Identification of Biomarkers for Meat Quality in Sichuan Goats Through 4D Label-Free Quantitative Proteomics
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Eating Quality
2.2.1. Meat pH and Color
2.2.2. Cooking Loss
2.2.3. Shear Force
2.3. Proximate Analysis
2.4. Histology Analysis
2.5. Transmission Electron Microscope
2.6. Proteomics
2.6.1. Protein Extraction and Digestion
2.6.2. Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
2.7. Bioinformatics Analysis
2.8. Data Analysis
3. Results and Discussion
3.1. Quality Traits
3.1.1. Eating Quality and Chemical Composition
3.1.2. Histological and Ultrastructural Analysis
3.1.3. Principal Component Analysis (PCA)
3.2. Proteomics Analysis
3.2.1. Protein Identification and Quantification
3.2.2. DEPs Analysis
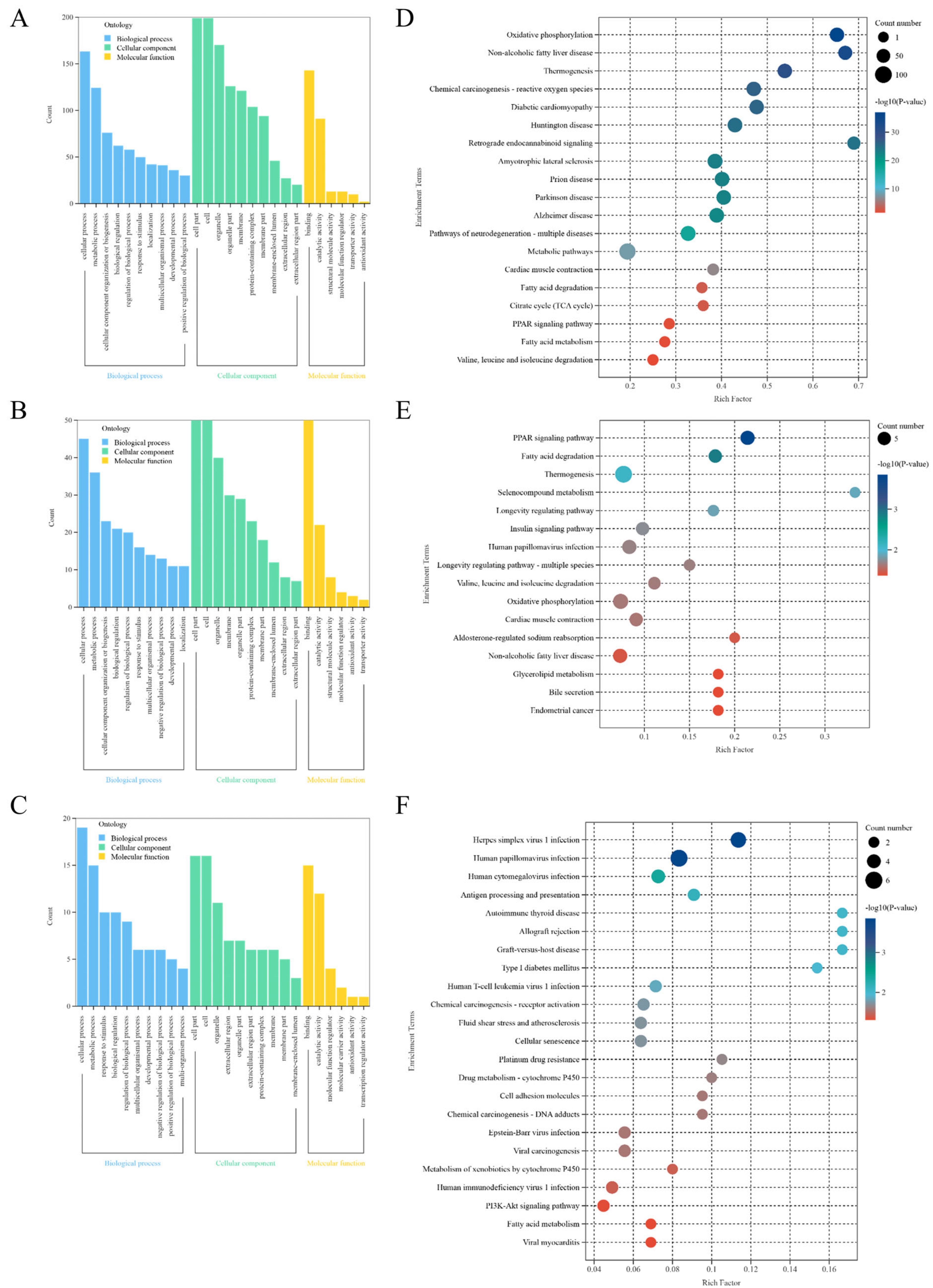
3.2.3. Enrichment Analysis
3.3. WPCNA
3.4. PPI Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mazhangara, I.R.; Chivandi, E.; Mupangwa, J.F.; Muchenje, V. The Potential of Goat Meat in the Red Meat Industry. Sustainability 2019, 11, 3671. [Google Scholar] [CrossRef]
- UNdata. Meat of Goat, Fresh or Chilled. Available online: https://data.un.org/Data.aspx?d=FAO&f=itemCode%3a1017 (accessed on 10 March 2025).
