Integrated lncRNA and mRNA Transcriptome Analyses of IGF1 and IGF2 Stimulated Ovaries Reveal Genes and Pathways Potentially Associated with Ovarian Development and Oocyte Maturation in Golden Pompano (Trachinotus ovatus)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
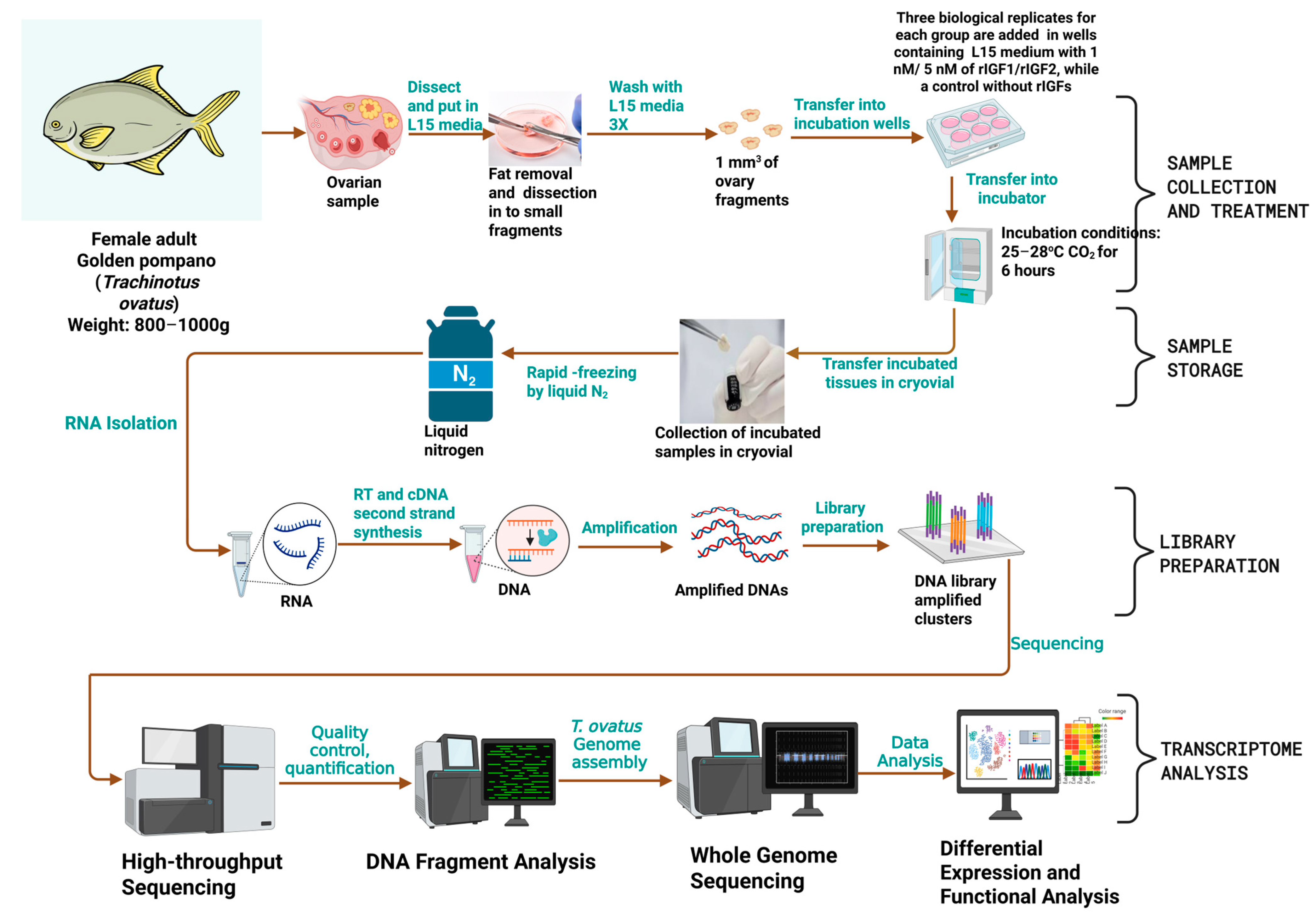
2.2. Sample Treatment
2.3. RNA Library Construction and Sequencing
2.4. Sequencing Quality Control and Analysis
2.5. lncRNA Identification and Prediction
2.6. Identification of DE lncRNAs and mRNAs
2.7. Functional Enrichment Analysis
2.8. PPI Network Construction and Hub Genes Analysis
2.9. lncRNA–mRNA Network Construction
2.10. Validation by Real-Time Quantitative PCR
2.11. Tissue Distribution of Candidate lncRNAs by RT-qPCR
2.12. Data Analysis
3. Results
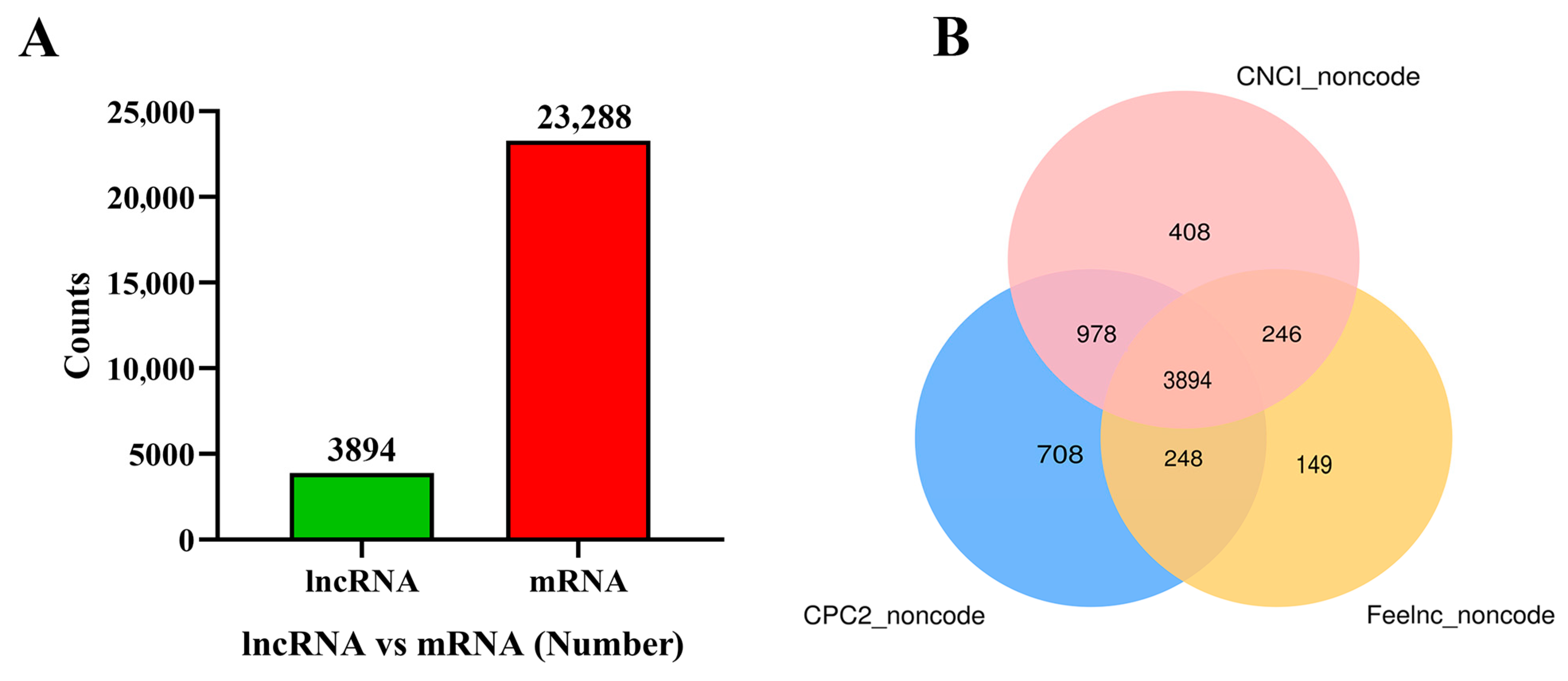
3.1. Overview of the RNA-Seq Results
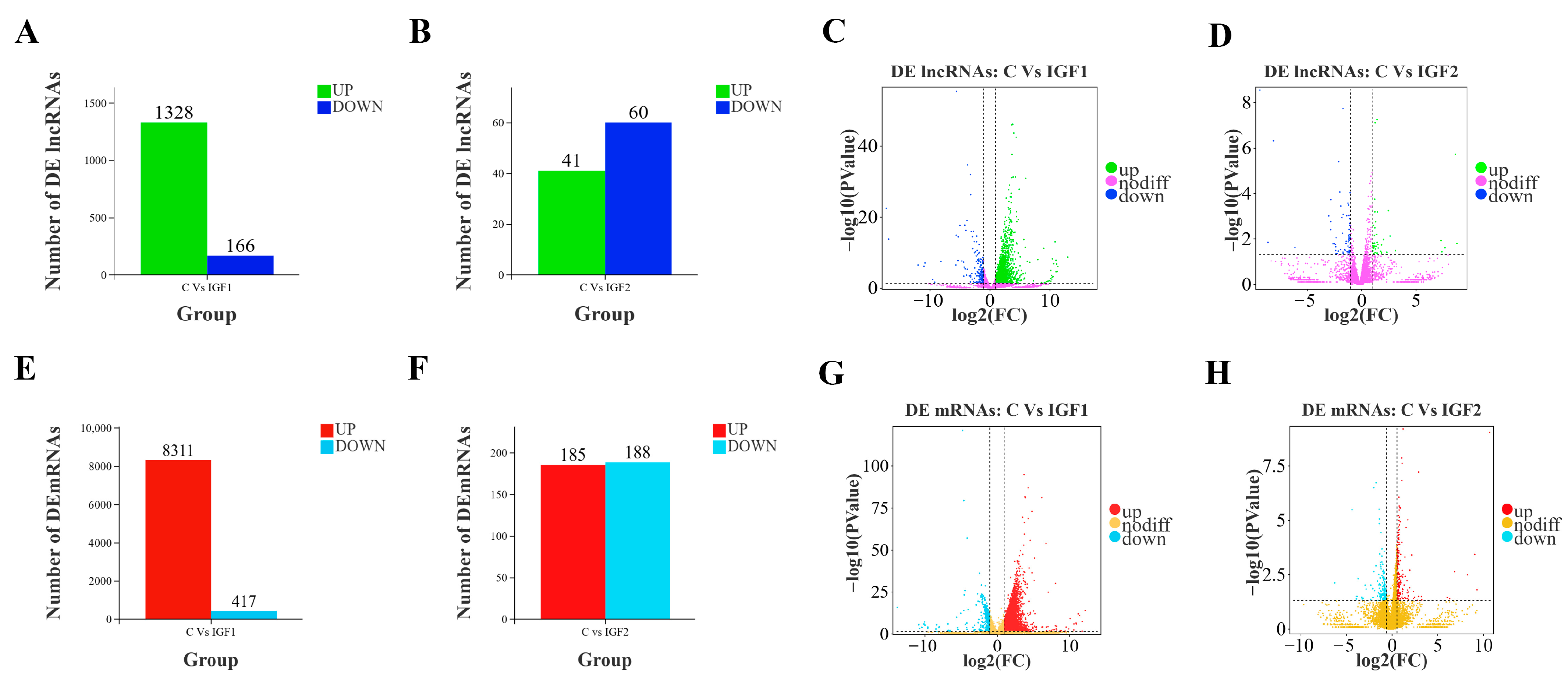
3.2. Identification of the DE lncRNAs and mRNAs
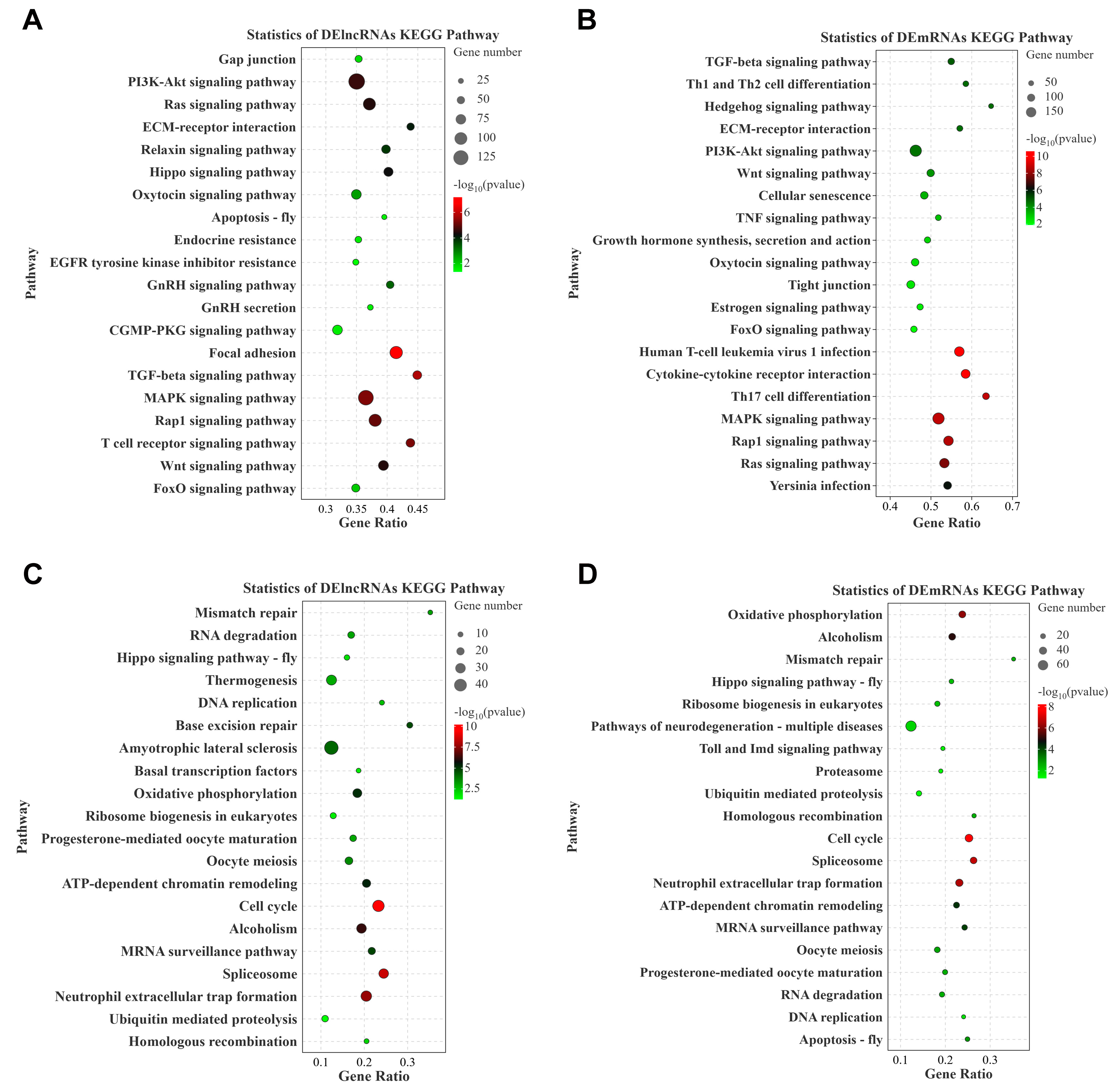
3.3. GO and KEGG Pathway Enrichment Analysis
3.4. Construction of the PPI Network and the Identification of Hub Genes
3.4.1. The PPI Network Following IGF1 Stimulation
3.4.2. The PPI Network Following IGF2 Stimulation
3.5. Construction of the lncRNA–mRNA Regulatory Network
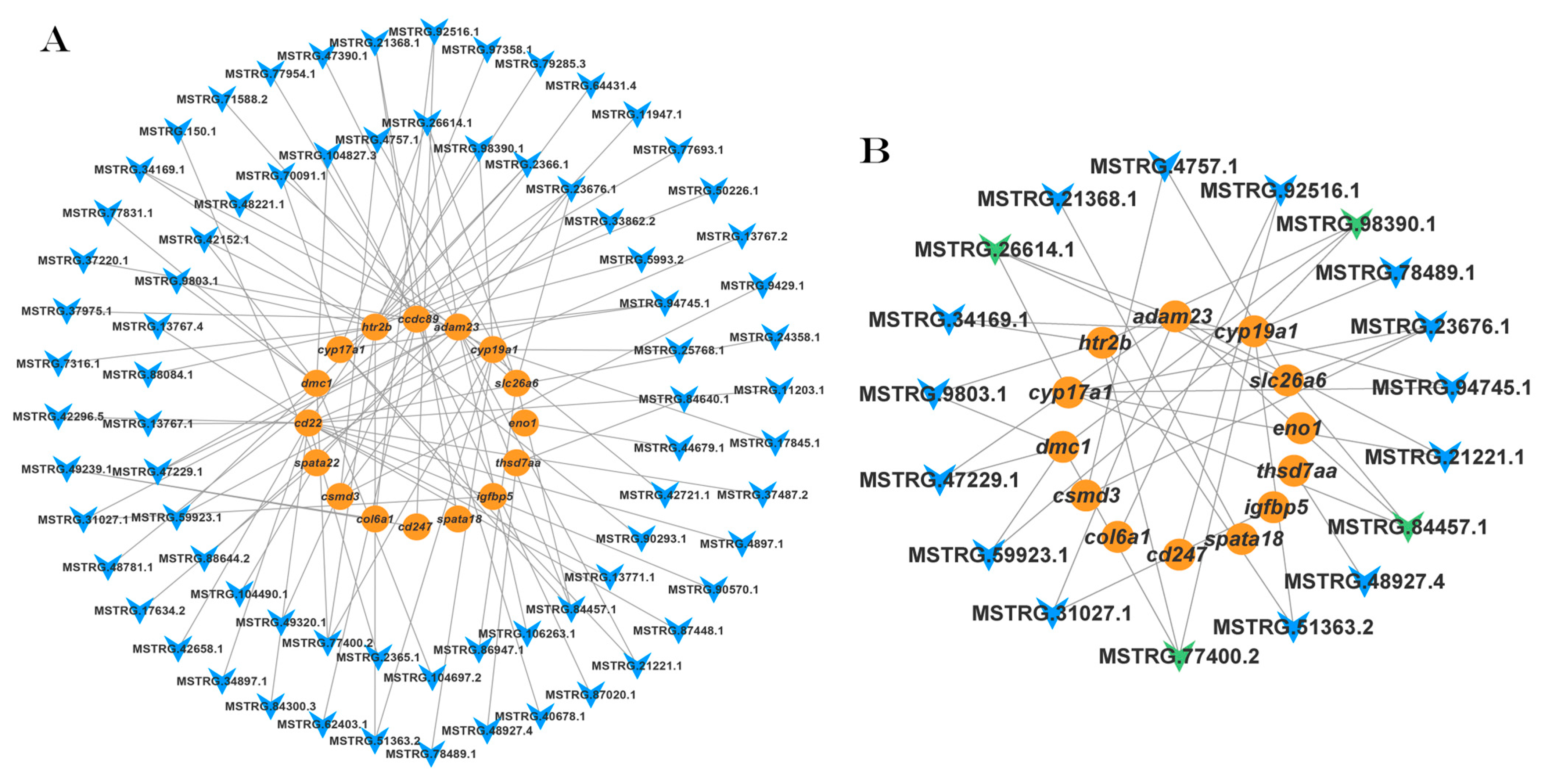
3.5.1. The IGF1 lncRNA–mRNA Regulatory Network
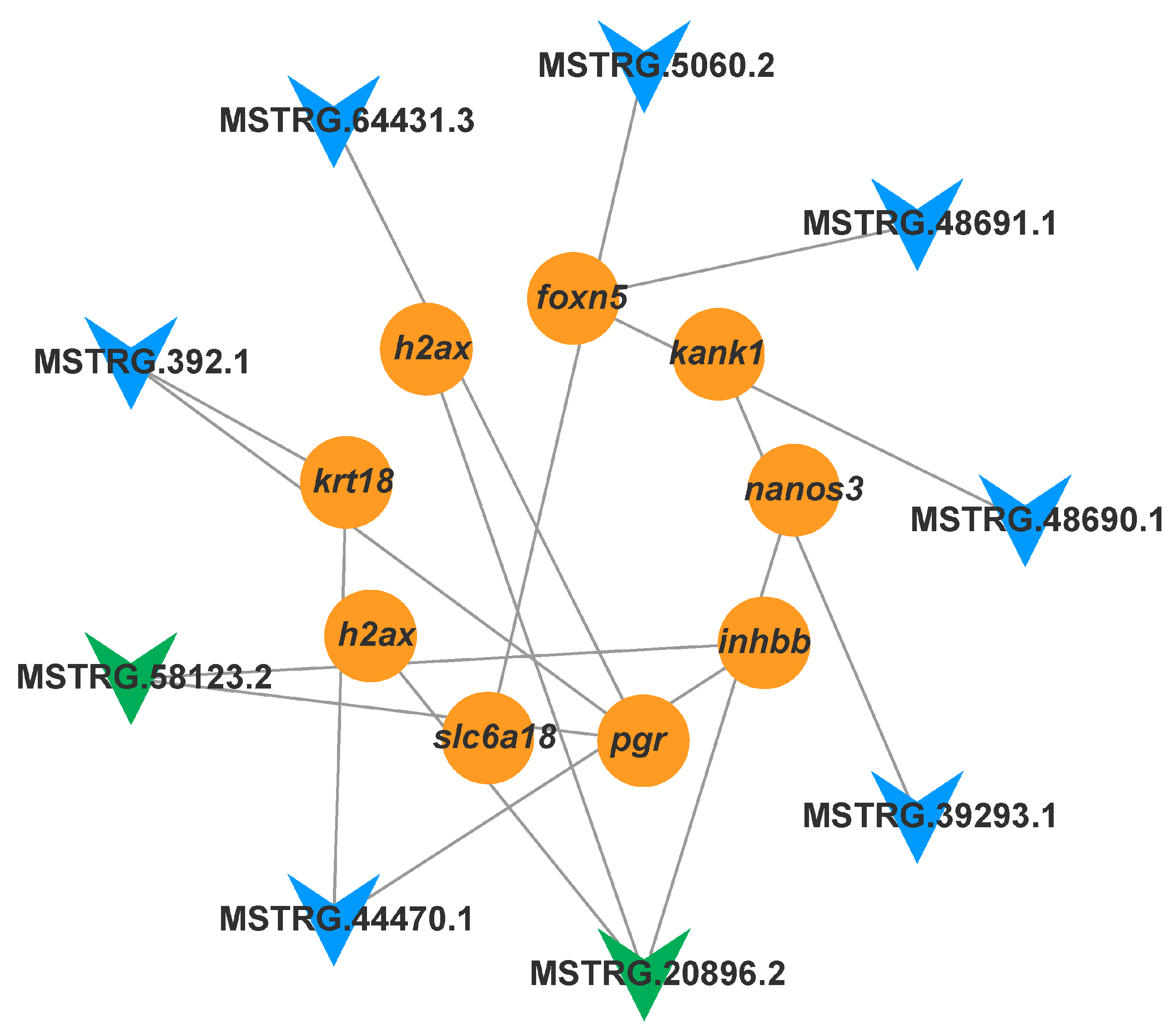
3.5.2. The IGF2 lncRNA-mRNA Regulatory Network
3.6. Validation by RT-qPCR
3.7. Tissue Distribution of the Candidate lncRNAs
4. Discussion
4.1. Differentially Expressed Genes and Their Regulatory Pathways
4.2. Co-Expression and Tissue Distribution Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jia, Y.; Nan, P.; Zhang, W.; Wang, F.; Zhang, R.; Liang, T.; Ji, X.; Du, Q.; Chang, Z. Transcriptome Analysis of Three Critical Periods of Ovarian Development in Yellow River Carp (Cyprinus carpio). Theriogenology 2018, 105, 15–26. [Google Scholar] [CrossRef]
- Li, Z.; Ren, X.; Guo, Y.; Ru, X.; Tian, C.; Shi, H.; Jiang, M.; Zhu, C.; Li, G.; Chen, H. Identification and Ovarian Developmental Regulation of Insulin-like Growth Factor 3 in Spotted Scat (Scatophagus argus). Aquac. Rep. 2021, 21, 100866. [Google Scholar] [CrossRef]
- Meijide, F.J.; Rey Vázquez, G.; Grier, H.J.; Lo Nostro, F.L.; Guerrero, G.A. Development of the Germinal Epithelium and Early Folliculogenesis during Ovarian Morphogenesis in the Cichlid Fish Cichlasoma dimerus (Teleostei, Perciformes). Acta Zool. 2016, 97, 18–33. [Google Scholar] [CrossRef]
- Yousefian, M.; Mousavi, S.E. The Mechanism of Reproduction and Hormonal Function in Finfish Species: A Review. Sci. Res. Essays 2011, 6, 3561–3570. [Google Scholar] [CrossRef]
- Gac, F.L.; Blaise, O.; Fostier, A.; Bail, P.L.; Maurice, L.; Mourot, B.; Weil, C. Growth Hormone (GH) and Reproduction: A Review. Fish Physiol. Biochem. 1993, 11, 219–232. [Google Scholar] [CrossRef]
- Reinecke, M. Insulin-like Growth Factors and Fish Reproduction. Biol. Reprod. 2010, 82, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Hu, Y.; Yu, F. A Review on Ghrelin and Fish Reproduction. Reprod. Breed. 2021, 1, 128–135. [Google Scholar] [CrossRef]
