Analysis of the Transcriptional Control of Bcl11b in Chicken: IRF1 and GATA1 as Negative Regulators
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Isolation and Culture of Cells
2.3. ALV-J Infection and Validation
2.4. Bioinformatics Analysis of Promoter
2.5. Vectors Construction
2.6. siRNA Design and In Vitro Transcription
2.7. Transfection and Dual Luciferase Assay
2.8. Promoter-Binding Transcription Factor Profiling Assay
2.9. Western Blot
2.10. ALV-J Intervention Study
2.11. qRT-PCR Analysis
2.12. Cell Apoptosis
2.13. Cell Proliferation Assay
2.14. BSP Methylation Sequencing
2.15. Statistical Analysis
3. Results
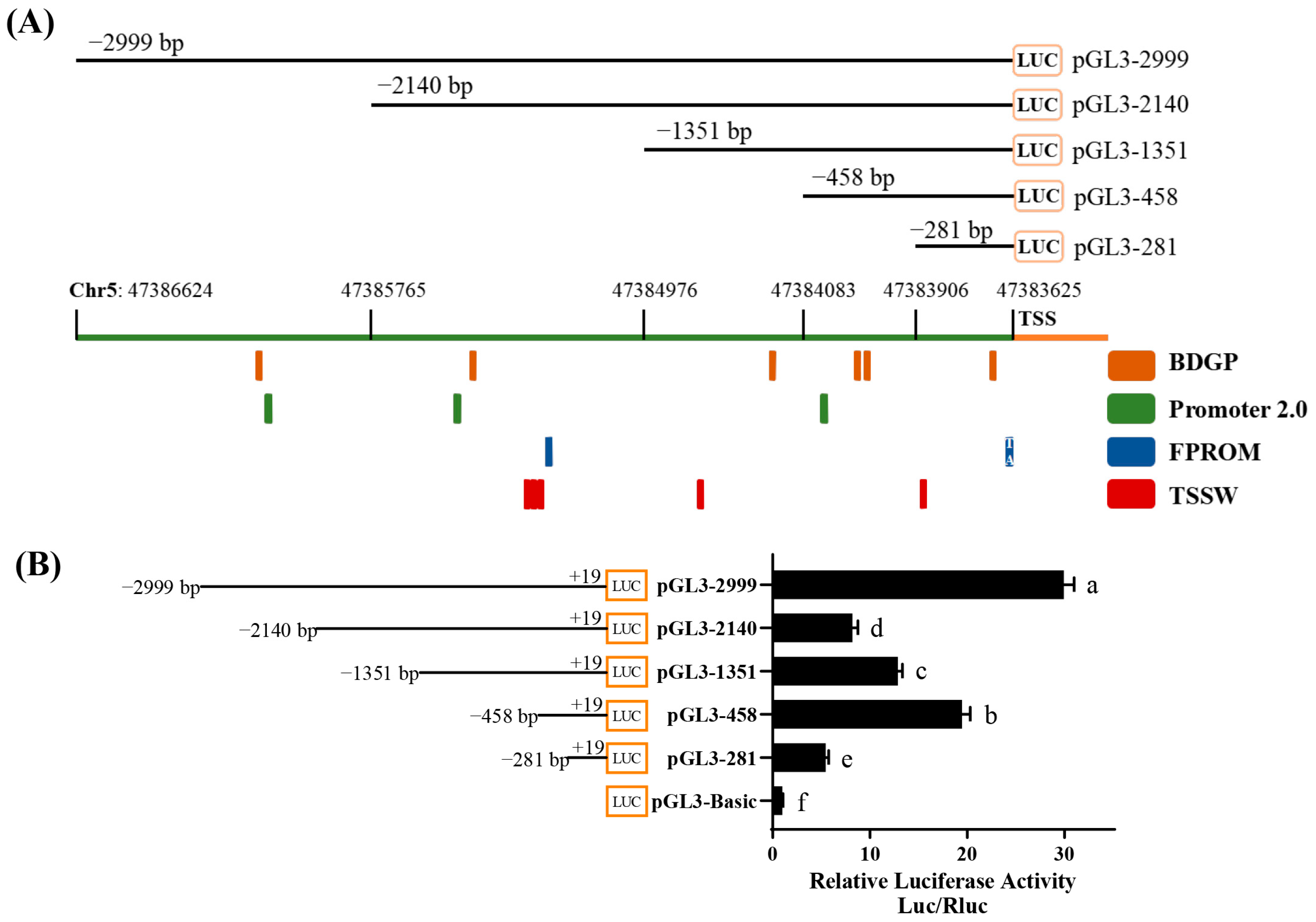
3.1. Promoter Region Analysis of Chicken Bcl11b Gene
3.2. Bioinformatics Analysis of the Promoter Region of Bcl11b
3.3. Identification of Core Regions and Key Transcription Factors of Chicken Bcl11b Promoter
3.4. Detection and Confirmation of Key Transcription Factors Binding Sites Within the Second Core Region of the Chicken Bcl11b Promoter
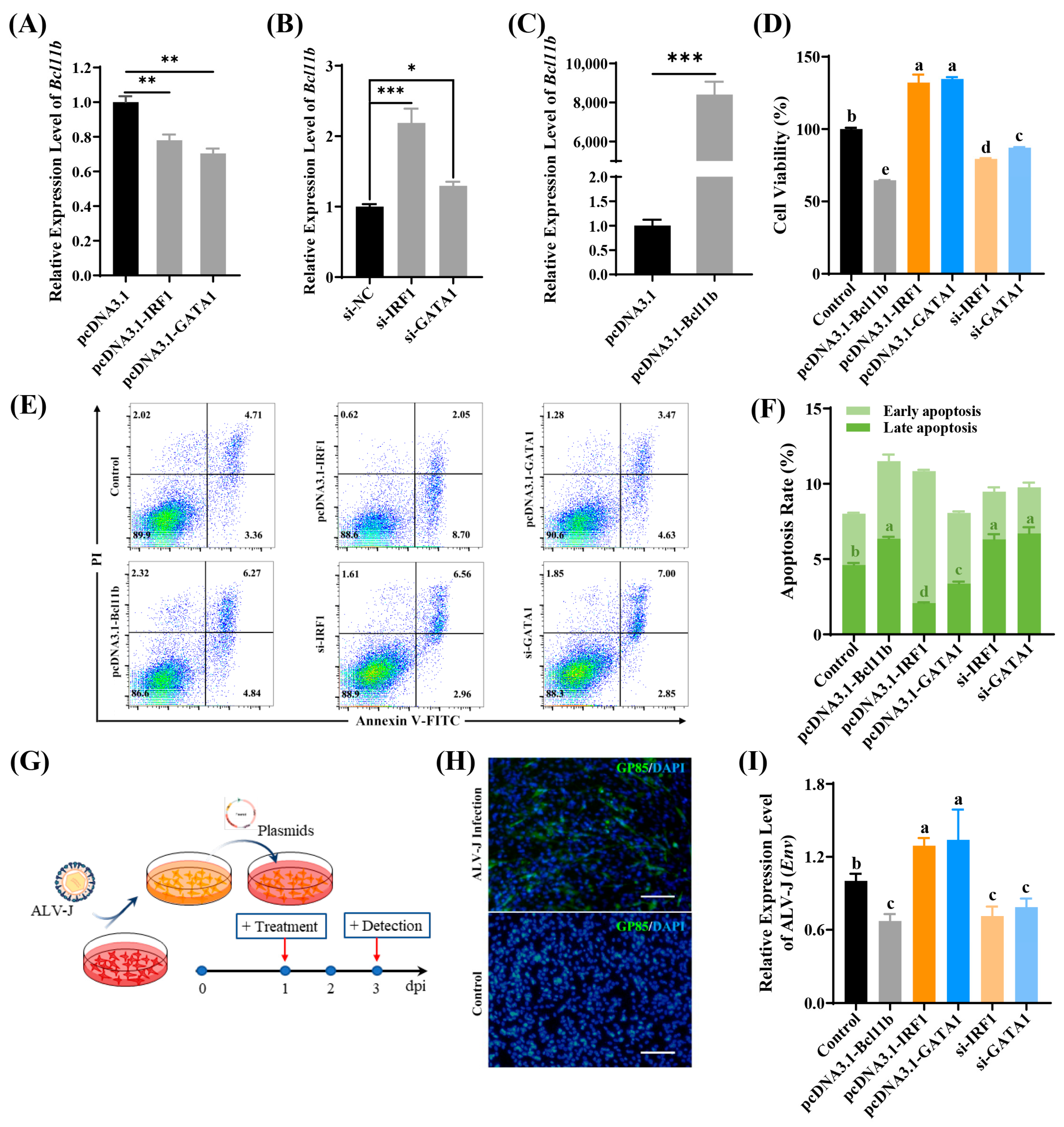
3.5. IRF1 and GATA1 Suppress Bcl11b Transcription and Promote ALV-J Replication
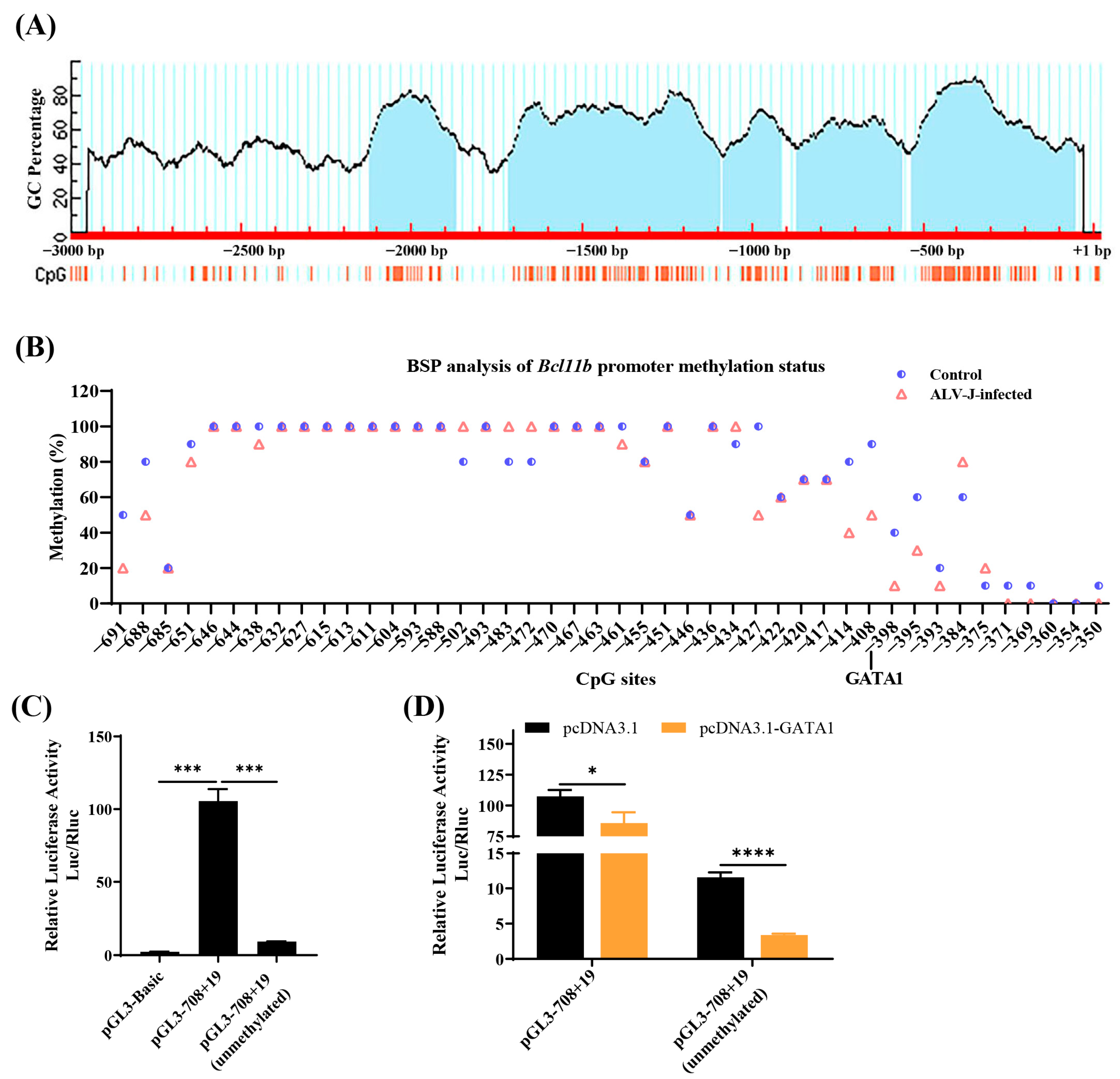
3.6. Methylation Analysis of the Second Core Region of the Chicken Bcl11b Promoter
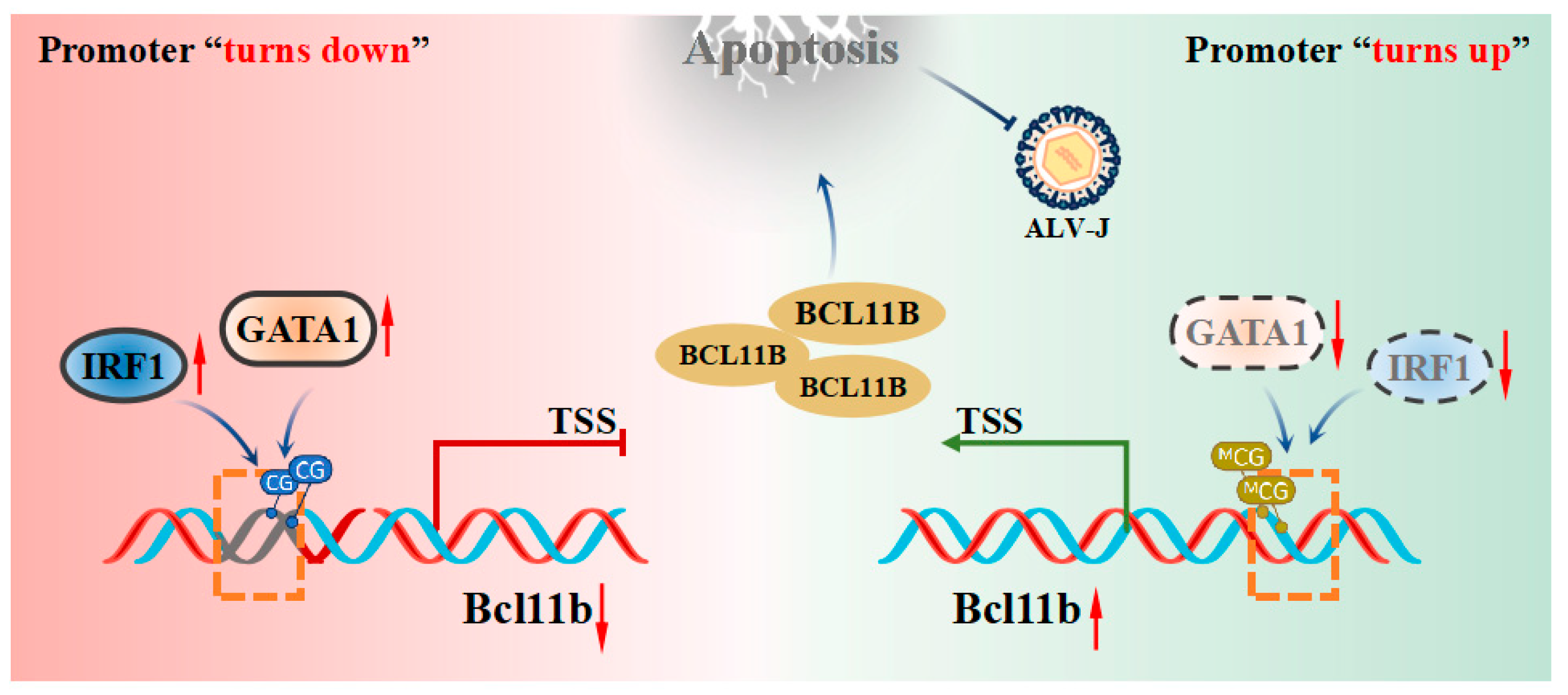
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Bcl11b | B-cell lymphoma/leukemia 11B |
| ALV-J | Avian leukosis virus subgroup J |
| IRF1 | Interferon regulatory factor-1 |
| GATA1 | GATA-binding protein 1 |
| TSS | Transcription start site |
| siRNA | Small Interfering RNA |
| hpi | Hours post-infection |
References
- Dong, X.; Ju, S.; Zhao, P.; Li, Y.; Meng, F.; Sun, P.; Cui, Z. Synergetic effects of subgroup J avian leukosis virus and reticuloendotheliosis virus co-infection on growth retardation and immunosuppression in SPF chickens. Vet. Microbiol. 2014, 172, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhao, P.; Chang, S.; Ju, S.; Li, Y.; Meng, F.; Sun, P.; Cui, Z. Synergistic pathogenic effects of co-infection of subgroup J avian leukosis virus and reticuloendotheliosis virus in broiler chickens. Avian Pathol. 2015, 44, 43–49. [Google Scholar] [CrossRef]
