Gut Microbiota of Ruminants and Monogastric Livestock: An Overview
Simple Summary
Abstract
1. Introduction
2. Gut Microbiota in Ruminants
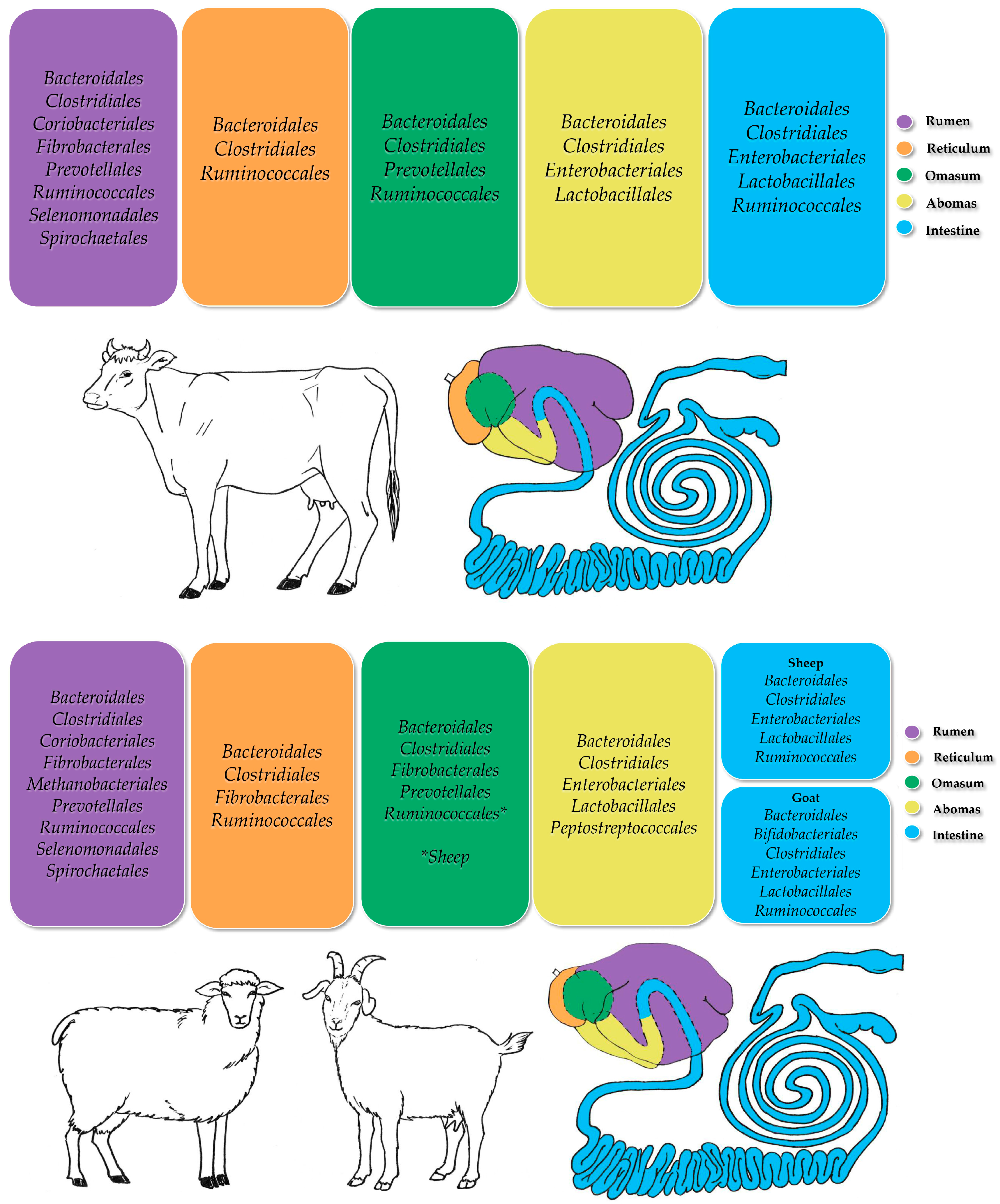
2.1. Cattle
2.2. Sheep
2.3. Goats
3. Gut Microbiota in Monogastric Species
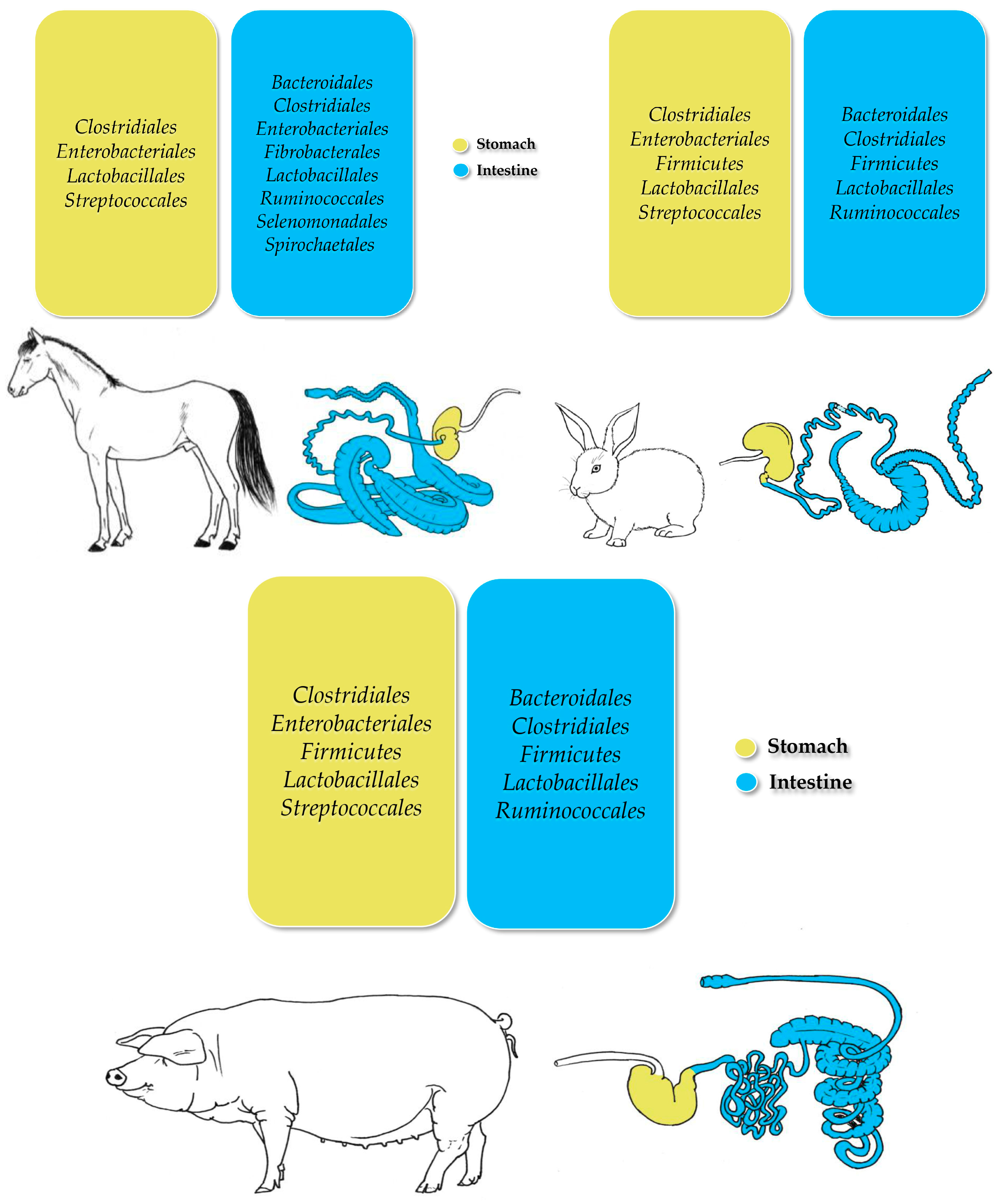
3.1. Horses
3.2. Pigs
3.3. Rabbits
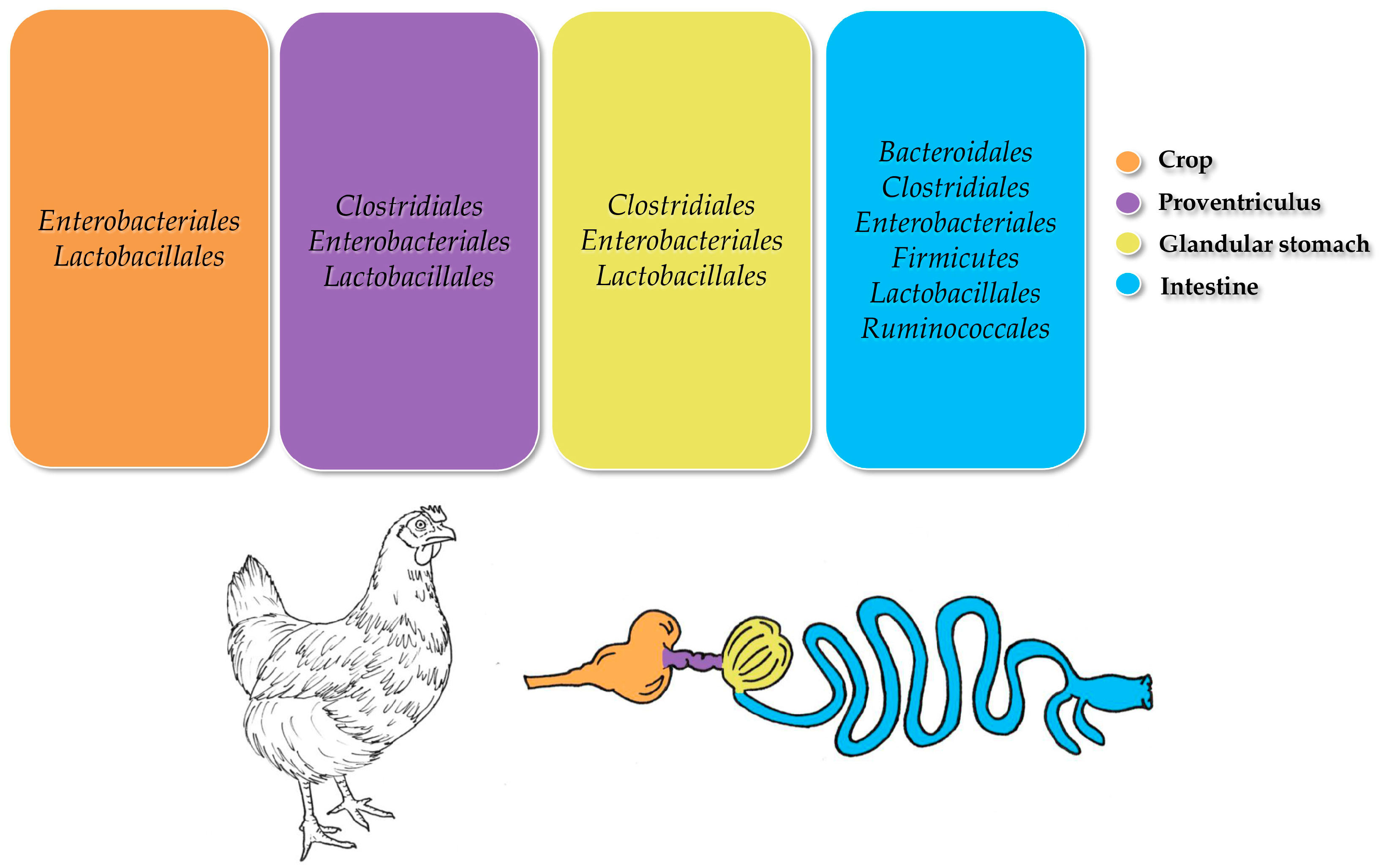
3.4. Chickens
4. Challenges in Sampling Gut Microbiota
5. Gut Microbiota in Nutrient Metabolism and Health
6. Gut Microbiota and Reproductive Endocrine Axis
7. Microbiota Manipulation: From Microbiome Characterization to Livestock Applications
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, W.; Dong, Y.; Guo, W.; Zhang, X.; Degen, A.A.; Bi, S.; Ding, L.; Chen, X.; Long, R. Linkages between Rumen Microbiome, Host, and Environment in Yaks, and Their Implications for Understanding Animal Production and Management. Front. Microbiol. 2024, 15, 1301258. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.Y.; Ji, S.K.; Yan, H.; Wang, Y.J.; Liu, J.J.; Cao, Z.J.; Yang, H.J.; Zhang, W.J.; Li, S.L. Dynamic Change of the Gastrointestinal Bacterial Ecology in Cows from Birth to Adulthood. MicrobiologyOpen 2020, 9, e1119. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.A.; Hassan, F.; Rehman, M.S.; Huws, S.A.; Cheng, Y.; Din, A.U. Gut Microbiome Colonization and Development in Neonatal Ruminants: Strategies, Prospects, and Opportunities. Anim. Nutr. 2021, 7, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Wegener Parfrey, L.; Walters, W.A.; Knight, R. Microbial Eukaryotes in the Human Microbiome: Ecology, Evolution, and Future Directions. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef]
- Gresse, R.; Chaucheyras Durand, F.; Dunière, L.; Blanquet-Diot, S.; Forano, E. Microbiota Composition and Functional Profiling throughout the Gastrointestinal Tract of Commercial Weaning Piglets. Microorganisms 2019, 7, 343. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Hill, C. Bacteriophages of the Human Gut: The “Known Unknown” of the Microbiome. Cell Host Microbe 2019, 25, 195–209. [Google Scholar] [CrossRef]
- Lin, Y.; Zhou, B.; Zhu, W. Pathogenic Escherichia Coli-Specific Bacteriophages and Polyvalent Bacteriophages in Piglet Guts with Increasing Coliphage Numbers after Weaning. Appl. Environ. Microbiol. 2021, 87, e00966-21. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and Cons of Phage Therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef]
- Hartinger, T.; Zebeli, Q. The Present Role and New Potentials of Anaerobic Fungi in Ruminant Nutrition. J. Fungi 2021, 7, 200. [Google Scholar] [CrossRef]
- Król, B.; Słupczyńska, M.; Wilk, M.; Asghar, M.; Cwynar, P. Anaerobic Rumen Fungi and Fungal Direct-Fed Microbials in Ruminant Feeding. J. Anim. Feed Sci. 2023, 32, 3–16. [Google Scholar] [CrossRef]
- Jyothi, C.; Muwel, N.; Nayak, S.; Khare, A.; Sharma, R.; Keshri, A.; Singour, S.; Mishra, R.; Tiwari, S. Anaerobic Rumen Fungi as a Feed Additive in Ruminants: A Review. J. Livest. Sci. 2024, 15, 78–85. [Google Scholar] [CrossRef]
- Hess, M.; Paul, S.S.; Puniya, A.K.; Van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerová, K. Anaerobic Fungi: Past, Present, and Future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef]
- Iliev, I.D.; Underhill, D.M. Striking a Balance: Fungal Commensalism versus Pathogenesis. Curr. Opin. Microbiol. 2013, 16, 366–373. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut Biogeography of the Bacterial Microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef]
- Bickhart, D.M.; McClure, J.C.; Schnabel, R.D.; Rosen, B.D.; Medrano, J.F.; Smith, T.P.L. Symposium Review: Advances in Sequencing Technology Herald a New Frontier in Cattle Genomics and Genome-Enabled Selection. J. Dairy Sci. 2020, 103, 5278–5290. [Google Scholar] [CrossRef]
- Russell, J.B.; Rychlik, J.L. Factors That Alter Rumen Microbial Ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef]
- Tugnoli, B.; Giovagnoni, G.; Piva, A.; Grilli, E. From Acidifiers to Intestinal Health Enhancers: How Organic Acids Can Improve Growth Efficiency of Pigs. Animals 2020, 10, 134. [Google Scholar] [CrossRef]
- McCallum, G.; Tropini, C. The Gut Microbiota and Its Biogeography. Nat. Rev. Microbiol. 2024, 22, 105–118. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Li, M.; Goonewardene, L.A.; Oba, M.; Guan, L.L. Effect of Calf Starter Feeding on Gut Microbial Diversity and Expression of Genes Involved in Host Immune Responses and Tight Junctions in Dairy Calves during Weaning Transition. J. Dairy. Sci. 2013, 96, 3189–3200. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Amos, G.C.A.; Logan, A.; Anwar, S.; Fritzsche, M.; Mate, R.; Bleazard, T.; Rijpkema, S. Developing Standards for the Microbiome Field. Microbiome 2020, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Wegl, G.; Grabner, N.; Köstelbauer, A.; Klose, V.; Ghanbari, M. Toward Best Practice in Livestock Microbiota Research: A Comprehensive Comparison of Sample Storage and DNA Extraction Strategies. Front. Microbiol. 2021, 12, 627539. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Guan, L.L. Gut Microbiome and Omics: A New Definition to Ruminant Production and Health. Anim. Front. 2016, 6, 8–12. [Google Scholar] [CrossRef]
- Alexander, T.W.; Plaizier, J.C. From the Editors: The Importance of Microbiota in Ruminant Production. Anim. Front. 2016, 6, 4–7. [Google Scholar] [CrossRef]
- Cholewińska, P.; Czyż, K.; Nowakowski, P.; Wyrostek, A. The Microbiome of the Digestive System of Ruminants—A Review. Anim. Health Res. Rev. 2020, 21, 3–14. [Google Scholar] [CrossRef]
- Alexandratos, N.; Bruinsma, J. (Eds.) World Agriculture Towards 2030/2050: The 2012 Revision; ESA Working Papers 12-03; FAO: Rome, Italy, 2012. [Google Scholar]
- Chen, B.; Li, D.; Leng, D.; Kui, H.; Bai, X.; Wang, T. Gut Microbiota and Meat Quality. Front. Microbiol. 2022, 13, 951726. [Google Scholar] [CrossRef]
- Celi, P.; Cowieson, A.J.; Fru-Nji, F.; Steinert, R.E.; Kluenter, A.-M.; Verlhac, V. Gastrointestinal Functionality in Animal Nutrition and Health: New Opportunities for Sustainable Animal Production. Anim. Feed Sci. Technol. 2017, 234, 88–100. [Google Scholar] [CrossRef]
- Denman, S.E.; Morgavi, D.P.; McSweeney, C.S. Review: The Application of Omics to Rumen Microbiota Function. Animal 2018, 12, s233–s245. [Google Scholar] [CrossRef]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and Comparison of Microbiota in the Gastrointestinal Tracts of the Goat (Capra Hircus) During Preweaning Development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef]