- Ndandala, C.B.; Dai, M.; Mustapha, U.F.; Li, X.; Liu, J.; Huang, H.; Li, G.; Chen, H. Current Research and Future Perspectives of GH and IGFs Family Genes in Somatic Growth and Reproduction of Teleost Fish. Aquac. Rep. 2022, 26, 101289. [Google Scholar] [CrossRef]
- Perrot, V.; Funkenstein, B. Cellular Distribution of Insulin-like Growth Factor II (IGF-II) MRNA and Hormonal Regulation of IGF-I and IGF-II MRNA Expression in Rainbow Trout Testis (Oncorhynchus mykiss). Fish Physiol. Biochem. 1999, 20, 219–229. [Google Scholar] [CrossRef]
- Yuan, Y.; Hong, Y. Medaka Insulin-like Growth Factor-2 Supports Self-Renewal of the Embryonic Stem Cell Line and Blastomeres in Vitro. Sci. Rep. 2017, 7, 78. [Google Scholar] [CrossRef]
- Higuchi, K.; Kazeto, Y.; Ozaki, Y.; Izumida, D.; Hotta, T.; Soyano, K.; Gen, K. Insulin-like Growth Factors 1 and 2 Regulate Gene Expression and Enzymatic Activity of Cyp17a1 in Ovarian Follicles of the Yellowtail, Seriola quinqueradiata. Heliyon 2020, 6, e04181. [Google Scholar] [CrossRef] [PubMed]
- Kwintkiewicz, J.; Giudice, L.C. The Interplay of Insulin-like Growth Factors, Gonadotropins, and Endocrine Disruptors in Ovarian Follicular Development and Function. Semin. Reprod. Med. 2009, 27, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.M. Effects of IGF1 and IGF2 on In Vitro Ovarian Follicle Maturation in Rainbow Trout, Oncorhynchus mykiss. Fishes 2023, 8, 367. [Google Scholar] [CrossRef]
- Moriyama, S.; Duguay, S.J.; Conlon, J.M.; Duan, C.; Dickhoff, W.W.; Plisetskaya, E.M. Recombinant Coho Salmon Insulin-like Growth Factor I: Expression in Escherichia coli, Purification and Characterization. Eur. J. Biochem. 1993, 218, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Swanson, P.; Dickhoff, W.W. Free and Protein-Bound Insulin-like Growth Factor-I (IGF-I) and IGF- Binding Proteins in Plasma of Coho Salmon, Oncorhynchus kisutch. Gen. Comp. Endocrinol. 1999, 115, 398–405. [Google Scholar] [CrossRef]
- Moreira, D.P.; Melo, R.M.C.; Weber, A.A.; Rizzo, E. Insulin-like Growth Factors 1 and 2 Are Associated with Testicular Germ Cell Proliferation and Apoptosis during Fish Reproduction. Reprod. Fertil. Dev. 2020, 32, 988–998. [Google Scholar] [CrossRef]
- Pierce, A.L.; Breves, J.P.; Moriyama, S.; Hirano, T.; Grau, E.G. Differential Regulation of Igf1 and Igf2 mRNA Levels in Tilapia Hepatocytes: Effects of Insulin and Cortisol on GH Sensitivity. J. Endocrinol. 2011, 211, 201–210. [Google Scholar] [CrossRef]
- Ahmed, S.J.; Mohammed, B.T. Biological Relationship between Insulin-like Growth Factors (Igf1 And Igf2) Genes And Steroidogenesis in Ovine Ovarian Antral Follicles. J. Univ. Duhok 2022, 25, 247–256. [Google Scholar] [CrossRef]
- Caruso, M.A.; Sheridan, M.A. New Insights into the Signaling System and Function of Insulin in Fish. Gen. Comp. Endocrinol. 2011, 173, 227–247. [Google Scholar] [CrossRef]