- Sun, S.; Guo, B.; Wei, Y.; Fan, M. Multi-element analysis for determining the geographical origin of mutton from different regions of China. Food Chem. 2011, 124, 1151–1156. [Google Scholar]
- Li, L.; Zhang, J.; Zhu, J.Q.; Gu, S.; Sun, Q.; Zhou, G.M.; Fu, C.X.; Li, Q.; Chen, L.Y.; Li, D.X.; et al. Genetic diversity of nine populations of the black goat (Capra hircus) in Sichuan, PR China. Zool. Sci. 2006, 23, 229–234. [Google Scholar]
- Guo, J.; Zhong, J.; Li, L.; Zhong, T.; Wang, L.; Song, T.; Zhang, H. Comparative genome analyses reveal the unique genetic composition and selection signals underlying the phenotypic characteristics of three Chinese domestic goat breeds. Genet. Sel. Evol. 2019, 51, 70. [Google Scholar]
- Wang, C.; Zhang, H.; Niu, L.; Guo, J.; Jia, X.; Wang, L.; Li, L.; Zhang, H.; Zhong, T. The novel SNPs of leptin gene and their associations with growth traits in Chinese Nanjiang Yellow goat. Gene 2015, 572, 35–41. [Google Scholar]
- Guo, J.; Tao, H.; Li, P.; Li, L.; Zhong, T.; Wang, L.; Ma, J.; Chen, X.; Song, T.; Zhang, H. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 2018, 8, 10405. [Google Scholar]
- Wei, Y.; Li, X.; Zhang, D.; Liu, Y. Comparison of protein differences between high- and low-quality goat and bovine parts based on iTRAQ technology. Food Chem. 2019, 289, 240–249. [Google Scholar]
- Patinho, I.; Antonelo, D.S.; Delgado, E.F.; Alessandroni, L.; Balieiro, J.C.C.; Contreras Castillo, C.J.; Gagaoua, M. In-depth exploration of the high and normal pH beef proteome: First insights emphasizing the dynamic protein changes in Longissimus thoracis muscle from pasture-finished Nellore bulls over different postmortem times. Meat Sci. 2024, 216, 109557. [Google Scholar]
- González-Blanco, L.; Oliván, M.; Diñeiro, Y.; Bravo, S.B.; Sierra, V.; Gagaoua, M. Sequential window acquisition of all theoretical mass spectra (SWATH-MS) as an emerging proteomics approach for the discovery of dark-cutting beef biomarkers. Meat Sci. 2024, 217, 109618. [Google Scholar] [PubMed]
- Wang, J.; Fu, Y.; Su, T.; Wang, Y.; Soladoye, O.P.; Huang, Y.; Zhao, Z.; Zhao, Y.; Wu, W. A Role of Multi-Omics Technologies in Sheep and Goat Meats: Progress and Way Ahead. Foods 2023, 12, 4069. [Google Scholar] [CrossRef]
- Sun, X.; Guo, J.; Li, L.; Zhong, T.; Wang, L.; Zhan, S.; Lu, J.; Wang, D.; Dai, D.; Liu, G.E.; et al. Genetic Diversity and Selection Signatures in Jianchang Black Goats Revealed by Whole-Genome Sequencing Data. Animals 2022, 12, 2365. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.Y.; Chen, L.; Li, L.; Wang, L.J.; Zhong, T.; Zhang, H.P. Molecular characterization and expression patterns of insulin-like growth factor-binding protein genes in postnatal Nanjiang brown goats. Genet. Mol. Res. 2015, 14, 12547–12560. [Google Scholar]