- Nair, V.; Gimeno, I.; Dunn, J.; Zavala, G.; Williams, S.M.; Reece, R.L.; Hafner, S. Neoplastic Diseases. In Diseases of Poultry, 14th ed.; Nair, V., Gimeno, I., Dunn, J., Zavala, G., Williams, S.M., Reece, R.L., Hafner, S., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 2020; Volume 1, pp. 548–715. [Google Scholar]
- Tang, S.; Li, J.; Chang, Y.-F.; Lin, W. Avian Leucosis Virus-Host Interaction: The Involvement of Host Factors in Viral Replication. Front. Immunol. 2022, 13, 907287. [Google Scholar] [CrossRef] [PubMed]
- Fandiño, S.; Gomez-Lucia, E.; Benítez, L.; Doménech, A. Avian Leukosis: Will We Be Able to Get Rid of It? Animals 2023, 13, 2358. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, C.; Deng, Y.; Pan, M.; Mo, G.; Liao, Z.; Zhang, X.; Zhang, D.; Li, H. PMAIP1 promotes J subgroup avian leukosis virus replication by regulating mitochondrial function. Poult. Sci. 2024, 103, 103617. [Google Scholar] [CrossRef] [PubMed]
- Mo, G.; Fu, H.; Hu, B.; Zhang, Q.; Xian, M.; Zhang, Z.; Lin, L.; Shi, M.; Nie, Q.; Zhang, X. SOCS3 Promotes ALV-J Virus Replication via Inhibiting JAK2/STAT3 Phosphorylation During Infection. Front. Cell. Infect. Microbiol. 2021, 11, 748795. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Liao, Z.; Dai, Z.; Yan, Y.; Yao, Z.; Chen, S.; Xie, Z.; Zhao, Q.; Lin, W.; et al. TCP1 mediates gp37 of avian leukosis virus subgroup J to inhibit autophagy through activating AKT in DF-1 cells. Vet. Microbiol. 2022, 271, 109472. [Google Scholar] [CrossRef]
- Li, T.; Xie, J.; Yao, X.; Zhang, J.; Li, C.; Ren, D.; Li, L.; Xie, Q.; Shao, H.; Qin, A.; et al. The tyrosine phosphatase SHP-2 dephosphorylated by ALV-J via its Env efficiently promotes ALV-J replication. Virulence 2021, 12, 1721–1731. [Google Scholar] [CrossRef]