- Mukherjee, D.; Majumder, S.; Roy Moulik, S.; Pal, P.; Gupta, S.; Guha, P.; Kumar, D. Membrane Receptor Cross Talk in Gonadotropin-, IGF-I-, and Insulin-Mediated Steroidogenesis in Fish Ovary: An Overview. Gen. Comp. Endocrinol. 2017, 240, 10–18. [Google Scholar] [CrossRef]
- Mahardini, A.; Yamauchi, C.; Takeuchi, Y.; Rizky, D.; Takekata, H.; Takemura, A. Changes in MRNA Abundance of Insulin-like Growth Factors in the Brain and Liver of a Tropical Damselfish, Chrysiptera cyanea, in Relation to Seasonal and Food-Manipulated Reproduction. Gen. Comp. Endocrinol. 2018, 269, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chu, L.; Sun, X.; Liu, Y.; Cheng, C.H.K. IGFS Mediate the Action of LH on Oocyte Maturation in Zebrafish. Mol. Endocrinol. 2015, 29, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Guo, Y.; Nong, C.; Ndandala, C.B.; Yang, H.; Huang, H.; Li, G.; Chen, H. Analysis on LncRNA and MRNA Expression Profiles of IGF3-Induced Ovarian Maturation in Spotted Scat (Scatophagus argus). Aquac. Rep. 2022, 27, 101367. [Google Scholar] [CrossRef]
- Dong, Y.; Lyu, L.; Zhang, D.; Li, J.; Wen, H.; Shi, B. Integrated LncRNA and MRNA Transcriptome Analyses in the Ovary of Cynoglossus semilaevis Reveal Genes and Pathways Potentially Involved in Reproduction. Front. Genet. 2021, 12, 671729. [Google Scholar] [CrossRef]
- García-Pérez, I.; Molsosa-Solanas, A.; Perelló-Amorós, M.; Sarropoulou, E.; Blasco, J.; Gutiérrez, J.; de la Serrana, D.G. The Emerging Role of Long Non-Coding RNAs in Development and Function of Gilthead Sea Bream (Sparus aurata) Fast Skeletal Muscle. Cells 2022, 11, 428. [Google Scholar] [CrossRef]
- Deng, Q.; Zhao, N.; Zhu, C.; Zhang, B. Long Non-Coding RNAs in the Physiology of Aquaculture Animals: A Perspective Update. Rev. Fish Biol. Fish. 2022, 32, 1103–1122. [Google Scholar] [CrossRef]
- Yang, Y.; Yujiao, W.; Fang, W.; Linhui, Y.; Ziqi, G.; Zhichen, W.; Zirui, W.; Shengwang, W. The Roles of MiRNA, LncRNA and CircRNA in the Development of Osteoporosis. Biol. Res. 2020, 53, 1–16. [Google Scholar] [CrossRef]
- Zhou, Z.; Leng, C.; Wang, Z.; Long, L.; Lv, Y.; Gao, Z.; Wang, Y.; Wang, S.; Li, P. The Potential Regulatory Role of the LncRNA-MiRNA-MRNA Axis in Teleost Fish. Front. Immunol. 2023, 14, 1065357. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, Y.; Zhao, W.; Huang, H.; Zhang, Q.; Liu, J. CeRNA Profiling and the Role in Regulating Gonadal Development in Gold Pompano. BMC Genom. 2025, 26, 43. [Google Scholar] [CrossRef]
- Hao, R.; Zhu, X.; Tian, C.; Jiang, M.; Huang, Y.; Li, G.; Zhu, C. LncRNA–MiRNA–MRNA CeRNA Network of Different Body Colors in Plectropomus leopardus. Front. Mar. Sci. 2023, 10, 1170762. [Google Scholar] [CrossRef]