- Odame, E.; Li, L.; Nabilla, J.A.; Cai, H.; Xiao, M.; Ye, J.; Chen, Y.; Kyei, B.; Dai, D.; Zhan, S.; et al. miR-145-3p Inhibits MuSCs Proliferation and Mitochondria Mass via Targeting MYBL1 in Jianzhou Big-Eared Goats. Int. J. Mol. Sci. 2023, 24, 8341. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Niu, Y.; Wang, J.; Yang, Z.; Cai, Z.; Dao, X.; Wang, C.; Wang, Y.; Lin, Y. Physicochemical property, bacterial diversity, and volatile profile during ripening of naturally fermented dry mutton sausage produced from Jianzhou big-eared goat. Front. Microbiol. 2022, 13, 961117. [Google Scholar]
- Miltenburg, G.A.; Wensing, T.; Smulders, F.J.; Breukink, H.J. Relationship between blood hemoglobin, plasma and tissue iron, muscle heme pigment, and carcass color of veal. J. Anim. Sci. 1992, 70, 2766–2772. [Google Scholar]
- Jo, K.; Lee, S.; Jeong, H.G.; Lee, D.-H.; Kim, H.B.; Seol, K.-H.; Kang, S.; Jung, S. Prediction of cooking loss of pork belly using quality properties of pork loin. Meat Sci. 2022, 194, 108957. [Google Scholar]
- Fu, L.; Du, L.; Sun, Y.; Fan, X.; Zhou, C.; He, J.; Pan, D. Effect of Lentinan on Lipid Oxidation and Quality Change in Goose Meatballs during Cold Storage. Foods 2022, 11, 1055. [Google Scholar] [CrossRef]
- GB 5009.6-2016; National Food Safety Standard—Determination of Fat in Foods. National Standard of the People’s Republic Of China: Beijing, China, 2016.
- GB 5009.5-2016; National Food Safety Standard—Determination of Protein in Foods. National Standard of the People’s Republic Of China: Beijing, China, 2016.
- GB 5009.238-2016; National Standard for food Safety—Determination of Water Activity in Foods. National Standard of the People’s Republic Of China: Beijing, China, 2016.
- GB 5009.4-2010; National Food Safety Standard—Determination of Ash in Foods. National Standard of the People’s Republic Of China: Beijing, China, 2010.
- Erdös, T.; Snellman, O. Electrophoretic investigations of crystallised myosin. Biochim. Biophys. Acta 1948, 2, 642–649. [Google Scholar]
- Huff-Lonergan, E.; Lonergan, S.M. Mechanisms of water-holding capacity of meat: The role of postmortem biochemical and structural changes. Meat Sci. 2005, 71, 194–204. [Google Scholar]
- Andrés-Bello, A.; Barreto-Palacios, V.; García-Segovia, P.; Mir-Bel, J.; Martínez-Monzó, J. Effect of pH on Color and Texture of Food Products. Food Eng. Rev. 2013, 5, 158–170. [Google Scholar]