- Vántus, V.B.; Kovács, M.; Zsolnai, A. The Rabbit Caecal Microbiota: Development, Composition and Its Role in the Prevention of Digestive Diseases–a Review on Recent Literature in the Light of Molecular Genetic Methods. Acta Agrar. Kaposváriensis 2014, 18, 55–65. [Google Scholar]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen Microbial Community Composition Varies with Diet and Host, but a Core Microbiome Is Found across a Wide Geographical Range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Estellé, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.Ø.; Kjeldsen, N.J.; Liu, C. A Reference Gene Catalogue of the Pig Gut Microbiome. Nat. Microbiol. 2016, 1, 16161. [Google Scholar] [CrossRef] [PubMed]
- Reece, W.O.; Rowe, E.W. Functional Anatomy and Physiology of Domestic Animals; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 978-1-119-27084-3. [Google Scholar]
- Bagwan, W.A.; Durgawale, P.; Jadhav, M.A.V.; Vidyapeeth, K.V. Exploring the Gut Microbiota of Ruminants and Its Impact on Digestive Efficiency and Methane Emissions in Livestock Production Systems. Afr. J. Biol. Sci. 2024, 6, 2226–2236. [Google Scholar]
- Na, S.W.; Guan, L.L. Understanding the Role of Rumen Epithelial Host-Microbe Interactions in Cattle Feed Efficiency. Anim. Nutr. 2022, 10, 41–53. [Google Scholar] [CrossRef]
- Palumbo, F.; Squartini, A.; Barcaccia, G.; Macolino, S.; Pornaro, C.; Pindo, M.; Sturaro, E.; Ramanzin, M. A Multi-Kingdom Metabarcoding Study on Cattle Grazing Alpine Pastures Discloses Intra-Seasonal Shifts in Plant Selection and Faecal Microbiota. Sci. Rep. 2021, 11, 889. [Google Scholar] [CrossRef]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the Bacterial Microbiota across the Gastrointestinal Tracts of Dairy Cattle: Membership and Potential Function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef]
- McGovern, E.; Kenny, D.A.; McCabe, M.S.; Fitzsimons, C.; McGee, M.; Kelly, A.K.; Waters, S.M. 16S rRNA Sequencing Reveals Relationship Between Potent Cellulolytic Genera and Feed Efficiency in the Rumen of Bulls. Front. Microbiol. 2018, 9, 01842. [Google Scholar] [CrossRef]
- Daghio, M.; Ciucci, F.; Buccioni, A.; Cappucci, A.; Casarosa, L.; Serra, A.; Conte, G.; Viti, C.; McAmmond, B.M.; Van Hamme, J.D.; et al. Correlation of Breed, Growth Performance, and Rumen Microbiota in Two Rustic Cattle Breeds Reared Under Different Conditions. Front. Microbiol. 2021, 12, 652031. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Li, S.; Tun, H.M.; Khafipour, E. Nutritional Models of Experimentally-Induced Subacute Ruminal Acidosis (SARA) Differ in Their Impact on Rumen and Hindgut Bacterial Communities in Dairy Cows. Front. Microbiol. 2017, 7, 02128. [Google Scholar] [CrossRef]
- Rudi, K.; Moen, B.; Sekelja, M.; Frisli, T.; Lee, M.R. An Eight-Year Investigation of Bovine Livestock Fecal Microbiota. Vet. Microbiol. 2012, 160, 369–377. [Google Scholar] [CrossRef]
- Jami, E.; Mizrahi, I. Composition and Similarity of Bovine Rumen Microbiota across Individual Animals. PLoS ONE 2012, 7, e33306. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Connor, E.E.; Li, C.; Baldwin, R.L., VI; Sparks, M.E. Characterization of the Rumen Microbiota of Pre-Ruminant Calves Using Metagenomic Tools. Environ. Microbiol. 2012, 14, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hagey, J.V.; Bhatnagar, S.; Heguy, J.M.; Karle, B.M.; Price, P.L.; Meyer, D.; Maga, E.A. Fecal Microbial Communities in a Large Representative Cohort of California Dairy Cows. Front. Microbiol. 2019, 10, 1093. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Sun, H.; Wu, X.; Guan, L.L.; Liu, J. Assessment of Rumen Microbiota from a Large Dairy Cattle Cohort Reveals the Pan and Core Bacteriomes Contributing to Varied Phenotypes. Appl. Environ. Microbiol. 2018, 84, e00970-18. [Google Scholar] [CrossRef]
- Taxis, T.M.; Wolff, S.; Gregg, S.J.; Minton, N.O.; Zhang, C.; Dai, J.; Schnabel, R.D.; Taylor, J.F.; Kerley, M.S.; Pires, J.C.; et al. The Players May Change but the Game Remains: Network Analyses of Ruminal Microbiomes Suggest Taxonomic Differences Mask Functional Similarity. Nucleic Acids Res. 2015, 43, 9600–9612. [Google Scholar] [CrossRef]
- Forcina, G.; Pérez-Pardal, L.; Carvalheira, J.; Beja-Pereira, A. Gut Microbiome Studies in Livestock: Achievements, Challenges, and Perspectives. Animals 2022, 12, 3375. [Google Scholar] [CrossRef]
- Rawal, S.; Kaur, H.; Bhathan, S.; Mittal, D.; Kaur, G.; Ali, S.A. Ruminant Gut Microbiota: Interplay, Implications, and Innovations for Sustainable Livestock Production. In Sustainable Agriculture Reviews: Animal Biotechnology for Livestock Production 4; Kumar Yata, V., Mohanty, A.K., Lichtfouse, E., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 205–228. ISBN 978-3-031-54372-2. [Google Scholar]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A Heritable Subset of the Core Rumen Microbiome Dictates Dairy Cow Productivity and Emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Walker, A.W.; Roehe, R.; Watson, M. Compendium of 4,941 Rumen Metagenome-Assembled Genomes for Rumen Microbiome Biology and Enzyme Discovery. Nat. Biotechnol. 2019, 37, 953–961. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Sun, H.-Z.; Wu, X.-H.; Liu, J.-X.; Guan, L.L. Multi-Omics Reveals That the Rumen Microbiome and Its Metabolome Together with the Host Metabolome Contribute to Individualized Dairy Cow Performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef]
- Scicutella, F.; Cucu, M.A.; Mannelli, F.; Pastorelli, R.; Daghio, M.; Paoli, P.; Pazzagli, L.; Turini, L.; Mantino, A.; Luti, S.; et al. Rumen Microbial Community and Milk Quality in Holstein Lactating Cows Fed Olive Oil Pomace as Part in a Sustainable Feeding Strategy. Animal 2023, 17, 100815. [Google Scholar] [CrossRef]
- Song, Y.C.; Holland, S.I.; Lee, M.; Chen, G.; Zaugg, J.; Löffler, F.E.; Manefield, M.J.; Hugenholtz, P.; Kappler, U. Corrigendum: ‘A Comparative Genome Analysis of the Bacillota (Firmicutes) Class Dehalobacteriia’. Microb. Genom. 2023, 9, 001092. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the Bovine Rumen Bacterial Community from Birth to Adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Nagata, R.; Ohkubo, A.; Ohtani, N.; Kushibiki, S.; Ichijo, T.; Sato, S. Changes in Ruminal and Reticular pH and Bacterial Communities in Holstein Cattle Fed a High-Grain Diet. BMC Vet. Res. 2018, 14, 310. [Google Scholar] [CrossRef] [PubMed]
- Myer, P.R.; Freetly, H.C.; Wells, J.E.; Smith, T.P.L.; Kuehn, L.A. Analysis of the Gut Bacterial Communities in Beef Cattle and Their Association with Feed Intake, Growth, and Efficiency. J. Anim. Sci. 2017, 95, 3215–3224. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, R.; Dicksved, J.; Sun, L.; Gonda, H.; Müller, B.; Schnürer, A.; Bertilsson, J. Methane Production in Dairy Cows Correlates with Rumen Methanogenic and Bacterial Community Structure. Front. Microbiol. 2017, 8, 226. [Google Scholar] [CrossRef]
- Danielsson, R.; Schnürer, A.; Arthurson, V.; Bertilsson, J. Methanogenic Population and CH4 Production in Swedish Dairy Cows Fed Different Levels of Forage. Appl. Environ. Microbiol. 2012, 78, 6172–6179. [Google Scholar] [CrossRef]