- Liu, B.; San, L.; Guo, H.; Zhu, K.; Zhang, N.; Yang, J.; Liu, B.; Hou, J.; Zhang, D. Transcriptomic Analysis Reveals Functional Interaction of MRNA-LncRNA-MiRNA in Trachinotus ovatus Infected by Cryptocaryon Irritans. Int. J. Mol. Sci. 2023, 24, 5886. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Gu, W.; Liu, E.; Shi, X.; Wang, B.; Wu, W.; Dong, F.; Xu, G. Comprehensive Analysis of MiRNA-MRNA/LncRNA during Gonadal Development of Triploid Female Rainbow Trout (Oncorhynchus mykiss). Genomics 2021, 113, 3533–3543. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Peng, H.; Ding, Y.; Zhang, L.; Yang, J.; Han, X. A Transcriptomic Regulatory Network among MiRNAs, PiRNAs, CircRNAs, LncRNAs and MRNAs Regulates Microcystin-Leucine Arginine (MC-LR)-Induced Male Reproductive Toxicity. Sci. Total Environ. 2019, 667, 563–577. [Google Scholar] [CrossRef]
- Ndandala, C.B.; Zhou, Q.; Li, Z.; Guo, Y.; Li, G. Identification of Insulin-like Growth Factor ( IGF ) Family Genes in the Golden Pompano, Trachinotus ovatus: Molecular Cloning, Characterization and Gene Expression. Int. J. Mol. Sci. 2024, 25, 2499. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zhang, Y.; Ji, C.; Zhao, Y.; Peng, J.; Xu, Y. Golden Pompano Genome Resource Enables Discovery of Valuable Gene Determining Growth Traits. bioRxiv 2021, 2021-05. [Google Scholar] [CrossRef]
- Liu, B.; Guo, H.Y.; Zhu, K.C.; Guo, L.; Liu, B.S.; Zhang, N.; Yang, J.W.; Jiang, S.G.; Zhang, D.C. Growth, Physiological, and Molecular Responses of Golden Pompano Trachinotus ovatus (Linnaeus, 1758) Reared at Different Salinities. Fish Physiol. Biochem. 2019, 45, 1879–1893. [Google Scholar] [CrossRef]
- Li, J.; Liu, Z.; Wang, D.; Cheng, C.H.K. Insulin-like Growth Factor 3 Is Involved in Oocyte Maturation in Zebrafish. Biol. Reprod. 2011, 84, 476–486. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg1, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements Daehwan HHS Public Access. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Hogue, C.W.; Groll, M. An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinform. 2001, 29, 137–140. [Google Scholar]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. CytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, T.J.; Daniel, R.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models. Genome Res. 1971, 13, 426. [Google Scholar] [CrossRef]
- Xu, R.; Li, Q.; Yu, H.; Kong, L. Oocyte Maturation and Origin of the Germline as Revealed by the Expression of Nanos-like in the Pacific Oyster Crassostrea gigas. Gene 2018, 663, 41–50. [Google Scholar] [CrossRef]
- Ma, H.; Gao, G.; Palti, Y.; Tripathi, V.; Birkett, J.E.; Weber, G.M. Transcriptomic Response of the Ovarian Follicle Complex in Post-Vitellogenic Rainbow Trout to 17α,20β-Dihdroxy-4-Pregnen-3-One In Vitro. Int. J. Mol. Sci. 2024, 25, 12683. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, G.; Sridevi, P.; Sahoo, P.K.; Swapna, I.; Ge, W.; Kirubagaran, R.; Dutta-Gupta, A.; Senthilkumaran, B. Cloning and Expression of StAR during Gonadal Cycle and HCG-Induced Oocyte Maturation of Air-Breathing Catfish, Clarias gariepinus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 154, 6–11. [Google Scholar] [CrossRef]
- Weber, G.M.; Sullivan, C.V. Effects of Insulin-like Growth Factor-I on in Vitro Final Oocyte Maturation and Ovarian Steroidogenesis in Striped Bass, Morone saxatilis. Biol. Reprod. 2000, 63, 1049–1057. [Google Scholar] [CrossRef]
