038 (accessed on 14 December 2024).
- Ministry of Agriculture and Livestock. MAG Ministerial Agreement 041: Authorize the Jersey Association of Ecuador to Manage the Genealogical Records of the Jersey Cattle Breed. Available online: https://app.vlex.com/vid/874235051 (accessed on 14 December 2024).
- Muñoz, E.C.; Andriamandroso, A.L.; Blaise, Y.; Ron, L.; Montufar, C.; Kinkela, P.M.; Lebeau, F.; Bindelle, J. How Do Management Practices and Farm Structure Impact Productive Performances of Dairy Cattle in the Province of Pichincha, Ecuador. J. Agric. Rural. Dev. Trop. Subtrop. 2020, 121, 233–241. [Google Scholar] [CrossRef]
- Felius, M.; Beerling, M.L.; Buchanan, D.S.; Theunissen, B.; Koolmees, P.A.; Lenstra, J.A. On the History of Cattle Genetic Resources. Diversity 2014, 6, 705–750. [Google Scholar] [CrossRef]
- France Genetique Elevage Montbéliarde Association—2022. Available online: https://www.montbeliarde.org/es-82-9.html (accessed on 15 December 2024).
- Magne, M.A.; Quénon, J. Dairy Crossbreeding Challenges the French Dairy Cattle Sociotechnical Regime. Agron. Sustain. Dev. 2021, 41, 1–15. [Google Scholar] [CrossRef]
- Hazel, A.R.; Heins, B.J.; Seykora, A.J.; Hansen, L.B. Montbéliarde-Sired Crossbreds Compared with Pure Holsteins for Dry Matter Intake, Production, and Body Traits during the First 150 Days of First Lactation. J. Dairy Sci. 2013, 96, 1915–1923. [Google Scholar] [CrossRef]
- Guagrabamba First Monbeliarde Cattle Farm in Ecuador. Available online: https://www.guagrabamba.com/ (accessed on 14 December 2024).
- MAGAP Ecuadorian Agricultural Policy: Towards Sustainable Rural Territorial Development: 2015–2025. Part I. Available online: https://www.fao.org/faolex/results/details/es/c/LEX-FAOC183434/ (accessed on 14 December 2024).
- Gutierrez-Reinoso, M.A.; Aponte, P.M.; Garcia-Herreros, M. Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- Sanchez, M.P.; Tribout, T.; Fritz, S.; Wolf, V.; Laithier, C.; Brochard, M.; Boichard, D. Opportunities for Genomic Selection of Cheese-Making Traits in Montbéliarde Cows. J. Dairy Sci. 2022, 105, 5206–5220. [Google Scholar] [CrossRef]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; García-Herreros, M. Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review. Animals 2023, 13, 1588. [Google Scholar] [CrossRef]
- Doublet, A.C.; Croiseau, P.; Fritz, S.; Michenet, A.; Hozé, C.; Danchin-Burge, C.; Laloë, D.; Restoux, G. The Impact of Genomic Selection on Genetic Diversity and Genetic Gain in Three French Dairy Cattle Breeds. Genet. Sel. Evol. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Cartuche Macas, L.F.; Camacho Vallejo, M.E.; González Ariza, A.; León Jurado, J.M.; Delgado Bermejo, J.V.; Marín Navas, C.; Navas González, F.J. Analysis of Endangered Andalusian Black Cattle (Negra Andaluza) Reveals Genetic Reservoir for Bovine Black Trunk. Animals 2024, 14, 1131. [Google Scholar] [CrossRef]
- Cervantes, I.; Goyache, F.; Molina, A.; Valera, M.; Gutiérrez, J.P. Application of Individual Increase in Inbreeding to Estimate Realized Effective Sizes from Real Pedigrees. J. Anim. Breed. Genet. 2008, 125, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, I.; Goyache, F.; Molina, A.; Valera, M.; Gutiérrez, J.P. Estimation of Effective Population Size from the Rate of Coancestry in Pedigreed Populations. J. Anim. Breed. Genet. 2011, 128, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; García-Herreros, M. A Review of Inbreeding Depression in Dairy Cattle: Current Status, Emerging Control Strategies, and Future Prospects. J. Dairy Res. 2022, 89, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Casanovas Arias, D.; León Jurado, J.M.; Bermejo Asensio, L.A.; Navas González, F.J.; Marín Navas, C.; Barba Capote, C.J. Genetic Diversity Evolution of a Sheep Breed Reintroduced after Extinction: Tracing Back Christopher Columbus’ First Imported Sheep. Res. Vet. Sci. 2020, 132, 207–216. [Google Scholar] [CrossRef]