- Maltin, C.; Balcerzak, D.; Tilley, R.; Delday, M. Determinants of meat quality: Tenderness. Proc. Nutr. Soc. 2003, 62, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Gawat, M.; Kaur, L.; Singh, J.; Boland, M. Physicochemical and quality characteristics of New Zealand goat meat and its ultrastructural features. Food Res. Int. 2022, 161, 111736. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M. Variation in proteolysis, sarcomere length, collagen content, and tenderness among major pork muscles. J. Anim. Sci. 2000, 78, 958–965. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Yin, Y.; Cao, Y.; Guo, L.; Li, P.; Jiang, J.; Huang, X.; Jiang, Y.; Wu, J. Proteomic and parallel reaction monitoring approaches to evaluate biomarkers of mutton tenderness. Food Chem. 2022, 397, 133746. [Google Scholar] [CrossRef]
- Zhu, Y.; Gagaoua, M.; Mullen, A.M.; Viala, D.; Rai, D.K.; Kelly, A.L.; Sheehan, D.; Hamill, R.M. Shotgun proteomics for the preliminary identification of biomarkers of beef sensory tenderness, juiciness and chewiness from plasma and muscle of young Limousin-sired bulls. Meat Sci. 2021, 176, 108488. [Google Scholar] [CrossRef]
- Li, X.; Ha, M.; Warner, R.D.; Dunshea, F.R. Meta-analysis of the relationship between collagen characteristics and meat tenderness. Meat Sci. 2022, 185, 108717. [Google Scholar] [CrossRef]
- Jeleníková, J.; Pipek, P.; Miyahara, M. The effects of breed, sex, intramuscular fat and ultimate pH on pork tenderness. Eur. Food Res. Technol. 2008, 227, 989–994. [Google Scholar] [CrossRef]
- Fortes, M.R.; Curi, R.A.; Chardulo, L.A.; Silveira, A.C.; Assumpção, M.E.; Visintin, J.A.; de Oliveira, H.N. Bovine gene polymorphisms related to fat deposition and meat tenderness. Genet. Mol. Biol. 2009, 32, 75–82. [Google Scholar] [CrossRef]
- Fiems, L.O.; Campeneere, S.D.; De Smet, S.; Van de Voorde, G.; Vanacker, J.M.; Boucqué, C.V. Relationship between fat depots in carcasses of beef bulls and effect on meat colour and tenderness. Meat Sci. 2000, 56, 41–47. [Google Scholar] [CrossRef]
- Della Malva, A.; Gagaoua, M.; Santillo, A.; di Corcia, M.; Natalello, A.; Sevi, A.; Albenzio, M. In-depth characterization of the sarcoplasmic muscle proteome changes in lambs fed with hazelnut skin by-products: Relationships with meat color. J. Proteom. 2023, 287, 104997. [Google Scholar]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar]
- Knoops, B.; Goemaere, J.; Van der Eecken, V.; Declercq, J.P. Peroxiredoxin 5: Structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid. Redox Signal 2011, 15, 817–829. [Google Scholar]