- Zhu, B.; Pardeshi, L.; Chen, Y.; Ge, W. Transcriptomic Analysis for Differentially Expressed Genes in Ovarian Follicle Activation in the Zebrafish. Front. Endocrinol. 2018, 9, 593. [Google Scholar] [CrossRef]
- Fan, Z.; Zou, Y.; Liang, D.; Tan, X.; Jiao, S.; Wu, Z.; Li, J.; Zhang, P.; You, F. Roles of Forkhead Box Protein L2 (Foxl2) during Gonad Differentiation and Maintenance in a Fish, the Olive Flounder (Paralichthys olivaceus). Reprod. Fertil. Dev. 2019, 31, 1742–1752. [Google Scholar] [CrossRef]
- Nóbrega, R.H.; De Souza Morais, R.D.V.; Crespo, D.; De Waal, P.P.; De França, L.R.; Schulz, R.W.; Bogerd, J. Fsh Stimulates Spermatogonial Proliferation and Differentiation in Zebrafish via Igf3. Endocrinology 2015, 156, 3804–3817. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.N.; Van Der Kraak, G. The Role of the Insulin-like Growth Factor (IGF) System in Zebrafish (Danio rerio) Ovarian Development. Gen. Comp. Endocrinol. 2010, 168, 103–110. [Google Scholar] [CrossRef]
- Hatef, A.; Unniappan, S. Metabolic Hormones and the Regulation of Spermatogenesis in Fishes; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 134, ISBN 3069667414. [Google Scholar]
- Rottmann, R.W.; Shireman, J.V.; Chapman, F.A. Hormonal Control of Reproduction in Fish for Induced Spawning; Southern Regional Aquaculture Center: Stoneville, MS, USA, 1991; Volume 424, pp. 1–4. [Google Scholar]
- Guo, S.; Zhong, Y.; Zhang, Y.; Zhu, Y.; Guo, J.; Fu, Y.; Li, M. Transcriptome Analysis Provides Insights into Long Noncoding RNAs in Medaka Gonads. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 39, 100842. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhao, Q.; Wang, H.; Wei, L.; Wang, S.; Li, S.; Yuan, D.; Wang, Z. Integrated Transcriptomic and Metabolomic Analyses Identify Key Factors in the Vitellogenesis of Juvenile Sichuan Bream (Sinibrama taeniatus). Front. Mar. Sci. 2023, 10, 1243767. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, B.; Liu, X.; Shi, B.; Li, B. Recombinant Expression and Comparative Bioactivity of Tongue Sole Insulin-like Growth Factor (IGF)-1 and IGF-2 in Pichia Pastoris. Aquac. Res. 2018, 49, 2193–2200. [Google Scholar] [CrossRef]
- Zhang, D.; Shi, B.; Shao, P.; Shao, C.; Wang, C.; Li, J.; Liu, X.; Ma, X.; Zhao, X. The Identification of MiRNAs That Regulate Ovarian Maturation in Cynoglossus semilaevis. Aquaculture 2022, 555, 738250. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, F.; Zhang, R.; Liang, T.; Zhang, W.W.; Ji, X.; Du, Q.; Chang, Z. Identification of Suh Gene and Evidence for Involvement of Notch Signaling Pathway on Gonadal Differentiation of Yellow River Carp (Cyprinus carpio). Fish Physiol. Biochem. 2018, 44, 375–386. [Google Scholar] [CrossRef]
- Lee, S.L.J.; Horsfield, J.A.; Black, M.A.; Rutherford, K.; Fisher, A.; Gemmell, N.J. Histological and Transcriptomic Effects of 17α-Methyltestosterone on Zebrafish Gonad Development. BMC Genom. 2017, 18, 557. [Google Scholar] [CrossRef] [PubMed]