- Navas, F.J.; Jordana, J.; León, J.M.; Barba, C.; Delgado, J.V. A Model to Infer the Demographic Structure Evolution of Endangered Donkey Populations. Animal 2017, 11, 2129–2138. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Marmi, J.; Goyache, F.; Jordana, J. Pedigree Information Reveals Moderate to High Levels of Inbreeding and a Weak Population Structure in the Endangered Catalonian Donkey Breed. J. Anim. Breed. Genet. 2005, 122, 378–386. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Cervantes, I.; Goyache, F. Improving the Estimation of Realized Effective Population Sizes in Farm Animals. J. Anim. Breed. Genet. 2009, 126, 327–332. [Google Scholar] [CrossRef]
- James, J.W. Computation of Genetic Contributions from Pedigrees. TAG Theor. Appl. Genet. 1972, 42, 272–273. [Google Scholar] [CrossRef]
- Vassallo, J.M.; Diaz, C.; Garcia-Medina, J.R. A Note on the Population Structure of the Avileña Breed of Cattle in Spain. Livest. Prod. Sci. 1986, 15, 285–288. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Luo, Z. Computing Inbreeding Coefficients in Large Populations. Genet. Sel. Evol. 1992, 24, 305–313. [Google Scholar] [CrossRef]
- Lindgren, D.; Gea, L.; Jefferson, P. Loss of Genetic Diversity Monitored by Status Number. Silvae Genet. 1996, 45, 52–58. [Google Scholar]
- Leroy, G.; Mary-Huard, T.; Verrier, E.; Danvy, S.; Charvolin, E.; Danchin-Burge, C. Methods to Estimate Effective Population Size Using Pedigree Data: Examples in Dog, Sheep, Cattle and Horse. Genet. Sel. Evol. 2013, 45, 1. [Google Scholar] [CrossRef] [PubMed]
- Colleau, J.J.; Sargolzaei, M. A Proximal Decomposition of Inbreeding, Coancestry and Contributions. Genet. Res. 2008, 90, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, P.M.; Wright, S. Evolution and the Genetics of Populations. Vol. 2. The Theory of Gene Frequencies. J. Anim. Ecol. 1971, 40, 266. [Google Scholar] [CrossRef]
- Caballero, A.; Toro, M.A. Interrelations between Effective Population Size and Other Pedigree Tools for the Management of Conserved Populations. Genet. Res. 2000, 75, 331–343. [Google Scholar] [CrossRef]
- Hill, W. Estimation of Effective Population Size from Data on Linkage Disequilibrium. Hill Genet. Res. 1981, 38, 209–216. [Google Scholar] [CrossRef]
- Hill, W.G.; Mackay, T. Falconer and Introduction to Quantitative Genetics. Genetics 2004, 167, 1529–1536. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Cervantes, I.; Molina, A.; Valera, M.; Goyache, F. Individual Increase in Inbreeding Allows Estimating Effective Sizes from Pedigrees. Genet. Sel. Evol. 2008, 40, 359–378. [Google Scholar] [CrossRef]
- Alderson, G.L. A System to Maximize the Maintenance of Genetic Variability in Small Populations. Genet. Conserv. Domest. Livest. 1992, 2, 18–29. [Google Scholar]
- Lacy, R.C. Analysis of Founder Representation in Pedigrees: Founder Equivalents and Founder Genome Equivalents. Zoo Biol. 1989, 8, 111–123. [Google Scholar] [CrossRef]
- Boichard, D.; Maignel, L.; Verrier, É. The Value of Using Probabilities of Gene Origin to Measure Genetic Variability in a Population. Genet. Sel. Evol. 1997, 29, 5–23. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Iwaisaki, H.; Colleau, J.J. CFC: A Tool for Monitoring Genetic Diversity. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Brazil, 13–18 August 2006. [Google Scholar]
- Gutiérrez, J.P.; Goyache, F. A Note on ENDOG: A Computer Program for Analysing Pedigree Information. J. Anim. Breed. Genet. 2005, 122, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, E.; Westhuizen, B.D.; Maiwashe, A.; Voordewind, F.; Ferraz, J.B. POPREP: A Generic Report for Population Management. Genet. Mol. Res. GMR 2009, 8, 1158–1178. [Google Scholar] [CrossRef] [PubMed]
- Mehdi Sargolzaei, H.; Iwaisaki, J.J.; Colleau, J. CFC: A Tool for Monitoring Genetic Diversity; Request PDF. In Proceedings of the Conference 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Brazil, 13–18 August 2006. [Google Scholar]
- IDELE—Institut de L´Elevage. Bilan de Variabilité Génétique à Partir Des Données de Généalogies—Races Bovines Laitières. Volume 23. Available online: https://idele.fr/ (accessed on 15 July 2024).