- Janssen, P.H.; Kirs, M. Structure of the Archaeal Community of the Rumen. Appl. Environ. Microbiol. 2008, 74, 3619–3625. [Google Scholar] [CrossRef]
- Toyber, I.; Kumar, R.; Jami, E. Rumen Protozoa Are a Hub for Diverse Hydrogenotrophic Functions. Environ. Microbiol. Rep. 2024, 16, e13298. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Forano, E.; Martin, C.; Newbold, C.J. Microbial Ecosystem and Methanogenesis in Ruminants. Animal 2010, 4, 1024–1036. [Google Scholar] [CrossRef]
- Foggi, G.; Terranova, M.; Daghio, M.; Amelchanka, S.L.; Conte, G.; Ineichen, S.; Agnolucci, M.; Viti, C.; Mantino, A.; Buccioni, A.; et al. Evaluation of Ruminal Methane and Ammonia Formation and Microbiota Composition as Affected by Supplements Based on Mixtures of Tannins and Essential Oils Using Rusitec. J. Anim. Sci. Biotechnol. 2024, 15, 48. [Google Scholar] [CrossRef]
- Palangi, V.; Taghizadeh, A.; Abachi, S.; Lackner, M. Strategies to Mitigate Enteric Methane Emissions in Ruminants: A Review. Sustainability 2022, 14, 13229. [Google Scholar] [CrossRef]
- Popova, M.; McGovern, E.; McCabe, M.S.; Martin, C.; Doreau, M.; Arbre, M.; Meale, S.J.; Morgavi, D.P.; Waters, S.M. The Structural and Functional Capacity of Ruminal and Cecal Microbiota in Growing Cattle Was Unaffected by Dietary Supplementation of Linseed Oil and Nitrate. Front. Microbiol. 2017, 8, 937. [Google Scholar] [CrossRef] [PubMed]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Characterization of the Core Rumen Microbiome in Cattle during Transition from Forage to Concentrate as Well as during and after an Acidotic Challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, L.; He, Y.; Luo, X.; Zhao, S.; Jia, X. Composition of Fecal Microbiota in Grazing and Feedlot Angus Beef Cattle. Animals 2021, 11, 3167. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Z.; Guo, P.; Li, F.; Chang, S.; Yan, T.; Zheng, H.; Hou, F. Shift of Feeding Strategies from Grazing to Different Forage Feeds Reshapes the Rumen Microbiota To Improve the Ability of Tibetan Sheep (Ovis Aries) To Adapt to the Cold Season. Microbiol. Spectr. 2023, 11, e02816-22. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.; Han, Y.; Zhao, J.; Zhou, Z. Characterization of the Microbial Communities along the Gastrointestinal Tract of Sheep by 454 Pyrosequencing Analysis. Asian-Australas J. Anim. Sci. 2017, 30, 100–110. [Google Scholar] [CrossRef]
- Wang, X.; Hu, L.; Liu, H.; Xu, T.; Zhao, N.; Zhang, X.; Geng, Y.; Kang, S.; Xu, S. Characterization of the Bacterial Microbiota across the Different Intestinal Segments of the Qinghai Semi-Fine Wool Sheep on the Qinghai-Tibetan Plateau. Anim. Biosci. 2021, 34, 1921–1929. [Google Scholar] [CrossRef]
- Cortés, A.; Wills, J.; Su, X.; Hewitt, R.E.; Robertson, J.; Scotti, R.; Price, D.R.G.; Bartley, Y.; McNeilly, T.N.; Krause, L.; et al. Infection with the Sheep Gastrointestinal Nematode Teladorsagia Circumcincta Increases Luminal Pathobionts. Microbiome 2020, 8, 60. [Google Scholar] [CrossRef]
- McLoughlin, S.; Spillane, C.; Claffey, N.; Smith, P.E.; O’Rourke, T.; Diskin, M.G.; Waters, S.M. Rumen Microbiome Composition Is Altered in Sheep Divergent in Feed Efficiency. Front. Microbiol. 2020, 11, 1981. [Google Scholar] [CrossRef]
- Chang, J.; Yao, X.; Zuo, C.; Qi, Y.; Chen, D.; Ma, W. The Gut Bacterial Diversity of Sheep Associated with Different Breeds in Qinghai Province. BMC Vet. Res. 2020, 16, 254. [Google Scholar] [CrossRef]
- Minozzi, G.; Biscarini, F.; Dalla Costa, E.; Chincarini, M.; Ferri, N.; Palestrini, C.; Minero, M.; Mazzola, S.; Piccinini, R.; Vignola, G. Analysis of Hindgut Microbiome of Sheep and Effect of Different Husbandry Conditions. Animals 2020, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.K.; Zhang, X.X.; Li, F.D.; Li, C.; Li, G.Z.; Zhang, D.Y.; Song, Q.Z.; Li, X.L.; Zhao, Y.; Wang, W.M. Characterization of the Rumen Microbiota and Its Relationship with Residual Feed Intake in Sheep. Animal 2021, 15, 100161. [Google Scholar] [CrossRef]
- Jewell, K.A.; McCormick, C.A.; Odt, C.L.; Weimer, P.J.; Suen, G. Ruminal Bacterial Community Composition in Dairy Cows Is Dynamic over the Course of Two Lactations and Correlates with Feed Efficiency. Appl. Environ. Microbiol. 2015, 81, 4697–4710. [Google Scholar] [CrossRef] [PubMed]
- Shabat, S.K.B.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific Microbiome-Dependent Mechanisms Underlie the Energy Harvest Efficiency of Ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zeng, D.; Ni, X.; Zhu, H.; Jian, P.; Zhou, Y.; Xu, S.; Lin, Y.; Li, Y.; Yin, Z.; et al. Microbial Community Compositions in the Gastrointestinal Tract of Chinese Mongolian Sheep Using Illumina MiSeq Sequencing Revealed High Microbial Diversity. AMB Express 2017, 7, 75. [Google Scholar] [CrossRef]
- Xie, F.; Jin, W.; Si, H.; Yuan, Y.; Tao, Y.; Liu, J.; Wang, X.; Yang, C.; Li, Q.; Yan, X. An Integrated Gene Catalog and over 10,000 Metagenome-Assembled Genomes from the Gastrointestinal Microbiome of Ruminants. Microbiome 2021, 9, 137. [Google Scholar] [CrossRef]
- Mani, S.; Aiyegoro, O.A.; Adeleke, M.A. Association between Host Genetics of Sheep and the Rumen Microbial Composition. Trop. Anim. Health Prod. 2022, 54, 109. [Google Scholar] [CrossRef]
- Lv, W.; Liu, X.; Sha, Y.; Shi, H.; Wei, H.; Luo, Y.; Wang, J.; Li, S.; Hu, J.; Guo, X. Rumen Fermentation—Microbiota—Host Gene Expression Interactions to Reveal the Adaptability of Tibetan Sheep in Different Periods. Animals 2021, 11, 3529. [Google Scholar] [CrossRef]
- Tanca, A.; Fraumene, C.; Manghina, V.; Palomba, A.; Abbondio, M.; Deligios, M.; Pagnozzi, D.; Addis, M.F.; Uzzau, S. Diversity and Functions of the Sheep Faecal Microbiota: A Multi-Omic Characterization. Microb. Biotechnol. 2017, 10, 541–554. [Google Scholar] [CrossRef]
- Guo, H.; Cui, J.; Li, Q.; Liang, X.; Li, J.; Yang, B.; Kalds, P.; Chen, Y.; Yang, Y. A Multi-Omic Assessment of the Mechanisms of Intestinal Microbes Used to Treat Diarrhea in Early-Weaned Lambs. Msystems 2024, 9, e00953-23. [Google Scholar] [CrossRef]
- Perea, K.; Perz, K.; Olivo, S.K.; Williams, A.; Lachman, M.; Ishaq, S.L.; Thomson, J.; Yeoman, C.J. Feed Efficiency Phenotypes in Lambs Involve Changes in Ruminal, Colonic, and Small-Intestine-Located Microbiota. J. Anim. Sci. 2017, 95, 2585–2592. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Xu, X.; Zhang, J.; Zhang, L. Effect of Different Feeding Methods on Rumen Microbes in Growing Chinese Tan Sheep. Rev. Bras. Zootec. 2020, 49, e20190258. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, K.; Zhang, C.; Feng, Y.; Zhang, X.; Wang, X.; Wu, G. Dynamics and Stabilization of the Rumen Microbiome in Yearling Tibetan Sheep. Sci. Rep. 2019, 9, 19620. [Google Scholar] [CrossRef]