- He, F.X.; Jiang, D.N.; Huang, Y.Q.; Mustapha, U.F.; Yang, W.; Cui, X.F.; Tian, C.X.; Chen, H.P.; Shi, H.J.; Deng, S.P.; et al. Comparative Transcriptome Analysis of Male and Female Gonads Reveals Sex-Biased Genes in Spotted Scat (Scatophagus argus). Fish Physiol. Biochem. 2019, 45, 1963–1980. [Google Scholar] [CrossRef]
- Liu, M.; Ni, H.; Rong, Z.; Wang, Z.; Yan, S.; Liao, X.; Dong, Z. Gonad Transcriptome Analysis Reveals the Differences in Gene Expression Related to Sex-Biased and Reproduction of Clam Cyclina sinensis. Front. Mar. Sci. 2023, 9, 1110587. [Google Scholar] [CrossRef]
- Li, J.; Zhou, Y.; Zhou, Z.; Lin, C.; Wei, J.; Qin, Y.; Xiang, Z.; Ma, H.; Zhang, Y.; Zhang, Y.; et al. Comparative Transcriptome Analysis of Three Gonadal Development Stages Reveals Potential Genes Involved in Gametogenesis of the Fluted Giant Clam (Tridacna squamosa). BMC Genom. 2020, 21, 872. [Google Scholar] [CrossRef]
- Wang, F.; Jia, Y.; Wang, P.; Yang, Q.; Du, Q.Y.; Chang, Z.J. Identification and Profiling of Cyprinus carpio MicroRNAs during Ovary Differentiation by Deep Sequencing. BMC Genom. 2017, 18, 333. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, Y.; Ndandala, C.B.; Chen, H.; Huang, C.; Zhao, G.; Huang, H.; Li, G.; Chen, H. Analysis of CircRNA and MiRNA Expression Profiles in IGF3-Induced Ovarian Maturation in Spotted Scat (Scatophagus argus). Front. Endocrinol. 2022, 13, 998207. [Google Scholar] [CrossRef]
- Zayed, Y. Characterization of Microrna Expression Profiles and Role of Nodal-Related Genes in Zebrafish Ovarian Follicles. Ph.D. Thesis, York University, Toronto, ON, Canada, 2019. Available online: http://hdl.handle.net/10315/40946 (accessed on 10 March 2025).
- Abdellaoui, N.; Sun, M. Transcriptome Profiling of Gene Expression in Atlantic Salmon ( Salmo salar ) at Early Stage of Development. Mar. Biotechnol. 2024, 26, 964–974. [Google Scholar] [CrossRef]
- Tenugu, S.; Pranoty, A.; Mamta, S.K.; Senthilkumaran, B. Development and Organisation of Gonadal Steroidogenesis in Bony Fishes—A Review. Aquac. Fish. 2021, 6, 223–246. [Google Scholar] [CrossRef]
- Li, L.; Shi, X.; Shi, Y.; Wang, Z. The Signaling Pathways Involved in Ovarian Follicle Development. Front. Physiol. 2021, 12, 730196. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Wang, Q.; Zhang, J.; Zhao, R.; Cao, Y.; Yu, S.; Wang, K.; Chen, Y.; Ma, Z.; Sun, X.; et al. Differential Expression of MiRNAs, LncRNAs, and CircRNAs between Ovaries and Testes in Common Carp (Cyprinus carpio). Cells 2023, 12, 2631. [Google Scholar] [CrossRef]
- Safian, D.; Bogerd, J.; Schulz, R.W. Regulation of Spermatogonial Development by Fsh: The Complementary Roles of Locally Produced Igf and Wnt Signaling Molecules in Adult Zebrafish Testis. Gen. Comp. Endocrinol. 2019, 284, 113244. [Google Scholar] [CrossRef]
- Sobinoff, A.P.; Sutherland, J.M.; Mclaughlin, E.A. Intracellular Signalling during Female Gametogenesis. Mol. Hum. Reprod. 2013, 19, 265–278. [Google Scholar] [CrossRef]
- Reindl, K.M.; Kittilson, J.D.; Bergan, H.E.; Sheridan, M.A. Growth Hormone-Stimulated Insulin-like Growth Factor-1 Expression in Rainbow Trout (Oncorhynchus mykiss) Hepatocytes Is Mediated by ERK, PI3K-AKT, and JAK-STAT. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R236–R243. [Google Scholar] [CrossRef]