- Wu, W.; Gao, X.-G.; Dai, Y.; Fu, Y.; Li, X.-M.; Dai, R.-T. Post-mortem changes in sarcoplasmic proteome and its relationship to meat color traits in M. semitendinosus of Chinese Luxi yellow cattle. Food Res. Int. 2015, 72, 98–105. [Google Scholar]
- Mahmood, S.; Turchinsky, N.; Paradis, F.; Dixon, W.T.; Bruce, H.L. Proteomics of dark cutting longissimus thoracis muscle from heifer and steer carcasses. Meat Sci. 2018, 137, 47–57. [Google Scholar]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar]
- Wirth, C.; Brandt, U.; Hunte, C.; Zickermann, V. Structure and function of mitochondrial complex I. Biochim. Biophys. Acta 2016, 1857, 902–914. [Google Scholar] [PubMed]
- Lenaz, G. Mitochondria and reactive oxygen species. Which role in physiology and pathology? Adv. Exp. Med. Biol. 2012, 942, 93–136. [Google Scholar]
- Maranzana, E.; Barbero, G.; Falasca, A.I.; Lenaz, G.; Genova, M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal 2013, 19, 1469–1480. [Google Scholar] [CrossRef]
- Zhang, W.; Xiao, S.; Ahn, D.U. Protein oxidation: Basic principles and implications for meat quality. Crit. Rev. Food Sci. Nutr. 2013, 53, 1191–1201. [Google Scholar] [CrossRef]
- Hughes, J.M.; Oiseth, S.K.; Purslow, P.P.; Warner, R.D. A structural approach to understanding the interactions between colour, water-holding capacity and tenderness. Meat Sci. 2014, 98, 520–532. [Google Scholar] [PubMed]
- Killilea, D.W.; Killilea, A.N. Mineral requirements for mitochondrial function: A connection to redox balance and cellular differentiation. Free Radic. Biol. Med. 2022, 182, 182–191. [Google Scholar] [CrossRef]
- Shapiro, I.M.; Risbud, M.V.; Landis, W.J. Toward understanding the cellular control of vertebrate mineralization: The potential role of mitochondria. Bone 2024, 185, 117112. [Google Scholar] [PubMed]
- England, E.M.; Matarneh, S.K.; Mitacek, R.M.; Abraham, A.; Ramanathan, R.; Wicks, J.C.; Shi, H.; Scheffler, T.L.; Oliver, E.M.; Helm, E.T.; et al. Presence of oxygen and mitochondria in skeletal muscle early postmortem. Meat Sci. 2018, 139, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Neath, K.E.; Del Barrio, A.N.; Lapitan, R.M.; Herrera, J.R.V.; Cruz, L.C.; Fujihara, T.; Muroya, S.; Chikuni, K.; Hirabayashi, M.; Kanai, Y. Difference in tenderness and pH decline between water buffalo meat and beef during postmortem aging. Meat Sci. 2007, 75, 499–505. [Google Scholar] [CrossRef]
- Boudon, S.; Ounaissi, D.; Viala, D.; Monteils, V.; Picard, B.; Cassar-Malek, I. Label free shotgun proteomics for the identification of protein biomarkers for beef tenderness in muscle and plasma of heifers. J. Proteom. 2020, 217, 103685. [Google Scholar]
