Phylogenetic Position of Hungarian Grey Cattle Breed Based on Total-Representation Sample
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
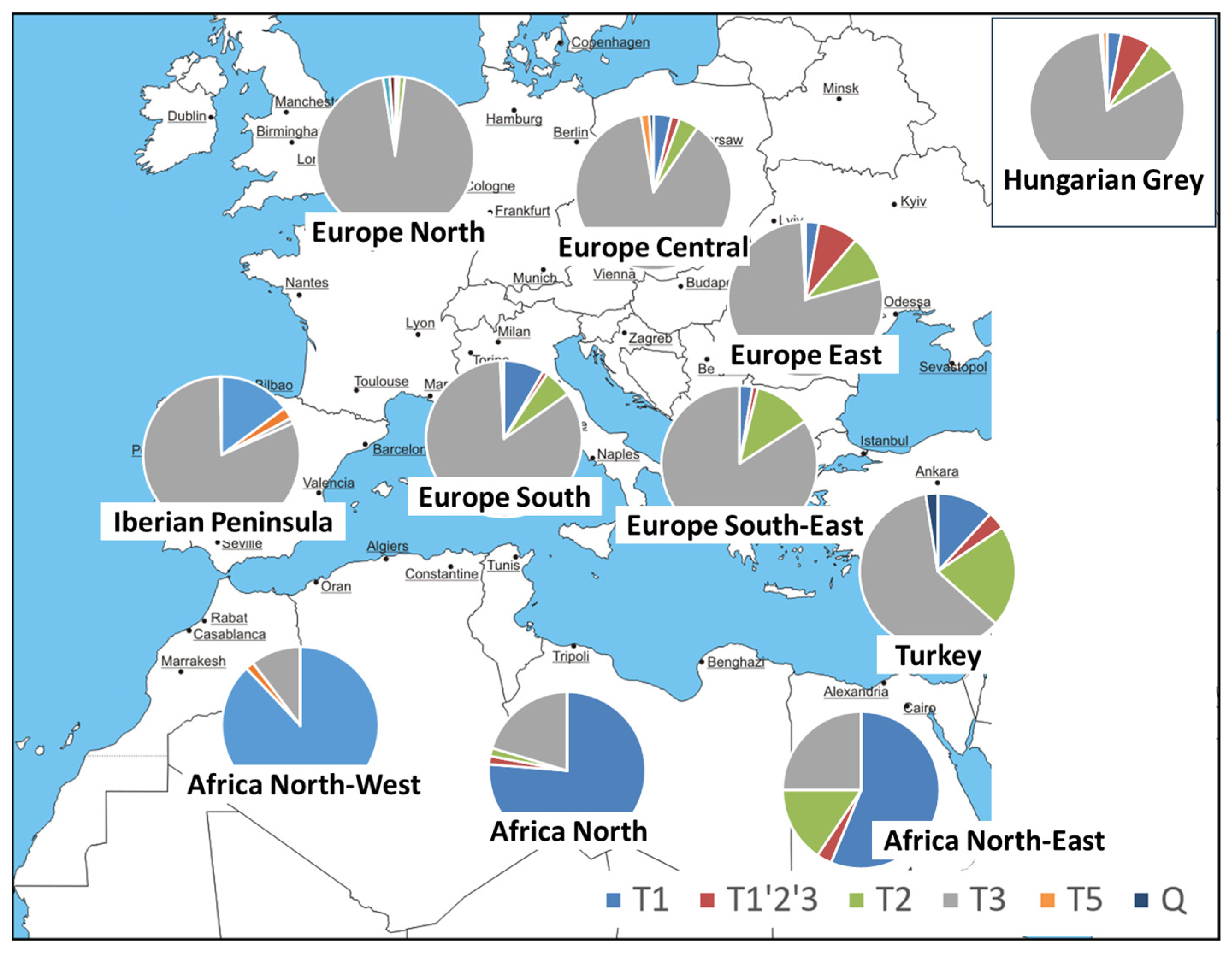
3.1. Founder Sampling
3.2. Mitochondrial Control Region Sequence Diversity
3.3. Mitochondrial DNA Haplotypes and Phylogeny
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bodó, I.; István, G.; Gábor, K. The Hungarian Grey Cattle Breed, 2004th ed.; Association of the Hungarian Grey Cattle breeders: Budapest, Hungary, 2004; ISBN 2399971316110. [Google Scholar]
- Di Lorenzo, P.; Lancioni, H.; Ceccobelli, S.; Colli, L.; Cardinali, I.; Karsli, T.; Capodiferro, M.R.; Sahin, E.; Ferretti, L.; Ajmone Marsan, P.; et al. Mitochondrial DNA Variants of Podolian Cattle Breeds Testify for a Dual Maternal Origin. PLoS ONE 2018, 13, e0192567. [Google Scholar] [CrossRef] [PubMed]
- Hankó, B. A Magyar Szarvasmarha Eredete; Tisia: Debrecen, Hungary, 1936; Volume 1. [Google Scholar]
- Wagner, M.; Brandt, J.F.; Erdl, M.P.; Erichson, W.F.; Koch, K.L.; von Nathusius, H.E.; Roßmäßler, E.A.; Schlegel, H.; Wagner, J.A.; Wagner, R. Reisen in Der Regentschaft Algier in Den Jahren 1836, 1837 und 1838. Dritter Band; Verlag Von Leopold Voss: St. Petersburg, Russia, 1841. [Google Scholar]
- Darwin, C.R. The Variation of Animals and Plants Under Domestication, 1st ed.; First Issue; John Murray: London, UK, 1868. [Google Scholar]
- Radácsi, A.; Bodó, I.; Béri, B. Horn and Coat Color Varieties of the Hungarian Grey Cattle. Acta Agrar. Debreceniensis 2006, 21, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Bartosiewicz, L. The Hungarian Grey Cattle: A Traditional European Breed. Anim. Genet. Resour. Génétiques Anim. Genéticos Anim. 1997, 21, 49–60. [Google Scholar] [CrossRef]
- Bartosiewicz, L. A Millennium of Migrations: Proto-Historic Mobile Pastoralism in Hungary. Bull. Fla. Mus. Nat. Hist. 2003, 44, 101–130. [Google Scholar] [CrossRef]
- Jankovich, M. Adatok a magyar szarvasmarha eredetének és hasznositásának kérdéséhez. Agrár. Szle. 1967, 3, 420–431. [Google Scholar]
- Rossi, C.; Sinding, M.-H.S.; Mullin, V.E.; Scheu, A.; Erven, J.A.M.; Verdugo, M.P.; Daly, K.G.; Ciucani, M.M.; Mattiangeli, V.; Teasdale, M.D.; et al. The Genomic Natural History of the Aurochs. Nature 2024, 635, 136–141. [Google Scholar] [CrossRef]
- Achilli, A.; Olivieri, A.; Pellecchia, M.; Uboldi, C.; Colli, L.; Al-Zahery, N.; Accetturo, M.; Pala, M.; Kashani, B.H.; Perego, U.A.; et al. Mitochondrial Genomes of Extinct Aurochs Survive in Domestic Cattle. Curr. Biol. 2008, 18, R157–R158. [Google Scholar] [CrossRef]
- Loftus, R.T.; MacHugh, D.E.; Bradley, D.G.; Sharp, P.M.; Cunningham, P. Evidence for Two Independent Domestications of Cattle. Proc. Natl. Acad. Sci. USA 1994, 91, 2757–2761. [Google Scholar] [CrossRef]