- Barclay, R.D.; Burd, N.A.; Tyler, C.; Tillin, N.A.; Mackenzie, R.W. The Role of the IGF-1 Signaling Cascade in Muscle Protein Synthesis and Anabolic Resistance in Aging Skeletal Muscle. Front. Nutr. 2019, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Arur, S. Conserved Insulin Signaling in the Regulation of Oocyte Growth, Development, and Maturation. Mol. Reprod. Dev. 2017, 84, 444–459. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhao, C.; Zhu, B.; Zhang, Z.; Ge, W. Loss of Inhibin Advances Follicle Activation and Female Puberty Onset but Blocks Oocyte Maturation in Zebrafish. Endocrinology 2020, 161, bqaa184. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhai, Y.; Geng, R.; Wu, K.; Song, W.; Ai, N.; Ge, W. Genetic Analysis of Activin/Inhibin β Subunits in Zebrafish Development and Reproduction. PLosGenet. 2022, 18, e1010523. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Duan, S.H.; Wang, G.L.; Li, J.L. Integrated MRNA and MiRNA Expression Profile Analysis of Female and Male Gonads in Hyriopsis cumingii. Sci. Rep. 2021, 11, 665. [Google Scholar] [CrossRef]
- Sukhan, Z.P.; Cho, Y.; Hossen, S.; Yang, S.W.; Hwang, N.Y.; Lee, W.K.; Kho, K.H. Functional Characterization of Three GnRH Isoforms in Small Yellow Croaker Larimichthys polyactis Maintained in Captivity: Special Emphasis on Reproductive Dysfunction. Biology 2022, 11, 1200. [Google Scholar] [CrossRef]





| ID Description | Gene Name | Primers (5′–3′) |
|---|---|---|
| SCSFRI_TO_T_00091676 | cyp19a1 | F: TGGCTCATGTGGATGTCCTCAGT |
| R: TGCTGTCCTGTGCTGCTGGTTA | ||
| SCSFRI_TO_T_00132967 | cyp17a1 | F: ACGCTCTGCTTCAACTCCTCCT |
| R: AAGATGTCCACCAGGCTGTCCTT | ||
| SCSFRI_TO_T_00065781 | adam23 | F: GTGGTGCTCGTTGCTGTGGA |
| R: GGCGTGGTGCTTGATGCTCT | ||
| SCSFRI_TO_T_00083864 | hsd17b7 | F: AGCCAGCTTCGCCTGTGTCT |
| R: ACCTCCTGAGCAGCAGTGAACA | ||
| SCSFRI_TO_T_00068502 | hsd17b3 | F: TGGTGACTGGTGCTTCAGAAGG |
| R: TGGCTACCTGGTCCAATGTTACTT | ||
| SCSFRI_TO_T_00052923 | sox3 | F: GAGCCAACGCCGTGAACAACT |
| R: GTAGGTCTGAGCCGAGGACATCA | ||
| SCSFRI_TO_T_00073261 | foxn5 | F: ACGCTGGGAGCCTCAAAGTC |
| R: CAGGTTGTGTCGGATGGTGTTC | ||
| SCSFRI_TO_T_00072390 | inhbb | F: GCAACGAGGTTCTAGCGGAGAC |
| R: ACACGGAAGTAGAGCCACAGGTT | ||
| SCSFRI_TO_T_00126075 | krt18 | F: ACCGCATCAGCATCTCCTCCA |
| R: CTCCAGGTTCCTCACTGTCTCCA | ||
| SCSFRI_TO_T_00025866 | h2ax | F: GTCGGTCGTGTTCACAGGCT |
| R: CCAGGATCTCAGCGGTCAGGTA | ||
| SCSFRI_TO_T_00061632 | nanos3 | F: ACCGCAAGAAGACGCCCAAG |
| R: CACAACACATCTCCTGCCTGGT | ||
| SCSFRI_TO_T_00003998 | kank1 | F: TGGCTCCAACAAGGCAACGAA |
| R: GGCTGGACACAGAGAACCACTC | ||
| SCSFRI_TO_T_00026350 | slc6a18 | F: CCCTGTGCTTTCCATCCCTCTG |
| R: CGGTTCCTCTGTTCCTGTGCTT | ||
| SCSFRI_TO_T_00012814 | pgr | F: GTTCAGAGGACGAATGTGGCTACG |
| R: TGCTGCTTCCTGTCGGTGTCA | ||
| SCSFRI_TO_T_00113132 | cdk15 | F: GCTGGTGGCTCTGAAGGTGATC |
| R: AGTCTCTGGCGTGGATGATGTCA | ||
| MSTRG.57329 | star | F: GAGGATGGATACAGCGAGGAGGAG |