- Chen, S.; Wang, D.; Liu, Y.; Zhao, R.; Wu, T.; Hu, X.; Pan, Z.; Cui, H. Targeting the Histone Methyltransferase Disruptor of Telomeric Silencing 1-Like Restricts Avian Leukosis Virus Subgroup J Replication by Restoring the Innate Immune Response in Chicken Macrophages. Front. Microbiol. 2020, 11, 603131. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, F.; Yin, H.; He, Q.; Lu, Y.; Zhu, Q.; Lan, X.; Zhao, X.; Li, D.; Liu, Y.; et al. Chicken interferon regulatory factor 7 (IRF7) can control ALV-J virus infection by triggering type I interferon production through affecting genes related with innate immune signaling pathway. Dev. Comp. Immunol. 2021, 119, 104026. [Google Scholar] [CrossRef]
- Li, L.; Feng, W.; Cheng, Z.; Yang, J.; Bi, J.; Wang, X.; Wang, G. TRIM62-mediated restriction of avian leukosis virus subgroup J replication is dependent on the SPRY domain. Poult. Sci. 2019, 98, 6019–6025. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Feng, M.; Zhang, X.; Li, X.; Mo, G.; Shi, M.; Zhang, X. Chicken CH25H inhibits ALV-J replication by promoting cellular autophagy. Front. Immunol. 2023, 14, 1093289. [Google Scholar] [CrossRef] [PubMed]
- Koslová, A.; Trefil, P.; Mucksová, J.; Reinišová, M.; Plachý, J.; Kalina, J.; Kučerová, D.; Geryk, J.; Krchlíková, V.; Lejčková, B.; et al. Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus. Proc. Natl. Acad. Sci. USA 2020, 117, 2108–2112. [Google Scholar] [CrossRef] [PubMed]
- Chai, N.; Bates, P. Na+ H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc. Natl. Acad. Sci. USA 2006, 103, 5531–5536. [Google Scholar] [CrossRef]
- Bangham, C.R.M.; Matoušková, M.; Plachý, J.; Kučerová, D.; Pecnová, Ľ.; Reinišová, M.; Geryk, J.; Karafiát, V.; Hron, T.; Hejnar, J. Rapid adaptive evolution of avian leukosis virus subgroup J in response to biotechnologically induced host resistance. PLoS Pathog. 2024, 20, e1012468. [Google Scholar]
- Qiu, L.; Yang, T.; Guo, Q.; Hua, T.; Bi, Y.; Chu, P.; Bai, H.; Chen, S.; Chang, G. C2H2-type zinc-finger protein BCL11B suppresses avian Leukosis virus subgroup J replication by regulating apoptosis. Int. J. Biol. Macromol. 2024, 275, 133644. [Google Scholar] [CrossRef]
- Qiu, L.; Chang, G.; Li, Z.; Bi, Y.; Liu, X.; Chen, G. Comprehensive Transcriptome Analysis Reveals Competing Endogenous RNA Networks During Avian Leukosis Virus, Subgroup J-Induced Tumorigenesis in Chickens. Front. Physiol. 2018, 9, 996. [Google Scholar] [CrossRef]