- Daghio, M.; Viti, C.; Mannelli, F.; Pauselli, M.; Natalello, A.; Luciano, G.; Valenti, B.; Buccioni, A. A Diet Supplemented with Hazelnut Skin Changes the Microbial Community Composition and the Biohydrogenation Pattern of Linoleic Acid in the Rumen of Growing Lambs. Ital. J. Anim. Sci. 2021, 20, 1256–1263. [Google Scholar] [CrossRef]
- Lima, J.; Ingabire, W.; Roehe, R.; Dewhurst, R.J. Estimating Microbial Protein Synthesis in the Rumen—Can ‘Omics’ Methods Provide New Insights into a Long-Standing Question? Vet. Sci. 2023, 10, 679. [Google Scholar] [CrossRef]
- Zhou, G.; Zhou, X.; He, Y.; Shao, J.; Hu, Z.; Liu, R.; Zhou, H.; Hosseinibai, S. Grazing Intensity Significantly Affects Belowground Carbon and Nitrogen Cycling in Grassland Ecosystems: A Meta-Analysis. Glob. Change Biol. 2017, 23, 1167–1179. [Google Scholar] [CrossRef]
- Liu, X.; Sha, Y.; Lv, W.; Cao, G.; Guo, X.; Pu, X.; Wang, J.; Li, S.; Hu, J.; Luo, Y. Multi-Omics Reveals That the Rumen Transcriptome, Microbiome, and Its Metabolome Co-Regulate Cold Season Adaptability of Tibetan Sheep. Front. Microbiol. 2022, 13, 859601. [Google Scholar] [CrossRef]
- Asanuma, N.; Yokoyama, S.; Hino, T. Effects of Nitrate Addition to a Diet on Fermentation and Microbial Populations in the Rumen of Goats, with Special Reference to Elenomonas Ruminantium Having the Ability to Reduce Nitrate and Nitrite. Anim. Sci. J. 2015, 86, 378–384. [Google Scholar] [CrossRef]
- Cremonesi, P.; Conte, G.; Severgnini, M.; Turri, F.; Monni, A.; Capra, E.; Rapetti, L.; Colombini, S.; Chessa, S.; Battelli, G.; et al. Evaluation of the Effects of Different Diets on Microbiome Diversity and Fatty Acid Composition of Rumen Liquor in Dairy Goat. Animal 2018, 12, 1856–1866. [Google Scholar] [CrossRef]
- Fliegerova, K.O.; Podmirseg, S.M.; Vinzelj, J.; Grilli, D.J.; Kvasnová, S.; Schierová, D.; Sechovcová, H.; Mrázek, J.; Siddi, G.; Arenas, G.N. The Effect of a High-Grain Diet on the Rumen Microbiome of Goats with a Special Focus on Anaerobic Fungi. Microorganisms 2021, 9, 157. [Google Scholar] [CrossRef]
- Peng, X.; Wilken, S.E.; Lankiewicz, T.S.; Gilmore, S.P.; Brown, J.L.; Henske, J.K.; Swift, C.L.; Salamov, A.; Barry, K.; Grigoriev, I.V. Genomic and Functional Analyses of Fungal and Bacterial Consortia That Enable Lignocellulose Breakdown in Goat Gut Microbiomes. Nat. Microbiol. 2021, 6, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Min, B.R.; Solaiman, S.; Shange, R.; Eun, J.-S. Gastrointestinal Bacterial and Methanogenic Archaea Diversity Dynamics Associated with Condensed Tannin-Containing Pine Bark Diet in Goats Using 16S rDNA Amplicon Pyrosequencing. Int. J. Microbiol. 2014, 2014, 141909. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Liu, G.; Meng, F.; Hong, L.; Li, Y.; Lian, Z.; Yang, Z.; Luo, C.; Liu, D. Exploring the Rumen and Cecum Microbial Community from Fetus to Adulthood in Goat. Animals 2020, 10, 1639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cai, K.; Mishra, R.; Jha, R. In Ovo Supplementation of Chitooligosaccharide and Chlorella Polysaccharide Affects Cecal Microbial Community, Metabolic Pathways, and Fermentation Metabolites in Broiler Chickens. Poult. Sci. 2020, 99, 4776–4785. [Google Scholar] [CrossRef]
- Palma-Hidalgo, J.M.; Jiménez, E.; Popova, M.; Morgavi, D.P.; Martín-García, A.I.; Yáñez-Ruiz, D.R.; Belanche, A. Inoculation with Rumen Fluid in Early Life Accelerates the Rumen Microbial Development and Favours the Weaning Process in Goats. Anim. Microbiome 2021, 3, 11. [Google Scholar] [CrossRef]
- Bu, D.; Zhang, X.; Ma, L.; Park, T.; Wang, L.; Wang, M.; Xu, J.; Yu, Z. Repeated Inoculation of Young Calves With Rumen Microbiota Does Not Significantly Modulate the Rumen Prokaryotic Microbiota Consistently but Decreases Diarrhea. Front. Microbiol. 2020, 11, 1403. [Google Scholar] [CrossRef]
- Mammeri, M.; Obregón, D.A.; Chevillot, A.; Polack, B.; Julien, C.; Pollet, T.; Cabezas-Cruz, A.; Adjou, K.T. Cryptosporidium Parvum Infection Depletes Butyrate Producer Bacteria in Goat Kid Microbiome. Front. Microbiol. 2020, 11, 548737. [Google Scholar] [CrossRef]
- Tong, J.; Ma, W.; Yang, R.; Wang, T.; Chen, X.; Zhang, X.; Tang, X.; Wen, Y.; Chang, J.; Chen, D. Dysbiosis of the Gut Microbiota Maybe Exacerbate Orf Pathology by Promoting Inflammatory Immune Responses. Vet. Microbiol. 2020, 251, 108884. [Google Scholar] [CrossRef]
- Jiang, S.; Huo, D.; You, Z.; Peng, Q.; Ma, C.; Chang, H.; Lin, X.; Wang, L.; Zhang, J. The Distal Intestinal Microbiome of Hybrids of Hainan Black Goats and Saanen Goats. PLoS ONE 2020, 15, e0228496. [Google Scholar] [CrossRef]
- Wang, L.; Shah, A.M.; Liu, Y.; Jin, L.; Wang, Z.; Xue, B.; Peng, Q. Relationship between True Digestibility of Dietary Phosphorus and Gastrointestinal Bacteria of Goats. PLoS ONE 2020, 15, e0225018. [Google Scholar] [CrossRef]
- Ren, Z.; Yao, R.; Liu, Q.; Deng, Y.; Shen, L.; Deng, H.; Zuo, Z.; Wang, Y.; Deng, J.; Cui, H. Effects of Antibacterial Peptides on Rumen Fermentation Function and Rumen Microorganisms in Goats. PLoS ONE 2019, 14, e0221815. [Google Scholar] [CrossRef] [PubMed]
- Shabana, I.; Bouqellah, N.A.; Albakri, N.N. Comparative Metagenomic Analysis of Small Ruminants’ Fecal Microbiota. Res. Sq. 2020, 10. [Google Scholar] [CrossRef]
- Wang, Z.; Yin, L.; Liu, L.; Lan, X.; He, J.; Wan, F.; Shen, W.; Tang, S.; Tan, Z.; Yang, Y. Tannic Acid Reduced Apparent Protein Digestibility and Induced Oxidative Stress and Inflammatory Response without Altering Growth Performance and Ruminal Microbiota Diversity of Xiangdong Black Goats. Front. Vet. Sci. 2022, 9, 1004841. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Feng, T.; Wu, Y.; Xu, Y.; Du, L.; Wang, T.; Luo, Y.; Wang, Y.; Li, Z.; Xuan, Z.; et al. The Multi-Kingdom Microbiome of the Goat Gastrointestinal Tract. Microbiome 2023, 11, 219. [Google Scholar] [CrossRef]
- Dal Pont, G.C.; Eyng, C.; Bortoluzzi, C.; Kogut, M.H. Enzymes and Gut Health in Monogastric Animals: Effects Beyond Digestibility. In Gut Microbiota, Immunity, and Health in Production Animals; Kogut, M.H., Zhang, G., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 33–55. ISBN 978-3-030-90303-9. [Google Scholar]
- Zhao, G.; Xiang, Y.; Wang, X.; Dai, B.; Zhang, X.; Ma, L.; Yang, H.; Lyu, W. Exploring the Possible Link between the Gut Microbiome and Fat Deposition in Pigs. Oxidative Med. Cell. Longev. 2022, 2022, 1098892. [Google Scholar] [CrossRef]
- Williams, A.R.; Myhill, L.J.; Stolzenbach, S.; Nejsum, P.; Mejer, H.; Nielsen, D.S.; Thamsborg, S.M. Emerging Interactions between Diet, Gastrointestinal Helminth Infection, and the Gut Microbiota in Livestock. BMC Vet. Res. 2021, 17, 62. [Google Scholar] [CrossRef]
- Jha, R.; Berrocoso, J.F.D. Dietary Fiber and Protein Fermentation in the Intestine of Swine and Their Interactive Effects on Gut Health and on the Environment: A Review. Anim. Feed Sci. Technol. 2016, 212, 18–26. [Google Scholar] [CrossRef]