- Gangotena Castro, C.J.; Landázuri Benítez, J.E.; Troncozo Montes, R.P. Nuevos Aportes a La Economía Especialidad: Economía Social de Mercado; Pontificia Universidad Católica del Ecuador: Quito, Ecuador, 2021; ISBN 978-9978-77-511-0. [Google Scholar]
- ESPAE Graduate School of Management. Industrial Studies: Strategic Orientation for Decision Making—Beef Cattle Industry—ESPAE Business School. Available online: https://www.espae.edu.ec/publicaciones/estudios-industriales-orientacion-estrategica-para-la-toma-de-decisiones-industria-de-ganaderia-de-carne/ (accessed on 14 December 2024).
- MAGAP Ministry of Agriculture, Livestock, Aquaculture and Fisheries, Resolution 60—The Applicable Technical Regulations Are Issued to Regulate and Control the Entry of Imported Genetic Material (Semen, Embryos and Live Animals) of Livestock Species of Commerci. Available online: https://app.vlex.com/vid/637657541 (accessed on 14 December 2024).
- Chanfrau, J.M.P.; Pérez, J.N.; Fiallos, M.V.L.; Intriago, L.M.R.; Toledo, L.E.T.; Guerrero, M.J.C. Milk Whey Valorization: An Overview from Biotechnology. Bionatura 2017, 2, 468–476. [Google Scholar] [CrossRef]
- SCE No. 012 The Superintendence of Economic Competition Concluded the Market Study on the Agri-Food Chains of Ecuador—Superintendence of Economic Competition: Report on the Dairy Sector in Ecuador. Available online: https://www.sce.gob.ec/sitio/no-012-la-superintendencia-de-competencia-economica-concluyo-el-estudio-de-mercado-sobre-las-cadenas-agroalimentarias-del-ecuador/ (accessed on 14 December 2024).
- González, A.R.M.; Navas González, F.J.; Crudeli, G.Á.; Delgado Bermejo, J.V.; Camacho Vallejo, M.E.; Quirino, C.R. Process of Introduction of Australian Braford Cattle to South America: Configuration of Population Structure and Genetic Diversity Evolution. Animals 2022, 12, 275. [Google Scholar] [CrossRef]
- de Fátima Sieklicki, M.; Mulim, H.A.; Pinto, L.F.B.; Valloto, A.A.; Pedrosa, V.B. Population Structure and Inbreeding of Holstein Cattle in Southern Brazil. Rev. Bras. Zootec. 2020, 49, e20190052. [Google Scholar] [CrossRef]
- Vásquez-Loaiza, M.; Cruz-Méndez, A.; Castro-Vásquez, R.; Cascante-Segura, S.; Camacho-Sandoval, J.; Domínguez-Viveros, J.; Vásquez-Loaiza, M.; Cruz-Méndez, A.; Castro-Vásquez, R.; Cascante-Segura, S.; et al. Pedigree analysis and population structure of brahman cattle from costa rica. Chil. J. Agric. Amp; Anim. Sci. 2021, 37, 177–183. [Google Scholar] [CrossRef]
- Maccluer, J.W.; Boyce, A.J.; Dyke, B.; Weitkamp, L.R.; Pfenning, D.W.; Parsons, C.J. Inbreeding and Pedigree Structure in Standardbred Horses. J. Hered. 1983, 74, 394–399. [Google Scholar] [CrossRef]
- Forneris, N.S.; Garcia-Baccino, C.A.; Cantet, R.J.C.; Vitezica, Z.G. Estimating Inbreeding Depression for Growth and Reproductive Traits Using Pedigree and Genomic Methods in Argentinean Brangus Cattle. J. Anim. Sci. 2021, 99, skab289. [Google Scholar] [CrossRef]