- Satterwhite, E.; Sonoki, T.; Willis, T.G.; Harder, L.; Nowak, R.; Arriola, E.L.; Liu, H.; Price, H.P.; Gesk, S.; Steinemann, D.; et al. The BCL11 gene family: Involvement of BCL11A in lymphoid malignancies. Blood 2001, 98, 3413–3420. [Google Scholar] [CrossRef]
- Dubuissez, M.; Loison, I.; Paget, S.; Vorng, H.; Ait-Yahia, S.; Rohr, O.; Tsicopoulos, A.; Leprince, D. PKC-mediated phosphorylation of BCL11B at Serine 2 negatively regulates its interaction with NuRD complexes during CD4+ T cell activation. Mol. Cell. Biol. 2016, 36, 1881–1898. [Google Scholar] [CrossRef]
- Vickridge, E.; Faraco, C.C.F.; Lo, F.Y.; Rahimian, H.; Liu, Z.Y.; Tehrani, P.S.; Djerir, B.; Ramdzan, Z.M.; Leduy, L.; Marechal, A.; et al. The function of BCL11B in base excision repair contributes to its dual role as an oncogene and a haplo-insufficient tumor suppressor gene. Nucleic Acids Res. 2024, 52, 223–242. [Google Scholar] [CrossRef]
- Dimopoulos, Y.P.; Loghavi, S. Acute leukemia of ambiguous lineage with BCL11B rearrangement. Blood 2024, 143, 183. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Okuyama, K.; Sidwell, T.; Yamashita, M.; Endo, T.; Satoh-Takayama, N.; Ohno, H.; Morio, T.; Rothenberg, E.V.; Taniuchi, I. A Bcl11bN797K variant isolated from an immunodeficient patient inhibits early thymocyte development in mice. Front. Immunol. 2024, 15, 1363704. [Google Scholar] [CrossRef] [PubMed]
- Koumoundourou, A.; Rannap, M.; De Bruyckere, E.; Nestel, S.; Reissner, C.; Egorov, A.; Liu, P.T.; Missler, M.; Heimrich, B.; Draguhn, A.; et al. Regulation of hippocampal mossy fiber-CA3 synapse function by a Bcl11b/C1ql2/Nrxn3(25b+) pathway. eLife 2024, 12, 89854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; M, R.; Verma, R.K.; Albu, D.I.; Califano, D.; VanValkenburgh, J.; Merchant, A.; Rangel-Moreno, J.; Randall, T.D.; Jenkins, N.A.; et al. Antigen-specific clonal expansion and cytolytic effector function of CD8+ T lymphocytes depend on the transcription factor Bcl11b. J. Exp. Med. 2010, 207, 1687–1699. [Google Scholar] [CrossRef]