| R: CCAATGTCAGGCAACACCTTACTCA | ||
| SCSFRI_TO_T_00138572 | axin2 | F: CGCAGAGAGTCGCCAAAGCAAT |
| R: TCCAGGTAGATGTCAGAGGTCAGAA | ||
| SCSFRI_TO_T_00073261 | β-actin | F: GAGAGGTTCCGTTGCCCAGAG |
| R: CAGACAGCACAGTGTTGGCGT | ||
| MSTRG.26614.1 | F: AACATCACGGCTCTTGGCTCTC | |
| R: GCTTGTGCTGGAGGCTGAAGT | ||
| MSTRG.98390.1 | F: CGCTGGCATTGGTCACATTCAC | |
| R: GAGAACGGTGTCCACTCCATCG | ||
| MSTRG.77400.2 | F: GACATGCCTGCTCTGTGATTGA | |
| R: CCACTCTGTCAGTACCGAGGAA | ||
| MSTRG.84457.1 | F: ATCGCCTTCCGTCCCTTCCA | |
| R: CCTCGCCTGCTGTTTGGTCAT | ||
| MSTRG.20896.2 | F: TGTCTGCCTGCTCCACATCCT | |
| R: TGTCCTCCCAGCCCTGTCATT | ||
| MSTRG.58123.2 | F: AGATAAGGAGTGCAGCCTGTGT | |
| R: CGCAAGGAGAACGGACAGAGA |
| Sample | Clean Read Bases | Q20 (%) | Q30 (%) | Total Clean Reads | Mapped Reads | Mapping Ratio (%) |
|---|---|---|---|---|---|---|
| C1 | 15,630,440,161 | 98.17 | 94.96 | 1.11 × 108 | 1,332,878 | 91.75 |
| C2 | 13,264,457,783 | 98.37 | 95.34 | 94,414,990 | 1,897,056 | 90.71 |
| C3 | 12,982,764,898 | 98.16 | 94.91 | 91,848,966 | 1,367,594 | 91.24 |
| IGF1T1 | 13,110,122,504 | 98.31 | 95.26 | 93,054,020 | 1,202,186 | 92.21 |
| IGF1T2 | 13,516,459,173 | 98.25 | 95.13 | 98,391,424 | 3,977,422 | 90.13 |
| IGF1T3 | 12,471,670,121 | 98.38 | 95.44 | 91,910,782 | 3,211,596 | 91.53 |
| IGF2T1 | 13,269,497,776 | 98.33 | 95.28 | 97,543,940 | 1,117,334 | 92.94 |
| IGF2T2 | 14,829,071,350 | 98.29 | 95.14 | 1.09 × 108 | 971,076 | 93.06 |
| IGF2T3 | 14,431,477,858 | 98.42 | 95.47 | 1.05 × 108 | 1,243,074 | 93.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndandala, C.B.; Guo, Y.; Ju, Z.; Fachri, M.; Mwemi, H.M.; Chen, H. Integrated lncRNA and mRNA Transcriptome Analyses of IGF1 and IGF2 Stimulated Ovaries Reveal Genes and Pathways Potentially Associated with Ovarian Development and Oocyte Maturation in Golden Pompano (Trachinotus ovatus). Animals 2025, 15, 1134. https://doi.org/10.3390/ani15081134
Ndandala CB, Guo Y, Ju Z, Fachri M, Mwemi HM, Chen H. Integrated lncRNA and mRNA Transcriptome Analyses of IGF1 and IGF2 Stimulated Ovaries Reveal Genes and Pathways Potentially Associated with Ovarian Development and Oocyte Maturation in Golden Pompano (Trachinotus ovatus). Animals. 2025; 15(8):1134. https://doi.org/10.3390/ani15081134
Chicago/Turabian StyleNdandala, Charles Brighton, Yuwen Guo, Zhimin Ju, Muhammad Fachri, Happiness Moses Mwemi, and Huapu Chen. 2025. "Integrated lncRNA and mRNA Transcriptome Analyses of IGF1 and IGF2 Stimulated Ovaries Reveal Genes and Pathways Potentially Associated with Ovarian Development and Oocyte Maturation in Golden Pompano (Trachinotus ovatus)" Animals 15, no. 8: 1134. https://doi.org/10.3390/ani15081134
APA StyleNdandala, C. B., Guo, Y., Ju, Z., Fachri, M., Mwemi, H. M., & Chen, H. (2025). Integrated lncRNA and mRNA Transcriptome Analyses of IGF1 and IGF2 Stimulated Ovaries Reveal Genes and Pathways Potentially Associated with Ovarian Development and Oocyte Maturation in Golden Pompano (Trachinotus ovatus). Animals, 15(8), 1134. https://doi.org/10.3390/ani15081134