- Hagger, C. Estimates of Genetic Diversity in the Brown Cattle Population of Switzerland Obtained from Pedigree Information. J. Anim. Breed. Genet. 2005, 122, 405–413. [Google Scholar] [CrossRef]
- Honda, T.; Nomura, T.; Yamaguchi, Y.; Mukai, F. Monitoring of Genetic Diversity in the Japanese Black Cattle Population by the Use of Pedigree InformationMonitoring Genetischer Diversität in Der Population Des Japanischen Schwarzen Rindes Unter Verwendung von Pedigreeinformation. J. Anim. Breed. Genet. 2004, 121, 242–252. [Google Scholar] [CrossRef]
- Danchin-Burge, C.; Leroy, G.; Brochard, M.; Moureaux, S.; Verrier, E. Evolution of the Genetic Variability of Eight French Dairy Cattle Breeds Assessed by Pedigree Analysis. J. Anim. Breed. Genet. 2012, 129, 206–217. [Google Scholar] [CrossRef] [PubMed]
- IDELE—Institut de L´Elevage. Bilan de Variabilité Génétique à Partir Des Données de Généalogies—Races Bovines Laitières. Volume 21. Available online: https://idele.fr/ (accessed on 30 July 2024).
- Utrera, Á.R.; Murillo, V.E.V.; Bermúdez, M.M.; Velázquez, G.M.; Ponce, S.I.R. Genetic Diversity Assessment of the Mexican Simmental Population through Pedigree Analysis. Rev. Bras. Zootec. 2018, 47, e20160088. [Google Scholar] [CrossRef]
- Uemoto, Y.; Suzuki, K.; Yasuda, J.; Roh, S.; Satoh, M. Evaluation of Inbreeding and Genetic Diversity in Japanese Shorthorn Cattle by Pedigree Analysis. Anim. Sci. J. 2021, 92, e13643. [Google Scholar] [CrossRef]
- Interbull Centre. INTERBULL Executive Summary. Available online: https://interbull.org/ib/executivesummary (accessed on 15 December 2024).
- Rodríguez-Ramilo, S.T.; Fernández, J.; Toro, M.A.; Hernández, D.; Villanueva, B. Genome-Wide Estimates of Coancestry, Inbreeding and Effective Population Size in the Spanish Holstein Population. PLoS ONE 2015, 10, e0124157. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in Genetic Selection Differentials and Generation Intervals in US Holstein Dairy Cattle as a Result of Genomic Selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef]
- Magalhães Araújo da Silva, M.H.; Malhado, C.H.M.; Costa, J.L.; Cobuci, J.A.; Costa, C.N.; Carneiro, P.L.S. Population Genetic Structure in the Holstein Breed in Brazil. Trop. Anim. Health Prod. 2016, 48, 331–336. [Google Scholar] [CrossRef]
- Núñez-Domínguez, R.; Martínez-Rocha, R.E.; Hidalgo-Moreno, J.A.; Ramírez-Valverde, R.; García-Muñiz, J.G. Evaluation of the Romosinuano Cattle Population Structure in Mexico Using Pedigree Analysis. Rev. Colomb. Cienc. Pecu. 2020, 33, 44–59. [Google Scholar] [CrossRef]
- González-Cano, R.; González-Martínez, A.; Muñoz-Mejías, M.E.; Valera, P.; Rodero, E. Analyses of Genetic Diversity in the Endangered ”Berrenda” Spanish Cattle Breeds Using Pedigree Data. Animals 2022, 12, 249. [Google Scholar] [CrossRef]
- Tenhunen, S.; Thomasen, J.R.; Sørensen, L.P.; Berg, P.; Kargo, M. Genomic Analysis of Inbreeding and Coancestry in Nordic Jersey and Holstein Dairy Cattle Populations. J. Dairy Sci. 2024, 107, 5897–5912. [Google Scholar] [CrossRef]