- Cysique, L.A.; Juge, L.; Lennon, M.J.; Gates, T.M.; Brew, B.J. HIV brain latency as measured by CSF BcL11b relates to disrupted brain cellular energy in virally suppressed HIV infection. AIDS 2019, 33, 441–443. [Google Scholar] [CrossRef]
- Grabarczyk, P.; Winkler, P.; Delin, M.; Sappa, P.K.; Bekeschus, S.; Hildebrandt, P.; Przybylski, G.K.; Völker, U.; Hammer, E.; Schmidt, C.A. The N-Terminal CCHC Zinc Finger Motif Mediates Homodimerization of Transcription Factor BCL11B. Mol. Cell. Biol. 2018, 38, e00368-17. [Google Scholar] [CrossRef]
- Zhao, C.; Li, X.; Han, B.; You, Z.; Qu, L.; Liu, C.; Song, J.; Lian, L.; Yang, N. Gga-miR-219b targeting BCL11B suppresses proliferation, migration and invasion of Marek’s disease tumor cell MSB1. Sci. Rep. 2017, 7, 4247. [Google Scholar] [CrossRef]
- Nobuyuki, K.; Rika, F.; Tatsuhiko, O.; Takahiro, I.; Naoki, S.; Masaharu, I.; Martinez, C.J.A. Reduced Level of the BCL11B Protein Is Associated with Adult T-Cell Leukemia/Lymphoma. PLoS ONE 2013, 8, e55147. [Google Scholar]
- Jin, P.; Chen, H.; Xie, J.; Zhou, C.; Zhu, X. Essential role of microRNA-650 in the regulation of B-cell CLL/lymphoma 11B gene expression following transplantation: A novel mechanism behind the acute rejection of renal allografts. Int. J. Mol. Med. 2017, 40, 1840–1850. [Google Scholar] [CrossRef]
- He, Z.; Liao, Z.; Chen, S.; Li, B.; Yu, Z.; Luo, G.; Yang, L.; Zeng, C.; Li, Y. Downregulated miR-17, miR-29c, miR-92a and miR-214 may be related to BCL11B overexpression in T cell acute lymphoblastic leukemia. Asia-Pac. J. Clin. Oncol. 2018, 14, e259–e265. [Google Scholar] [CrossRef]
- Dolens, A.C.; Durinck, K.; Lavaert, M.; Van der Meulen, J.; Velghe, I.; De Medts, J.; Weening, K.; Roels, J.; De Mulder, K.; Volders, P.J.; et al. Distinct Notch1 and BCL11B requirements mediate human γδ/αβ T cell development. EMBO Rep. 2020, 21, e49006. [Google Scholar] [CrossRef]