- Kauter, A.; Epping, L.; Semmler, T.; Antao, E.-M.; Kannapin, D.; Stoeckle, S.D.; Gehlen, H.; Lübke-Becker, A.; Günther, S.; Wieler, L.H.; et al. The Gut Microbiome of Horses: Current Research on Equine Enteral Microbiota and Future Perspectives. Anim. Microbiome 2019, 1, 14. [Google Scholar] [CrossRef]
- Pan, D.; Yu, Z. Intestinal Microbiome of Poultry and Its Interaction with Host and Diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef]
- Metcalf, J.L.; Song, S.J.; Morton, J.T.; Weiss, S.; Seguin-Orlando, A.; Joly, F.; Feh, C.; Taberlet, P.; Coissac, E.; Amir, A.; et al. Evaluating the Impact of Domestication and Captivity on the Horse Gut Microbiome. Sci. Rep. 2017, 7, 15497. [Google Scholar] [CrossRef]
- Boucher, L.; Leduc, L.; Leclère, M.; Costa, M.C. Current Understanding of Equine Gut Dysbiosis and Microbiota Manipulation Techniques: Comparison with Current Knowledge in Other Species. Animals 2024, 14, 758. [Google Scholar] [CrossRef]
- Lara, F.; Castro, R.; Thomson, P. Changes in the Gut Microbiome and Colic in Horses: Are They Causes or Consequences? Open Vet. J. 2022, 12, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Garber, A.; Hastie, P.; Murray, J.-A. Factors Influencing Equine Gut Microbiota: Current Knowledge. J. Equine Vet. Sci. 2020, 88, 102943. [Google Scholar] [CrossRef]
- Sorensen, R.J.; Drouillard, J.S.; Douthit, T.L.; Ran, Q.; Marthaler, D.G.; Kang, Q.; Vahl, C.I.; Lattimer, J.M. Effect of Hay Type on Cecal and Fecal Microbiome and Fermentation Parameters in Horses. J. Anim. Sci. 2021, 99, skaa407. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, A.C.; Johnson, P.J.; Lopes, M.A.; Perry, S.C.; Lanter, H.R. A Microbiological Map of the Healthy Equine Gastrointestinal Tract. PLoS ONE 2016, 11, e0166523. [Google Scholar] [CrossRef]
- Mach, N.; Foury, A.; Kittelmann, S.; Reigner, F.; Moroldo, M.; Ballester, M.; Esquerré, D.; Rivière, J.; Sallé, G.; Gérard, P.; et al. The Effects of Weaning Methods on Gut Microbiota Composition and Horse Physiology. Front. Physiol. 2017, 8, 535. [Google Scholar] [CrossRef]
- Mura, E.; Edwards, J.; Kittelmann, S.; Kaerger, K.; Voigt, K.; Mrázek, J.; Moniello, G.; Fliegerova, K. Anaerobic Fungal Communities Differ along the Horse Digestive Tract. Fungal Biol. 2019, 123, 240–246. [Google Scholar] [CrossRef]
- Costa, M.C.; Silva, G.; Ramos, R.V.; Staempfli, H.R.; Arroyo, L.G.; Kim, P.; Weese, J.S. Characterization and Comparison of the Bacterial Microbiota in Different Gastrointestinal Tract Compartments in Horses. Vet. J. 2015, 205, 74–80. [Google Scholar] [CrossRef]
- McKinney, C.A.; Oliveira, B.C.; Bedenice, D.; Paradis, M.-R.; Mazan, M.; Sage, S.; Sanchez, A.; Widmer, G. The Fecal Microbiota of Healthy Donor Horses and Geriatric Recipients Undergoing Fecal Microbial Transplantation for the Treatment of Diarrhea. PLoS ONE 2020, 15, e0230148. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Sacy, A.; Karges, K.; Apper, E. Gastro-Intestinal Microbiota in Equines and Its Role in Health and Disease: The Black Box Opens. Microorganisms 2022, 10, 2517. [Google Scholar] [CrossRef]
- Álvarez Narváez, S.; Beaudry, M.S.; Norris, C.G.; Bartlett, P.B.; Glenn, T.C.; Sanchez, S. Improved Equine Fecal Microbiome Characterization Using Target Enrichment by Hybridization Capture. Animals 2024, 14, 445. [Google Scholar] [CrossRef] [PubMed]
- Cooke, C.G.; Gibb, Z.; Grupen, C.G.; Schemann, K.; Deshpande, N.; Harnett, J.E. Effect of Probiotics and Prebiotics on the Composition of the Equine Fecal and Seminal Microbiomes and Sperm Quality: A Pilot Study. J. Equine Vet. Sci. 2024, 135, 105032. [Google Scholar] [CrossRef] [PubMed]
- Dou, S.; Gadonna-Widehem, P.; Rome, V.; Hamoudi, D.; Rhazi, L.; Lakhal, L.; Larcher, T.; Bahi-Jaber, N.; Pinon-Quintana, A.; Guyonvarch, A. Characterisation of Early-Life Fecal Microbiota in Susceptible and Healthy Pigs to Post-Weaning Diarrhoea. PLoS ONE 2017, 12, e0169851. [Google Scholar] [CrossRef] [PubMed]
- Sutera, A.M.; Arfuso, F.; Tardiolo, G.; Riggio, V.; Fazio, F.; Aiese Cigliano, R.; Paytuví, A.; Piccione, G.; Zumbo, A. Effect of a Co-Feed Liquid Whey-Integrated Diet on Crossbred Pigs’ Fecal Microbiota. Animals 2023, 13, 1750. [Google Scholar] [CrossRef]
- Tardiolo, G.; Romeo, O.; Zumbo, A.; Di Marsico, M.; Sutera, A.M.; Cigliano, R.A.; Paytuví, A.; D’Alessandro, E. Characterization of the Nero Siciliano Pig Fecal Microbiota after a Liquid Whey-Supplemented Diet. Animals 2023, 13, 642. [Google Scholar] [CrossRef]
- De Rodas, B.; Youmans, B.P.; Danzeisen, J.L.; Tran, H.; Johnson, T.J. Microbiome Profiling of Commercial Pigs from Farrow to Finish. J. Anim. Sci. 2018, 96, 1778–1794. [Google Scholar] [CrossRef]
- Motta, V.; Trevisi, P.; Bertolini, F.; Ribani, A.; Schiavo, G.; Fontanesi, L.; Bosi, P. Exploring Gastric Bacterial Community in Young Pigs. PLoS ONE 2017, 12, e0173029. [Google Scholar] [CrossRef]
- Verbeek, E.; Keeling, L.; Landberg, R.; Lindberg, J.E.; Dicksved, J. The Gut Microbiota and Microbial Metabolites Are Associated with Tail Biting in Pigs. Sci. Rep. 2021, 11, 20547. [Google Scholar] [CrossRef]
- Borewicz, K.A.; Kim, H.B.; Singer, R.S.; Gebhart, C.J.; Sreevatsan, S.; Johnson, T.; Isaacson, R.E. Changes in the Porcine Intestinal Microbiome in Response to Infection with Salmonella Enterica and Lawsonia Intracellularis. PLoS ONE 2015, 10, e0139106. [Google Scholar] [CrossRef]
- Azziz, G.; Giménez, M.; Carballo, C.; Espino, N.; Barlocco, N.; Batista, S. Characterization of the Fecal Microbiota of Pampa Rocha Pigs, a Genetic Resource Endemic to Eastern Uruguay. Heliyon 2023, 9, e16643. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Gan, M.; Chen, L.; Zhao, Y.; Zhu, Y.; Niu, L.; Zhang, S.; Zhu, L.; Shen, L. Gut Microbiota Composition and Diversity in Different Commercial Swine Breeds in Early and Finishing Growth Stages. Animals 2022, 12, 1607. [Google Scholar] [CrossRef] [PubMed]
- Han, G.G.; Lee, J.-Y.; Jin, G.-D.; Park, J.; Choi, Y.H.; Chae, B.J.; Kim, E.B.; Choi, Y.-J. Evaluating the Association between Body Weight and the Intestinal Microbiota of Weaned Piglets via 16S rRNA Sequencing. Appl. Microbiol. Biotechnol. 2017, 101, 5903–5911. [Google Scholar] [CrossRef] [PubMed]
- Holman, D.B.; Gzyl, K.E.; Mou, K.T.; Allen, H.K. Weaning Age and Its Effect on the Development of the Swine Gut Microbiome and Resistome. Msystems 2021, 6, e00682-21. [Google Scholar] [CrossRef] [PubMed]
- Gaio, D.; DeMaere, M.Z.; Anantanawat, K.; Chapman, T.A.; Djordjevic, S.P.; Darling, A.E. Post-Weaning Shifts in Microbiome Composition and Metabolism Revealed by over 25,000 Pig Gut Metagenome-Assembled Genomes. Microb. Genom. 2021, 7, 000501. [Google Scholar]
- Guevarra, R.B.; Hong, S.H.; Cho, J.H.; Kim, B.-R.; Shin, J.; Lee, J.H.; Kang, B.N.; Kim, Y.H.; Wattanaphansak, S.; Isaacson, R.E.; et al. The Dynamics of the Piglet Gut Microbiome during the Weaning Transition in Association with Health and Nutrition. J. Anim. Sci. Biotechnol. 2018, 9, 54. [Google Scholar] [CrossRef]