- Xie, Y.; Zhang, C.; Qin, Q.; Li, X.; Guo, J.; Dai, D.; Wang, Z.; Zhao, Y.; Su, R.; Wang, Z.; et al. Proteomics Analysis of Meat to Identify Goat Intramuscular Fat Deposits Potential Biomarkers. Food Anal. Method. 2023, 16, 1191–1202. [Google Scholar] [CrossRef]
- Han, S.; Lim, S.; Yeo, S. Association Between Decreased ITGA7 Levels and Increased Muscle α-Synuclein in an MPTP-Induced Mouse Model of Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 5646. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G. The role of the extracellular matrix in skeletal muscle development. Poult. Sci. 1999, 78, 778–784. [Google Scholar] [CrossRef]
- Nishimura, T. Role of extracellular matrix in development of skeletal muscle and postmortem aging of meat. Meat Sci. 2015, 109, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Končitíková, R.; Vigouroux, A.; Kopečná, M.; Šebela, M.; Moréra, S.; Kopečný, D. Kinetic and structural analysis of human ALDH9A1. Biosci. Rep. 2019, 39, BSR20190558. [Google Scholar] [CrossRef] [PubMed]
- Puig-Oliveras, A.; Revilla, M.; Castelló, A.; Fernández, A.I.; Folch, J.M.; Ballester, M. Expression-based GWAS identifies variants, gene interactions and key regulators affecting intramuscular fatty acid content and composition in porcine meat. Sci. Rep. 2016, 6, 31803. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef]
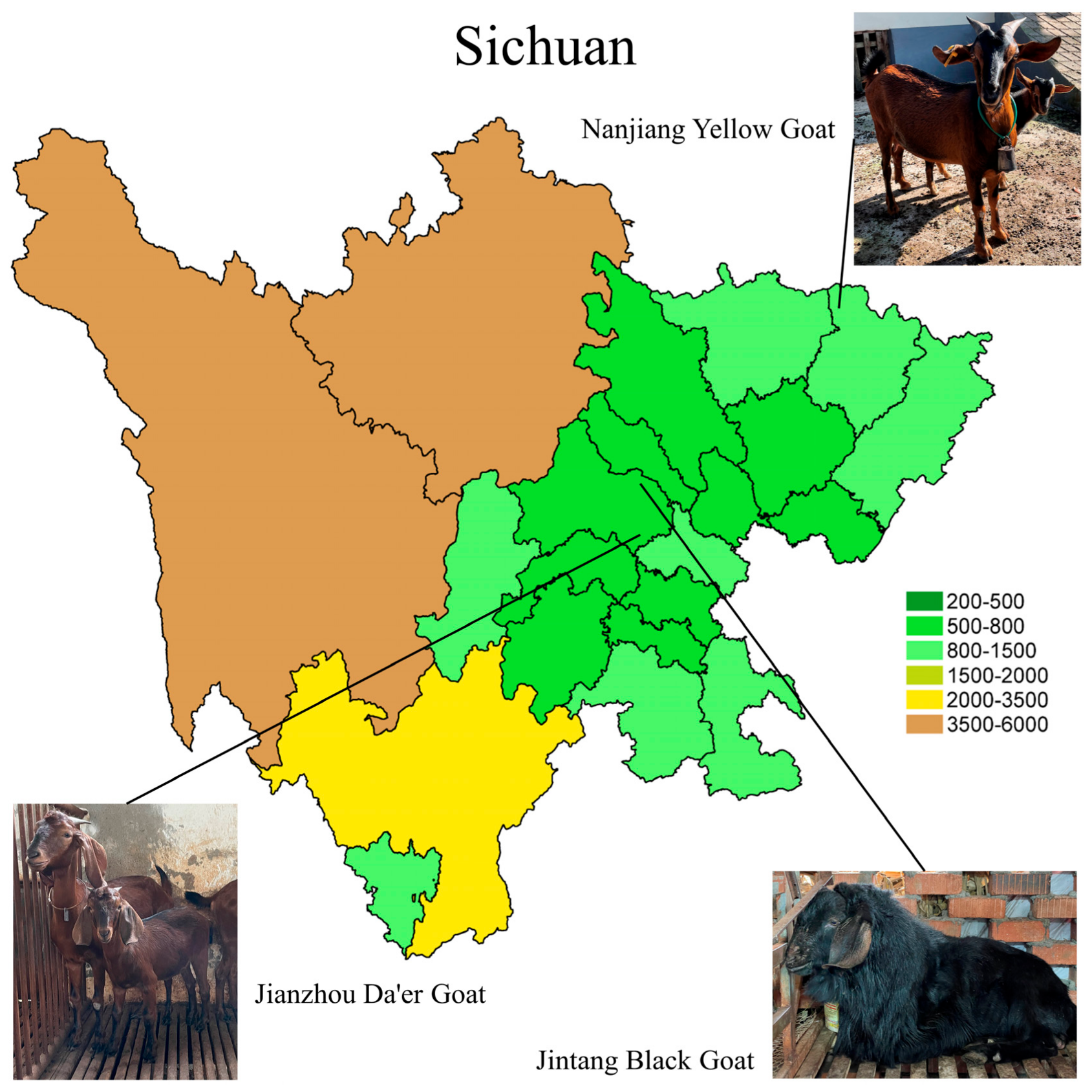





| Parameters | NJYG | JTBG | JZDEG |
|---|---|---|---|
| pH0.5h | 6.65 ± 0.19 | 6.59 ± 0.26 | 6.55 ± 0.38 |
| L*0.5h | 34.68 ± 4.47 | 34.64 ± 0.89 | 33.65 ± 2.02 |
| a*0.5h | 16.10 ± 1.82 | 15.40 ± 0.62 | 16.15 ± 1.09 |
| b*0.5h | 4.22 ± 0.72 ab | 3.80 ± 0.17 b | 4.70 ± 0.70 a |
| pH24h | 5.60 ± 0.06 b | 5.74 ± 0.20 ab | 5.85 ± 0.32 a |
| L*24h | 44.61 ± 5.15 a | 39.36 ± 2.54 b | 39.60 ± 2.91 b |
| a*24h | 12.43 ± 5.31 b | 17.73 ± 2.39 a | 17.35 ± 1.09 a |
| b*24h | 4.44 ± 4.39 b | 7.70 ± 2.79 a | 10.50 ± 2.00 a |
| Cooking loss (%) | 28.80 ± 3.90 | 30.00 ± 7.23 | 22.11 ± 8.26 |
| Shear force (N) | 122.70 ± 10.36 | 93.27 ± 13.49 | 100.13 ± 44.70 |
| Chemical composition | |||
| Ash (%) | 1.50 ± 0.12 a | 1.36 ± 0.11 b | 1.43 ± 0.05 ab |
| Fat (%) | 3.95 ± 0.17 b | 3.64 ± 0.16 c | 4.25 ± 0.06 a |
| Protein (%) | 24.48 ± 4.55 | 21.72 ± 2.56 | 24.61 ± 3.89 |
| Water activity (%) | 0.95 ± 0.00 b | 0.98 ± 0.01 a | 0.97 ± 0.20 a |
| Module | Protein | NJYG | JTBG | JZDEG | FC | |||
|---|---|---|---|---|---|---|---|---|
| NJYG vs. JTBG | NJYG vs. JZDEG | JTBG vs. JZDEG | ||||||
| MEbrown | GSTM3 | 6.386 | 5.293 | 10.106 | 0.524 * | |||
| SOD3 | 0.545 | 0.397 | 0.612 | 0.649 * | ||||
| PRDX5 | 1.548 | 1.195 | 1.824 | 1.295 * | ||||
| MEturquoise | B-I | NDUFS3 | 5.241 | 3.150 | 3.944 | 1.664 ** | ||