- Hong, G.H.; Guan, Q.; Peng, H.; Luo, X.H.; Mao, Q. Identification and validation of a T-cell-related MIR600HG/hsa-mir-21-5p competing endogenous RNA network in tuberculosis activation based on integrated bioinformatics approaches. Front. Genet. 2022, 13, 979213. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, J.; Bai, Y.; Liu, Z.; Wei, Y.; Guo, D.; Jia, X.; Shi, B.; Zhang, X.; Zhao, Z.; et al. Unlocking the Transcriptional Control of NCAPG in Bovine Myoblasts: CREB1 and MYOD1 as Key Players. Int. J. Mol. Sci. 2024, 25, 2506. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cheng, Y.; Wei, F.; Liu, Y.; Zhu, R.; Zhao, P.; Zhang, J.; Xiang, C.; Kang, E.; Shang, Z. Arabidopsis thaliana MYC2 and MYC3 Are Involved in Ethylene-Regulated Hypocotyl Growth as Negative Regulators. Int. J. Mol. Sci. 2024, 25, 8022. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.V.; Shahmuradov, I.A.; Salamov, A.A. Identification of Promoter Regions and Regulatory Sites. In Computational Biology of Transcription Factor Binding; Ladunga, I., Ed.; Humana Press: Totowa, NJ, USA, 2010; Volume 674, pp. 57–83. [Google Scholar]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Mao, Z.; Qian, Y.; Liu, Z.; Shi, Y.; Fan, L.; Zhang, Q. LINC00158 modulates the function of BEAS-2B cells via targeting BCL11B and ameliorates OVA-LPS-induced severe asthma in mice models. Int. Immunopharmacol. 2024, 130, 111739. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, D.; Li, X.; Ge, M.; Hou, Z. Research Note: Identification of core promoter region of the polyunsaturated fatty acid synthesis-related gene family in chicken. Poult. Sci. 2023, 102, 102857. [Google Scholar] [CrossRef]
- García-Ojeda, M.E.; Klein Wolterink, R.G.J.; Lemaître, F.; Richard-Le Goff, O.; Hasan, M.; Hendriks, R.W.; Cumano, A.; Di Santo, J.P. GATA-3 promotes T-cell specification by repressing B-cell potential in pro–T cells in mice. Blood 2013, 121, 1749–1759. [Google Scholar] [CrossRef]
- Li, L.; Zhang, J.A.; Dose, M.; Kueh, H.Y.; Mosadeghi, R.; Gounari, F.; Rothenberg, E.V. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 2013, 122, 902–911. [Google Scholar] [CrossRef]