- Munk, P.; Andersen, V.D.; de Knegt, L.; Jensen, M.S.; Knudsen, B.E.; Lukjancenko, O.; Mordhorst, H.; Clasen, J.; Agersø, Y.; Folkesson, A. A Sampling and Metagenomic Sequencing-Based Methodology for Monitoring Antimicrobial Resistance in Swine Herds. J. Antimicrob. Chemother. 2017, 72, 385–392. [Google Scholar] [CrossRef]
- Gweon, H.S.; Shaw, L.P.; Swann, J.; De Maio, N.; AbuOun, M.; Niehus, R.; Hubbard, A.; Bowes, M.J.; Bailey, M.J.; Peto, T.E. The Impact of Sequencing Depth on the Inferred Taxonomic Composition and AMR Gene Content of Metagenomic Samples. Environ. Microbiome 2019, 14, 1–15. [Google Scholar] [CrossRef]
- Fenske, G.J.; Ghimire, S.; Antony, L.; Christopher-Hennings, J.; Scaria, J. Integration of Culture-Dependent and Independent Methods Provides a More Coherent Picture of the Pig Gut Microbiome. FEMS Microbiol. Ecol. 2020, 96, fiaa022. [Google Scholar] [CrossRef]
- Wylezich, C.; Belka, A.; Hanke, D.; Beer, M.; Blome, S.; Höper, D. Metagenomics for Broad and Improved Parasite Detection: A Proof-of-Concept Study Using Swine Faecal Samples. Int. J. Parasitol. 2019, 49, 769–777. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Prenafeta-Boldú, F.; Zingaretti, L.M.; Gonzalez-Rodriguez, O.; Dalmau, A.; Quintanilla, R.; Ballester, M. Gut Eukaryotic Communities in Pigs: Diversity, Composition and Host Genetics Contribution. Anim. Microbiome 2020, 2, 18. [Google Scholar] [CrossRef]
- Mann, E.; Schmitz-Esser, S.; Zebeli, Q.; Wagner, M.; Ritzmann, M.; Metzler-Zebeli, B.U. Mucosa-Associated Bacterial Microbiome of the Gastrointestinal Tract of Weaned Pigs and Dynamics Linked to Dietary Calcium-Phosphorus. PLoS ONE 2014, 9, e86950. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wei, S.; Chen, N.; Xiang, Y.; Wang, Y.; Jin, M. Characteristics of Gut Microbiota in Pigs with Different Breeds, Growth Periods and Genders. Microb. Biotechnol. 2022, 15, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Combes, S.; Fortun-Lamothe, L.; Cauquil, L.; Gidenne, T. Engineering the Rabbit Digestive Ecosystem to Improve Digestive Health and Efficacy. Animal 2013, 7, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Han, S.; Wang, P.; Wen, B.; Jian, W.; Guo, W.; Yu, Z.; Du, D.; Fu, X.; Kong, F.; et al. The Bacterial Communities Associated with Fecal Types and Body Weight of Rex Rabbits. Sci. Rep. 2015, 5, 9342. [Google Scholar] [CrossRef]
- Velasco-Galilea, M.; Piles, M.; Viñas, M.; Rafel, O.; González-Rodríguez, O.; Guivernau, M.; Sánchez, J.P. Rabbit Microbiota Changes Throughout the Intestinal Tract. Front. Microbiol. 2018, 9, 2144. [Google Scholar] [CrossRef]
- Ye, D.; Ding, X.; Pang, S.; Gan, Y.; Li, Z.; Gan, Q.; Fang, S. Seasonal Variations in Production Performance, Health Status, and Gut Microbiota of Meat Rabbit Reared in Semi-Confined Conditions. Animals 2023, 14, 113. [Google Scholar] [CrossRef]
- Clemente, J.C.; Manasson, J.; Scher, J.U. The Role of the Gut Microbiome in Systemic Inflammatory Disease. BMJ 2018, 360, j5145. [Google Scholar] [CrossRef]
- Fang, S.; Chen, X.; Ye, X.; Zhou, L.; Xue, S.; Gan, Q. Effects of Gut Microbiome and Short-Chain Fatty Acids (SCFAs) on Finishing Weight of Meat Rabbits. Front. Microbiol. 2020, 11, 1835. [Google Scholar] [CrossRef]
- Wang, Z.; He, H.; Chen, M.; Ni, M.; Yuan, D.; Cai, H.; Chen, Z.; Li, M.; Xu, H. Impact of Coprophagy Prevention on the Growth Performance, Serum Biochemistry, and Intestinal Microbiome of Rabbits. BMC Microbiol. 2023, 23, 125. [Google Scholar] [CrossRef]
- De Cesare, A.; Sirri, F.; Manfreda, G.; Moniaci, P.; Giardini, A.; Zampiga, M.; Meluzzi, A. Effect of Dietary Supplementation with Lactobacillus Acidophilus D2/CSL (CECT 4529) on Caecum Microbioma and Productive Performance in Broiler Chickens. PLoS ONE 2017, 12, e0176309. [Google Scholar] [CrossRef]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and Shotgun Sequencing Data for the Taxonomic Characterization of the Gut Microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, R.; Ravi, A.; Getino, M.; Pursley, I.; Horton, D.L.; Alikhan, N.-F.; Baker, D.; Gharbi, K.; Hall, N.; Watson, M.; et al. Extensive Microbial Diversity within the Chicken Gut Microbiome Revealed by Metagenomics and Culture. PeerJ 2021, 9, e10941. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Hou, L.; Yang, Y. The Development of the Gut Microbiota and Short-Chain Fatty Acids of Layer Chickens in Different Growth Periods. Front. Vet. Sci. 2021, 8, 666535. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, G.A.; Richardson, E.; Clark, J.; Keshri, J.; Drechsler, Y.; Berrang, M.E.; Meinersmann, R.J.; Cox, N.A.; Oakley, B.B. Broiler Chickens and Early Life Programming: Microbiome Transplant-Induced Cecal Community Dynamics and Phenotypic Effects. PLoS ONE 2020, 15, e0242108. [Google Scholar] [CrossRef]
- Kabir, M.H.B.; Han, Y.; Lee, S.-H.; Nugraha, A.B.; Recuenco, F.; Murakoshi, F.; Xuan, X.; Kato, K. Prevalence and Molecular Characterization of Cryptosporidium Species in Poultry in Bangladesh. One Health 2020, 9, 100122. [Google Scholar] [CrossRef]
- Lee, S.-J.; Cho, S.; La, T.-M.; Lee, H.-J.; Lee, J.-B.; Park, S.-Y.; Song, C.-S.; Choi, I.-S.; Lee, S.-W. Comparison of Microbiota in the Cloaca, Colon, and Magnum of Layer Chicken. PLoS ONE 2020, 15, e0237108. [Google Scholar] [CrossRef]
- Wei, S.; Morrison, M.; Yu, Z. Bacterial Census of Poultry Intestinal Microbiome. Poult. Sci. 2013, 92, 671–683. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding Host-Microbial Interactions in Rumen: Searching the Best Opportunity for Microbiota Manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Yu, Z.; Xu, Q.; Zheng, N.; Zhao, S.; Huang, G.; Wang, J. Ruminal Microbiota–Host Interaction and Its Effect on Nutrient Metabolism. Anim. Nutr. 2021, 7, 49–55. [Google Scholar] [CrossRef]
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Durunna, O.N.; Moore, S.S.; Guan, L.L. Impact of Feed Efficiency and Diet on Adaptive Variations in the Bacterial Community in the Rumen Fluid of Cattle. Appl. Environ. Microbiol. 2012, 78, 1203–1214. [Google Scholar] [CrossRef]
- de Assis Lage, C.F.; Räisänen, S.E.; Melgar, A.; Nedelkov, K.; Chen, X.; Oh, J.; Fetter, M.E.; Indugu, N.; Bender, J.S.; Vecchiarelli, B.; et al. Comparison of Two Sampling Techniques for Evaluating Ruminal Fermentation and Microbiota in the Planktonic Phase of Rumen Digesta in Dairy Cows. Front. Microbiol. 2020, 11, 618032. [Google Scholar] [CrossRef]
- Amin, N.; Schwarzkopf, S.; Kinoshita, A.; Tröscher-Mußotter, J.; Dänicke, S.; Camarinha-Silva, A.; Huber, K.; Frahm, J.; Seifert, J. Evolution of Rumen and Oral Microbiota in Calves Is Influenced by Age and Time of Weaning. Anim. Microbiome 2021, 3, 31. [Google Scholar] [CrossRef]