- Wang, Y. Genetic and Geographic Diversity of Gyr (Bos Indicus) Cattle in Brazil; University of Natural Resources and Life Sciences: Vienna, Austria, 2015. [Google Scholar]
- Rosendo Ponce, A.; Palacios Jiménez, A.L.; Rosales Martínez, F.; Torres Hernández, G.; Ramírez Valverde, R.; Becerril Pérez, C.M. Genetic Variability of Tropical Milking Criollo Cattle of Mexico Estimated from Genealogy Information. Rev. Colomb. De Cienc. Pecu. 2018, 31, 196–203. [Google Scholar] [CrossRef]
- Peixoto, M.G.C.D.; Carvalho, M.R.S.; Egito, A.A.; Steinberg, R.S.; Bruneli, F.Â.T.; Machado, M.A.; Santos, F.C.; Rosse, I.C.; Fonseca, P.A.S. Genetic Diversity and Population Genetic Structure of a Guzerá (Bos Indicus) Meta-Population. Animals 2021, 11, 1125. [Google Scholar] [CrossRef] [PubMed]
- FAO; Scherf, B.D.; Pilling, D. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2019; ISBN 9789251088203. [Google Scholar]
- Kadlečík, O.; Pavlík, I.; Moravčíková, N.; Kasarda, R. Inbreeding and Genetic Diversity Loss of Four Cattle Beef Breeds in Slovakia. Acta Fytotech. Zootech. 2016, 19, 59–63. [Google Scholar] [CrossRef][Green Version]
- Santana, M.L.; Pereira, R.J.; Bignardi, A.B.; Ayres, D.R.; Menezes, G.R.O.; Silva, L.O.C.; Leroy, G.; Machado, C.H.C.; Josahkian, L.A.; Albuquerque, L.G. Structure and Genetic Diversity of Brazilian Zebu Cattle Breeds Assessed by Pedigree Analysis. Livest. Sci. 2016, 187, 6–15. [Google Scholar] [CrossRef]
- Honnay, O. Genetic Drift. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Academic Press: New York, NY, USA, 2013; pp. 251–253. [Google Scholar] [CrossRef]
- Chacón Marcheco, E.; Cartuche Macas, L.F.; Villavicencio Estrella, A.; Toro Molina, B.; Silva Déley, L.; Andrade Aulestia, P. Diversidad Genética de La Población de Toros Holstein Friesian Importados Al Ecuador Entre El 2000–2021. Rev. Cient. Tecnol. UPSE 2023, 10, 33–40. [Google Scholar] [CrossRef]
- Stachowicz, K.; Sargolzaei, M.; Miglior, F.; Schenkel, F.S. Rates of Inbreeding and Genetic Diversity in Canadian Holstein and Jersey Cattle. J. Dairy Sci. 2011, 94, 5160–5175. [Google Scholar] [CrossRef]
- Melka, M.G.; Stachowicz, K.; Miglior, F.; Schenkel, F.S. Analyses of Genetic Diversity in Five Canadian Dairy Breeds Using Pedigree Data. J. Anim. Breed. Genet. 2013, 130, 476–486. [Google Scholar] [CrossRef]
- Cartuche-Macas, L.F.; Lozada, E.F.; Gutiérrez-Reinoso, M.A.; Chacón, E.; Navas, F.J.; García-Herreros, M. Evolution of Population Structure, Reproductive Performance, Inbreeding, and Genetic Diversity in Ecuadorian Charolais Cattle. Vet. Sci. 2024, 11, 566. [Google Scholar] [CrossRef]
- Cedeño Quevedo, D.A.; Calpa Oliva, C.A.; Bravo, N.L.; Hernández, D.R. Clinical, Histopathological and Immunohistochemical Study of Bovine Ocular Squamous Cell Carcinoma in the Nariño Department, Colombia. Rev. Investig. Vet. Peru 2020, 31, 1–11. [Google Scholar] [CrossRef]
- Lacy, R.C. Extending Pedigree Analysis for Uncertain Parentage and Diverse Breeding Systems. J. Hered. 2012, 103, 197–205. [Google Scholar] [CrossRef]



| Demographic Parameter | Historical | 1999–2003 | 2004–2008 | 2009–2013 | 2014–2018 | 2019–2023 |