| NDUFA8 | 3.870 | 2.421 | 2.895 | 1.598 ** | ||||
| NDUFS6 | 3.030 | 1.903 | 2.362 | 1.592 *** | ||||
| NDUFS7 | 8.375 | 5.761 | 6.724 | 1.454 * | ||||
| NDUFAB1 | 4.332 | 2.275 | 3.282 | 1.904 ** | ||||
| NDUFV1 | 3.420 | 2.215 | 2.745 | 1.544 ** | ||||
| NDUFB7 | 2.797 | 1.705 | 2.134 | 1.641 ** | ||||
| NDUFC2 | 5.413 | 3.325 | 4.033 | 1.628 ** | ||||
| NDUFB4 | 4.523 | 2.750 | 3.262 | 1.644 ** | 1.386 * | |||
| NDUFB3 | 4.786 | 3.090 | 3.697 | 1.549 ** | ||||
| B-II | SUCLG2 | 3.575 | 1.654 | 2.061 | 2.161 * | |||
| SUCLG1 | 8.394 | 6.084 | 6.824 | 1.380 * | ||||
| OGDH | 5.607 | 4.253 | 4.882 | 1.319 * | ||||
| ACO2 | 15.219 | 8.030 | 10.182 | 1.895 * | 1.495 * | |||
| CS | 16.078 | 10.818 | 12.348 | 1.486 * | ||||
| B-III | HADH | 4.330 | 2.654 | 3.386 | 1.632 * | |||
| ACAT1 | 12.509 | 5.618 | 7.839 | 2.227 ** | ||||
| ACADS | 2.219 | 0.763 | 1.647 | 2.908 * | ||||
| ACAA2 | 2.785 | 1.239 | 1.609 | 2.248 * | ||||
| MEgreen | HSPG2 | 7.333 | 5.765 | 5.913 | 1.272 * | 1.240 ** | ||
| COL4A2 | 13.659 | 8.980 | 10.846 | 1.521 * | ||||
| LAMC1 | 3.253 | 2.790 | 2.638 | 1.233 * | ||||
| LAMA2 | 10.567 | 8.793 | 8.141 | 1.298 * | ||||
| ITGA7 | 0.532 | 0.414 | 0.457 | 1.285 * | ||||
| PARVB | 0.558 | 0.370 | 0.406 | 1.508 ** | 1.374 * | |||
| MEyellow | ALDH9A1 | 1.052 | 0.747 | 0.650 | 1.409 * | 1.619 * | ||
| ADH5 | 2.931 | 2.566 | 2.419 | 1.211 * | ||||
| LOC102190016 | 2.076 | 1.921 | 1.294 | 1.605 * | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Xu, M.; Xu, R.; Bai, T.; Liu, D.; Wang, X.; Pan, D.; Zhang, Y.; Zhang, L.; Pan, S.; et al. Identification of Biomarkers for Meat Quality in Sichuan Goats Through 4D Label-Free Quantitative Proteomics. Animals 2025, 15, 887. https://doi.org/10.3390/ani15060887
Zhang R, Xu M, Xu R, Bai T, Liu D, Wang X, Pan D, Zhang Y, Zhang L, Pan S, et al. Identification of Biomarkers for Meat Quality in Sichuan Goats Through 4D Label-Free Quantitative Proteomics. Animals. 2025; 15(6):887. https://doi.org/10.3390/ani15060887
Chicago/Turabian StyleZhang, Rui, Mengling Xu, Rui Xu, Ting Bai, Dayu Liu, Xinhui Wang, Daodong Pan, Yin Zhang, Lin Zhang, Shifeng Pan, and et al. 2025. "Identification of Biomarkers for Meat Quality in Sichuan Goats Through 4D Label-Free Quantitative Proteomics" Animals 15, no. 6: 887. https://doi.org/10.3390/ani15060887
APA StyleZhang, R., Xu, M., Xu, R., Bai, T., Liu, D., Wang, X., Pan, D., Zhang, Y., Zhang, L., Pan, S., & Zhang, J. (2025). Identification of Biomarkers for Meat Quality in Sichuan Goats Through 4D Label-Free Quantitative Proteomics. Animals, 15(6), 887. https://doi.org/10.3390/ani15060887