- Hagey, J.V.; Laabs, M.; Maga, E.A.; DePeters, E.J. Rumen Sampling Methods Bias Bacterial Communities Observed. PLoS ONE 2022, 17, e0258176. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Su, Q.; Li, Q.; Liu, J.; Du, Y.; Shi, H.; Li, Z.; Ma, Y.; Niu, Y.; Chen, L.; et al. Microbial Community Analyses Provide a Differential Diagnosis for the Antemortem and Postmortem Injury of Decayed Cadaver: An Animal Model. J. Forensic Leg. Med. 2023, 93, 102473. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, X.; Hu, S.; Nie, H.; Gui, P.; Zhong, Z.; Guo, Y.; Zhao, X. Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation. Microorganisms 2023, 11, 2811. [Google Scholar] [CrossRef]
- Hilal, M.G.; Yu, Q.; Zhou, R.; Wang, Y.; Feng, T.; Li, X.; Li, H. Exploring Microbial Communities, Assessment Methodologies and Applications of Animal’s Carcass Decomposition: A Review. FEMS Microbiol. Ecol. 2021, 97, fiab098. [Google Scholar] [CrossRef]
- Turner, P.V.; Kloeze, H.; Dam, A.; Ward, D.; Leung, N.; Brown, E.E.L.; Whiteman, A.; Chiappetta, M.E.; Hunter, D.B. Mass Depopulation of Laying Hens in Whole Barns with Liquid Carbon Dioxide: Evaluation of Welfare Impact. Poult. Sci. 2012, 91, 1558–1568. [Google Scholar] [CrossRef]
- Kittelmann, S.; Kirk, M.R.; Jonker, A.; McCulloch, A.; Janssen, P.H. Buccal Swabbing as a Noninvasive Method To Determine Bacterial, Archaeal, and Eukaryotic Microbial Community Structures in the Rumen. Appl. Environ. Microbiol. 2015, 81, 7470–7483. [Google Scholar] [CrossRef]
- Tapio, I.; Shingfield, K.J.; McKain, N.; Bonin, A.; Fischer, D.; Bayat, A.R.; Vilkki, J.; Taberlet, P.; Snelling, T.J.; Wallace, R.J. Oral Samples as Non-Invasive Proxies for Assessing the Composition of the Rumen Microbial Community. PLoS ONE 2016, 11, e0151220. [Google Scholar] [CrossRef]
- Young, J.; Skarlupka, J.H.; Cox, M.S.; Resende, R.T.; Fischer, A.; Kalscheur, K.F.; McClure, J.C.; Cole, J.B.; Suen, G.; Bickhart, D.M. Validating the Use of Bovine Buccal Sampling as a Proxy for the Rumen Microbiota by Using a Time Course and Random Forest Classification Approach. Appl. Environ. Microbiol. 2020, 86, e00861-20. [Google Scholar] [CrossRef]
- Miura, H.; Takeda, M.; Yamaguchi, M.; Ohtani, Y.; Endo, G.; Masuda, Y.; Ito, K.; Nagura, Y.; Iwashita, K.; Mitani, T. Application of MinION Amplicon Sequencing to Buccal SWAB Samples for Improving Resolution and Throughput of Rumen Microbiota Analysis. Front. Microbiol. 2022, 13, 783058. [Google Scholar] [CrossRef] [PubMed]
- Eisler, M.C.; Lee, M.R.F.; Tarlton, J.F.; Martin, G.B.; Beddington, J.; Dungait, J.A.J.; Greathead, H.; Liu, J.; Mathew, S.; Miller, H.; et al. Agriculture: Steps to Sustainable Livestock. Nature 2014, 507, 32–34. [Google Scholar] [CrossRef] [PubMed]
- Nkrumah, J.D.; Okine, E.K.; Mathison, G.W.; Schmid, K.; Li, C.; Basarab, J.A.; Price, M.A.; Wang, Z.; Moore, S.S. Relationships of Feedlot Feed Efficiency, Performance, and Feeding Behavior with Metabolic Rate, Methane Production, and Energy Partitioning in Beef Cattle. J. Anim. Sci. 2006, 84, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Dagar, S.S.; Puniya, A.K.; Upadhyay, R.C. Changes in Methane Emission, Rumen Fermentation in Response to Diet and Microbial Interactions. Res. Vet. Sci. 2013, 94, 263–268. [Google Scholar] [CrossRef]
- Curtis, H.; Blaser, M.J.; Dirk, G.; Kota, K.C.; Rob, K.; Liu, B.; Wang, L.; Sahar, A.; White, J.R.; Badger, J.H. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Elolimy, A.; Alharthi, A.; Zeineldin, M.; Parys, C.; Loor, J.J. Residual Feed Intake Divergence during the Preweaning Period Is Associated with Unique Hindgut Microbiome and Metabolome Profiles in Neonatal Holstein Heifer Calves. J. Anim. Sci. Biotechnol. 2020, 11, 13. [Google Scholar] [CrossRef]
- Diaz, J.; Reese, A.T. Possibilities and Limits for Using the Gut Microbiome to Improve Captive Animal Health. Anim. Microbiome 2021, 3, 89. [Google Scholar] [CrossRef]
- Lourenco, J.M.; Welch, C.B. Using Microbiome Information to Understand and Improve Animal Performance. Ital. J. Anim. Sci. 2022, 21, 899–913. [Google Scholar] [CrossRef]
- Du, W.; Wang, X.; Hu, M.; Hou, J.; Du, Y.; Si, W.; Yang, L.; Xu, L.; Xu, Q. Modulating Gastrointestinal Microbiota to Alleviate Diarrhea in Calves. Front. Microbiol. 2023, 14, 1181545. [Google Scholar] [CrossRef]
- Xiang, Q.; Wu, X.; Pan, Y.; Wang, L.; Guo, Y.; Cui, C.; Hu, L.; Zhu, L.; Peng, J.; Wei, H. Early Intervention Using Fecal Microbiota Transplantation Combined with Probiotics Influence the Growth Performance, Diarrhea, and Intestinal Barrier Function of Piglets. Appl. Sci. 2020, 10, 568. [Google Scholar] [CrossRef]
- Khalil, A.; Batool, A.; Arif, S. Healthy Cattle Microbiome and Dysbiosis in Diseased Phenotypes. Ruminants 2022, 2, 134–156. [Google Scholar] [CrossRef]
- Clemmons, B.A.; Voy, B.H.; Myer, P.R. Altering the Gut Microbiome of Cattle: Considerations of Host-Microbiome Interactions for Persistent Microbiome Manipulation. Microb. Ecol. 2019, 77, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, G.; Liu, Z.; Wu, P.; Yu, Z.; Wang, J. Repeated Inoculation with Fresh Rumen Fluid before or during Weaning Modulates the Microbiota Composition and Co-Occurrence of the Rumen and Colon of Lambs. BMC Microbiol. 2020, 20, 29. [Google Scholar] [CrossRef]
- Cox, A.; Bomstein, Z.; Jayaraman, A.; Allred, C. The Intestinal Microbiota as Mediators between Dietary Contaminants and Host Health. Exp. Biol. Med. 2023, 248, 2131–2150. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Kaoutari, A.E.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The Abundance and Variety of Carbohydrate-Active Enzymes in the Human Gut Microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial Degradation of Complex Carbohydrates in the Gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef]
- Berlemont, R.; Martiny, A.C. Genomic Potential for Polysaccharide Deconstruction in Bacteria. Appl. Environ. Microbiol. 2015, 81, 1513–1519. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Dao, T.-K.; Do, T.-H.; Le, N.-G.; Nguyen, H.-D.; Nguyen, T.-Q.; Le, T.-T.-H.; Truong, N.-H. Understanding the Role of Prevotella Genus in the Digestion of Lignocellulose and Other Substrates in Vietnamese Native Goats’ Rumen by Metagenomic Deep Sequencing. Animals 2021, 11, 3257. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Ma, Q.; Liu, Y.; Khan, M.Z.; Wu, B.; Chen, W.; Liu, X.; Wang, C.; Li, Y. Exploring the Effect of Gastrointestinal Prevotella on Growth Performance Traits in Livestock Animals. Animals 2024, 14, 1965. [Google Scholar] [CrossRef] [PubMed]
- Solden, L.; Lloyd, K.; Wrighton, K. The Bright Side of Microbial Dark Matter: Lessons Learned from the Uncultivated Majority. Curr. Opin. Microbiol. 2016, 31, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Toyama, H.; Matsumoto, T.; Qasimi, M.I.; Inoue, R.; Murase, H.; Yamamoto, Y.; Nagaoka, K. Changes in Fecal Microbiota during Estrous Cycle in Healthy Thoroughbred Mares. J. Equine Vet. Sci. 2024, 135, 105034. [Google Scholar] [CrossRef]