|---|---|---|---|---|---|---|
| Number of animals with pedigree | 2399 | 211 | 332 | 747 | 434 | 167 |
| Number of animals (reference population) | 1309 | 141 | 198 | 338 | 308 | 58 |
| Dams (total) | 2033 | 151 | 296 | 681 | 392 | 156 |
| Sires (total) | 366 | 60 | 36 | 66 | 42 | 11 |
| Individuals with progeny (offspring) | 1320 | 199 | 215 | 310 | 94 | 22 |
| Individuals without progeny (offspring) | 1079 | 12 | 117 | 437 | 340 | 145 |
| Number of animals with both known parents | 1309 | 141 | 198 | 338 | 308 | 58 |
| Number of animals only with known sire | 868 | 62 | 125 | 355 | 123 | 104 |
| Number of animals only with known dam | 14 | - | 2 | 9 | 2 | - |
| Number of animals with no known parents | 208 | 8 | 7 | 45 | 1 | 5 |
| Reproductive parameter | 1999–2003 | 2004–2008 | 2009–2013 | 2014–2018 | 2019–2023 | |
| Calvings per service (AI) ratio (%) | 39.27 | - | 34.38 | 39.24 | 39.77 | 36.39 |
| Number of calves born (offspring) | 3500 | 986 | 431 | 500 | 211 | 33 |
| Average number of calves per sire | 1.72 | 6.53 | 1.46 | 0.73 | 0.54 | 0.21 |
| Maximum number of calves per sire | 107 | 79 | 54 | 15 | 29 | 5 |
| Average number of calves per dam | 9.56 | 16.43 | 11.97 | 7.58 | 5.02 | 3.00 |
| Maximum number of calves per dam | 5 | 5 | 5 | 5 | 3 | 2 |
| Parameter | Historical * | 1999–2003 | 2004–2008 | 2009–2013 | 2014–2018 | 2019–2023 |
|---|---|---|---|---|---|---|
| Population of animals with pedigree | 2399 | 211 | 332 | 747 | 434 | 167 |
| Number of generations (n) | 13 | 7 | 8 | 10 | 11 | 13 |
| 1st generation (%) | 72.95 | 81.52 | 78.77 | 69.61 | 85.37 | 65.87 |
| 2nd generation (%) | 61.79 | 70.26 | 70.63 | 63.69 | 77.07 | 61.83 |
| 3rd generation (%) | 47.07 | 44.67 | 53.09 | 50.69 | 69.64 | 60.40 |
| 4th generation (%) | 28.23 | 19.82 | 30.23 | 28.75 | 48.44 | 54.83 |
| 5th generation (%) | 15.11 | 2.61 | 7.93 | 15.37 | 30.39 | 45.45 |
| Average GMax | 4.70 | 3.94 | 4.58 | 4.79 | 6.34 | 9.22 |
| Average GCom | 0.91 | 1.08 | 1.00 | 0.80 | 1.35 | 0.65 |
| Average GEqu | 2.36 | 2.19 | 2.42 | 2.32 | 3.30 | 3.67 |
| Parameter | Historical | 1999–2003 | 2004–2008 | 2009–2013 | 2014–2018 | 2019–2023 |
|---|---|---|---|---|---|---|
| Inbreeding coefficient (F, %) | 0.52 | 0.63 | 0.57 | 0.46 | 0.90 | 0.63 |
| Inbreeding increment (ΔF, %) | 0.15 | 0.24 | 0.17 | 0.13 | 0.22 | 0.12 |
| Maximum inbreeding coefficient (%) | 26.56 | 25.00 | 25.00 | 26.56 | 7.62 | 26.12 |
| Inbred animals (%) | 26.72 | 1.58 | 4.04 | 8.05 | 9.84 | 2.38 |
| Highly inbred animals (%) | 0.38 | 0.13 | 0.04 | 0.08 | 0.00 | 0.04 |
| Coancestry coefficient (C, %) | 1.29 | 1.36 | 1.40 | 1.34 | 1.57 | 1.03 |
| Average relatedness (AR, %) | 2.58 | 2.72 | 2.80 | 2.68 | 3.13 | 2.06 |
| Genetic conservation index (GCI) | 3.45 | 3.57 | 3.82 | 3.37 | 4.75 | 3.12 |
| Gene-Origin/Ancestral Contribution Parameters | Population | 1999–2003 | 2004–2008 | 2009–2013 | 2014–2018 | 2019–2023 |
|---|---|---|---|---|---|---|
| Historical population (n) | 2399 | 211 | 332 | 747 | 434 | 167 |