- Wang, F.; Qi, Z.; Yao, Y.; Yu, G.; Feng, T.; Zhao, T.; Xue, H.-H.; Zhao, Y.; Jiang, P.; Bao, L.; et al. Exploring the stage-specific roles of Tcf-1 in T cell development and malignancy at single-cell resolution. Cell. Mol. Immunol. 2021, 18, 644–659. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.-L.; Gupta, S.; Miele, A.; Shiffman, D.; Stein, J.L.; Stein, G.S.; van Wijnen, A.J. The Tumor Suppressor Interferon Regulatory Factor 1 Interferes with SP1 Activation to Repress the Human CDK2 Promoter. J. Biol. Chem. 2003, 278, 26589–26596. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, K.; Yokosawa, H. Degradation of transcription factor IRF-1 by the ubiquitin–proteasome pathway. Eur. J. Biochem. 2000, 267, 1680–1686. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhu, X.X.; Wei, R.; Zhao, L.; Zhang, Z.; Yin, X.Q.; Zhang, Y.H.; Chu, C.; Wang, B.; Li, X. miR-130b-3p regulates M1 macrophage polarization via targeting IRF1. J. Cell. Physiol. 2021, 236, 2008–2022. [Google Scholar] [CrossRef]
- Ramsauer, K.; Farlik, M.; Zupkovitz, G.; Seiser, C.; Kroger, A.; Hauser, H.; Decker, T. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-γ-inducible gbp2 gene. Proc. Natl. Acad. Sci. USA 2007, 104, 2849–2854. [Google Scholar] [CrossRef]
- Ren, G.; Cui, K.; Zhang, Z.; Zhao, K. Division of labor between IRF1 and IRF2 in regulating different stages of transcriptional activation in cellular antiviral activities. Cell Biosci. 2015, 5, 17. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, X.; Wang, W.; Wang, Y.; Yin, Y.; Laan, L.J.W.; Sprengers, D.; Metselaar, H.J.; Peppelenbosch, M.P.; Pan, Q. IFN regulatory factor 1 restricts hepatitis E virus replication by activating STAT1 to induce antiviral IFN-stimulated genes. FASEB J. 2016, 30, 3352–3367. [Google Scholar] [CrossRef]
- Purbey, P.K.; Seo, J.; Paul, M.K.; Iwamoto, K.S.; Daly, A.E.; Feng, A.-C.; Champhekar, A.S.; Langerman, J.; Campbell, K.M.; Schaue, D.; et al. Opposing tumor-cell-intrinsic and -extrinsic roles of the IRF1 transcription factor in antitumor immunity. Cell Rep. 2024, 43, 114289. [Google Scholar] [CrossRef]
- Xu, Y.-Y.; Jin, R.; Zhou, G.-P.; Xu, H.-G. Involvement of GATA1 and Sp3 in the activation of the murine STING gene promoter in NIH3T3 cells. Sci. Rep. 2017, 7, 2090. [Google Scholar] [CrossRef]