- Diviccaro, S.; FitzGerald, J.A.; Cioffi, L.; Falvo, E.; Crispie, F.; Cotter, P.D.; O’Mahony, S.M.; Giatti, S.; Caruso, D.; Melcangi, R.C. Gut Steroids and Microbiota: Effect of Gonadectomy and Sex. Biomolecules 2022, 12, 767. [Google Scholar] [CrossRef]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex Differences and Hormonal Effects on Gut Microbiota Composition in Mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Sovijit, W.N.; Sovijit, W.E.; Pu, S.; Usuda, K.; Inoue, R.; Watanabe, G.; Yamaguchi, H.; Nagaoka, K. Ovarian Progesterone Suppresses Depression and Anxiety-like Behaviors by Increasing the Lactobacillus Population of Gut Microbiota in Ovariectomized Mice. Neurosci. Res. 2021, 168, 76–82. [Google Scholar] [CrossRef]
- Gu, X.; Chen, J.; Li, H.; Song, Z.; Chang, L.; He, X.; Fan, Z. Isomaltooligosaccharide and Bacillus Regulate the Duration of Farrowing and Weaning-Estrous Interval in Sows during the Perinatal Period by Changing the Gut Microbiota of Sows. Anim. Nutr. 2021, 7, 72–83. [Google Scholar] [CrossRef]
- Diviccaro, S.; Giatti, S.; Borgo, F.; Falvo, E.; Caruso, D.; Garcia-Segura, L.M.; Melcangi, R.C. Steroidogenic Machinery in the Adult Rat Colon. J. Steroid Biochem. Mol. Biol. 2020, 203, 105732. [Google Scholar] [CrossRef]
- Sand, E.; Bergvall, M.; Ekblad, E.; D’Amato, M.; Ohlsson, B. Expression and Distribution of GnRH, LH, and FSH and Their Receptors in Gastrointestinal Tract of Man and Rat. Regul. Pept. 2013, 187, 24–28. [Google Scholar] [CrossRef]
- Schieren, A.; Koch, S.; Pecht, T.; Simon, M.-C. Impact of Physiological Fluctuations of Sex Hormones during the Menstrual Cycle on Glucose Metabolism and the Gut Microbiota. Exp. Clin. Endocrinol. Diabetes 2024, 132, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Mihajlovic, J.; Leutner, M.; Hausmann, B.; Kohl, G.; Schwarz, J.; Röver, H.; Stimakovits, N.; Wolf, P.; Maruszczak, K.; Bastian, M.; et al. Combined Hormonal Contraceptives Are Associated with Minor Changes in Composition and Diversity in Gut Microbiota of Healthy Women. Environ. Microbiol. 2021, 23, 3037–3047. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wang, C.; Simujide, H.; Liu, B.; Chen, Z.; Zhao, P.; Huangfu, M.; Liu, J.; Gao, X.; Wu, Y. Reproductive Hormones Mediate Intestinal Microbiota Shifts during Estrus Synchronization in Grazing Simmental Cows. Animals 2022, 12, 1751. [Google Scholar] [CrossRef] [PubMed]
- Menon, R.; Watson, S.E.; Thomas, L.N.; Allred, C.D.; Dabney, A.; Azcarate-Peril, M.A.; Sturino, J.M. Diet Complexity and Estrogen Receptor β Status Affect the Composition of the Murine Intestinal Microbiota. Appl. Environ. Microbiol. 2013, 79, 5763–5773. [Google Scholar] [CrossRef]
- Yoon, K.; Kim, N. Roles of Sex Hormones and Gender in the Gut Microbiota. J. Neurogastroenterol. Motil. 2021, 27, 314–325. [Google Scholar] [CrossRef]
- Hussain, T.; Murtaza, G.; Kalhoro, D.H.; Kalhoro, M.S.; Metwally, E.; Chughtai, M.I.; Mazhar, M.U.; Khan, S.A. Relationship between Gut Microbiota and Host-Metabolism: Emphasis on Hormones Related to Reproductive Function. Anim. Nutr. 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Gancz, N.N.; Levinson, J.A.; Callaghan, B.L. Sex and Gender as Critical and Distinct Contributors to the Human Brain-Gut-Microbiome Axis. Brain Res. Bull. 2023, 199, 110665. [Google Scholar] [CrossRef]
- García-Gómez, E.; González-Pedrajo, B.; Camacho-Arroyo, I. Role of Sex Steroid Hormones in Bacterial-Host Interactions. BioMed Res. Int. 2013, 2013, 928290. [Google Scholar] [CrossRef]
- Santos-Marcos, J.A.; Mora-Ortiz, M.; Tena-Sempere, M.; Lopez-Miranda, J.; Camargo, A. Interaction between Gut Microbiota and Sex Hormones and Their Relation to Sexual Dimorphism in Metabolic Diseases. Biol. Sex Differ. 2023, 14, 4. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, J.; Zhou, Y.; Xiong, S.; Zhou, M.; Wu, L.; Liu, Q.; Chen, Z.; Jiang, H.; Yang, J.; et al. Gut Microbiota Affects the Estrus Return of Sows by Regulating the Metabolism of Sex Steroid Hormones. J. Anim. Sci. Biotechnol. 2023, 14, 155. [Google Scholar] [CrossRef]
- Xu, H.; Lu, Y.; Li, D.; Yan, C.; Jiang, Y.; Hu, Z.; Zhang, Z.; Du, R.; Zhao, X.; Zhang, Y.; et al. Probiotic Mediated Intestinal Microbiota and Improved Performance, Egg Quality and Ovarian Immune Function of Laying Hens at Different Laying Stage. Front. Microbiol. 2023, 14, 1041072. [Google Scholar] [CrossRef]
- McCann, J.C.; Luan, S.; Cardoso, F.C.; Derakhshani, H.; Khafipour, E.; Loor, J.J. Induction of Subacute Ruminal Acidosis Affects the Ruminal Microbiome and Epithelium. Front. Microbiol. 2016, 7, 701. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Salminen, S.; Isolauri, E. Probiotics: An Overview of Beneficial Effects. In Lactic Acid Bacteria: Genetics, Metabolism and Applications: Proceedings of the Seventh Symposium on Lactic Acid Bacteria: Genetics, Metabolism and Applications, 1–5 September 2002, Egmond aan Zee, The Netherlands; Siezen, R.J., Kok, J., Abee, T., Schasfsma, G., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 279–289. ISBN 978-94-017-2029-8. [Google Scholar]
- Xu, Y.; Victor Curtasu, M.; Bendiks, Z.L.; Marco, M.; Nørskov, N.P.; Bach Knudsen, K.E.; Skou Hedemann, M.; Nygaard Lærke, H. Effects of Dietary Fibre and Protein Content on Intestinal Fibre Degradation, Short-Chain Fatty Acid and Microbiota Composition in a High-Fat Fructose-Rich Diet Induced Obese Göttingen Minipig Model. Food Funct. 2020, 11, 10758–10773. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Covington, A.; Pamer, E.G. The Intestinal Microbiota: Antibiotics, Colonization Resistance, and Enteric Pathogens. Immunol. Rev. 2017, 279, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, S.; Duan, H.; Shao, H.; Duan, Y. Bacteriophages and Their Potential for Treatment of Metabolic Diseases. J. Diabetes 2024, 16, e70024. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tardiolo, G.; La Fauci, D.; Riggio, V.; Daghio, M.; Di Salvo, E.; Zumbo, A.; Sutera, A.M. Gut Microbiota of Ruminants and Monogastric Livestock: An Overview. Animals 2025, 15, 758. https://doi.org/10.3390/ani15050758
Tardiolo G, La Fauci D, Riggio V, Daghio M, Di Salvo E, Zumbo A, Sutera AM. Gut Microbiota of Ruminants and Monogastric Livestock: An Overview. Animals. 2025; 15(5):758. https://doi.org/10.3390/ani15050758
Chicago/Turabian StyleTardiolo, Giuseppe, Deborah La Fauci, Valentina Riggio, Matteo Daghio, Eleonora Di Salvo, Alessandro Zumbo, and Anna Maria Sutera. 2025. "Gut Microbiota of Ruminants and Monogastric Livestock: An Overview" Animals 15, no. 5: 758. https://doi.org/10.3390/ani15050758
APA StyleTardiolo, G., La Fauci, D., Riggio, V., Daghio, M., Di Salvo, E., Zumbo, A., & Sutera, A. M. (2025). Gut Microbiota of Ruminants and Monogastric Livestock: An Overview. Animals, 15(5), 758. https://doi.org/10.3390/ani15050758