| Reference population (n) | 1309 | 141 | 198 | 338 | 308 | 58 |
| Base population (one or more unknown parents) | 1090 | 8 | 7 | 45 | 1 | 5 |
| Current base population (one unknown parent = half founder) | 649 | 62 | 127 | 364 | 125 | 104 |
| Number of founders contributing to the reference population (n) | 459 | 174 | 256 | 330 | 351 | 231 |
| Number of ancestors contributing to the reference population (n) | 439 | 145 | 202 | 273 | 228 | 52 |
| Effective number of non-founders (Nenf) | 34.82 | 28.49 | 28.63 | 25.71 | 20.62 | 13.52 |
| Effective number of founders (fe) | 71.64 | 59.21 | 64.49 | 66.06 | 68.69 | 90.64 |
| Effective number of ancestors (fa) | 37 | 31 | 33 | 31 | 28 | 23 |
| Founder genome equivalents (fg) | 23.43 | 19.24 | 19.83 | 18.51 | 15.86 | 11.76 |
| fe/fa ratio | 1.94 | 1.91 | 1.95 | 2.13 | 2.45 | 3.94 |
| fg/fe ratio | 0.33 | 0.32 | 0.31 | 0.28 | 0.23 | 0.13 |
| Number of ancestors to explain: | ||||||
| 25% of gene pool | 4 | 4 | 4 | 4 | 4 | 3 |
| 50% of gene pool | 14 | 11 | 11 | 11 | 9 | 10 |
| 75% of gene pool | 60 | 38 | 46 | 43 | 37 | 22 |
| 100% of gene pool | 145 | 202 | 273 | 228 | 52 | 145 |
| Genetic Diversity Parameters | 1999–2003 | 2004–2008 | 2009–2013 | 2014–2018 | 2019–2023 | % COT |
|---|---|---|---|---|---|---|
| GD (%) | 97.40 | 97.48 | 97.30 | 96.85 | 95.75 | 1.35 |
| 1 − GD (GD loss) | 2.60 | 2.52 | 2.70 | 3.15 | 4.25 | 1.65 |
| GD* (%) | 99.16 | 99.22 | 99.24 | 99.27 | 99.45 | 0.29 |
| Proportion of unequal contributions of the founders in GD loss (%) | 0.84 | 0.78 | 0.76 | 0.73 | 0.55 | −0.29 |
| Proportion of random genetic drift in GD loss (%) | 1.75 | 1.75 | 1.94 | 2.42 | 3.70 | 1.95 |
| Proportion of random genetic drift and bottle necks in GD loss (%) | 2.60 | 2.52 | 2.70 | 3.15 | 4.25 | 1.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cartuche-Macas, L.F.; Guaman Ilvay, O.J.; Chacón, E.; Gutierrez-Reinoso, M.A.; Garcia-Herreros, M. Reproductive Performance, Inbreeding, and Genetic Diversity in Montbeliarde Dairy Cattle Obtained by Absorption Crossing. Animals 2025, 15, 322. https://doi.org/10.3390/ani15030322
Cartuche-Macas LF, Guaman Ilvay OJ, Chacón E, Gutierrez-Reinoso MA, Garcia-Herreros M. Reproductive Performance, Inbreeding, and Genetic Diversity in Montbeliarde Dairy Cattle Obtained by Absorption Crossing. Animals. 2025; 15(3):322. https://doi.org/10.3390/ani15030322
Chicago/Turabian StyleCartuche-Macas, Luis F., Oscar J. Guaman Ilvay, Edilberto Chacón, Miguel A. Gutierrez-Reinoso, and Manuel Garcia-Herreros. 2025. "Reproductive Performance, Inbreeding, and Genetic Diversity in Montbeliarde Dairy Cattle Obtained by Absorption Crossing" Animals 15, no. 3: 322. https://doi.org/10.3390/ani15030322
APA StyleCartuche-Macas, L. F., Guaman Ilvay, O. J., Chacón, E., Gutierrez-Reinoso, M. A., & Garcia-Herreros, M. (2025). Reproductive Performance, Inbreeding, and Genetic Diversity in Montbeliarde Dairy Cattle Obtained by Absorption Crossing. Animals, 15(3), 322. https://doi.org/10.3390/ani15030322