- Atashi, A.; Jafaripour, L.; Froughi, K.; Behzadifard, M. GATA1 transcription factor targets the gene expression of B19 virus in HEK293 cell line. Ann. Med. Surg. 2024, 86, 7120–7124. [Google Scholar] [CrossRef]
- Seo, M.J.; Liu, X.; Chang, M.; Park, J.H. GATA-binding protein 1 is a novel transcription regulator of peroxiredoxin 5 in human breast cancer cells. Int. J. Oncol. 2012, 40, 655–664. [Google Scholar] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-H.; Kuo, H.-C.; Pan, C.-T.; Lin, Y.-S.; Huang, Y.-H.; Li, S.-C. Multiomics analyses identified epigenetic modulation of the S100A gene family in Kawasaki disease and their significant involvement in neutrophil transendothelial migration. Clin. Epigenetics 2018, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, eaaj2239. [Google Scholar] [CrossRef]
- Cui, T.T.; Huang, J.X.; Ning, B.L.; Mu, F.; Chen, H.Y.; Xing, T.Y.; Li, H.; Wang, N. DNA methylation promotes the expression of PPARγ transcript 1 at least in part by preventing NRF1 binding to the promoter P1 of chicken PPARγ gene. Poult. Sci. 2024, 103, 103559. [Google Scholar] [CrossRef]
- Bahar Halpern, K.; Vana, T.; Walker, M.D. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. J. Biol. Chem. 2014, 289, 23882–23892. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, L.; Wang, H.; Li, W.; Yang, T.; Bai, H.; Chang, G. Analysis of the Transcriptional Control of Bcl11b in Chicken: IRF1 and GATA1 as Negative Regulators. Animals 2025, 15, 665. https://doi.org/10.3390/ani15050665
Qiu L, Wang H, Li W, Yang T, Bai H, Chang G. Analysis of the Transcriptional Control of Bcl11b in Chicken: IRF1 and GATA1 as Negative Regulators. Animals. 2025; 15(5):665. https://doi.org/10.3390/ani15050665
Chicago/Turabian StyleQiu, Lingling, Haojie Wang, Wenhao Li, Ting Yang, Hao Bai, and Guobin Chang. 2025. "Analysis of the Transcriptional Control of Bcl11b in Chicken: IRF1 and GATA1 as Negative Regulators" Animals 15, no. 5: 665. https://doi.org/10.3390/ani15050665
APA StyleQiu, L., Wang, H., Li, W., Yang, T., Bai, H., & Chang, G. (2025). Analysis of the Transcriptional Control of Bcl11b in Chicken: IRF1 and GATA1 as Negative Regulators. Animals, 15(5), 665. https://doi.org/10.3390/ani15050665