- Troy, C.S.; MacHugh, D.E.; Bailey, J.F.; Magee, D.A.; Loftus, R.T.; Cunningham, P.; Chamberlain, A.T.; Sykes, B.C.; Bradley, D.G. Genetic Evidence for Near-Eastern Origins of European Cattle. Nature 2001, 410, 1088–1091. [Google Scholar] [CrossRef]
- Beja-Pereira, A.; Caramelli, D.; Lalueza-Fox, C.; Vernesi, C.; Ferrand, N.; Casoli, A.; Goyache, F.; Royo, L.J.; Conti, S.; Lari, M.; et al. The Origin of European Cattle: Evidence from Modern and Ancient DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 8113–8118. [Google Scholar] [CrossRef]
- Mannen, H.; Kohno, M.; Nagata, Y.; Tsuji, S.; Bradley, D.G.; Yeo, J.S.; Nyamsamba, D.; Zagdsuren, Y.; Yokohama, M.; Nomura, K.; et al. Independent Mitochondrial Origin and Historical Genetic Differentiation in North Eastern Asian Cattle. Mol. Phylogenet. Evol. 2004, 32, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Saliari, K.; Amory, C.; Draganits, E.; Ramsl, P.C.; Tobias, B.; Pucher, E.; Parson, W. Morphometric and Genetic Evidence for Cattle Imports from the Mediterranean into Present-Day Austria during the Iron Age. J. Archaeol. Sci. Rep. 2023, 48, 103842. [Google Scholar] [CrossRef]
- Cubric-Curik, V.; Novosel, D.; Brajkovic, V.; Rota Stabelli, O.; Krebs, S.; Sölkner, J.; Šalamon, D.; Ristov, S.; Berger, B.; Trivizaki, S.; et al. Large-Scale Mitogenome Sequencing Reveals Consecutive Expansions of Domestic Taurine Cattle and Supports Sporadic Aurochs Introgression. Evol. Appl. 2022, 15, 663–678. [Google Scholar] [CrossRef]
- Milhoffer, S. Magyarország Közgazdasága I; Franklin: Budapest, Hungary, 1904. [Google Scholar]
- Bartosiewicz, L. Cattle Trade Across the Danube at Vác (Hungary). Anim. Dans Espace Hum. Homme Dans Espace Anim. 1995, 21, 189–195. [Google Scholar]
- Bartosiewicz, L. Hungarian Grey Cattle: Parallels in Constituting Animal and Human Identities. In Interspecies Interactions; Routledge: London, UK, 2017; ISBN 978-1-315-10929-9. [Google Scholar]
- Davidescu, M.-A.; Simeanu, D.; Gorgan, D.-L.; Ciorpac, M.; Creanga, S. Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity. Agriculture 2022, 12, 2059. [Google Scholar] [CrossRef]
- Maróti-Agóts, Á. Effect of Different Sampling Methods on Cattle mtDNA Phylogenetic Studies. In Proceedings of the EAAP—59th Annual Meeting, Vilnius, Lithuania, 24–27 August 2008. [Google Scholar]
- Anderson, S.; de Bruijn, M.H.; Coulson, A.R.; Eperon, I.C.; Sanger, F.; Young, I.G. Complete Sequence of Bovine Mitochondrial DNA. Conserved Features of the Mammalian Mitochondrial Genome. J. Mol. Biol. 1982, 156, 683–717. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Eltsov, N.P.; Volodko, N.V.; Starikovskaya, E.B.; Mazunin, I.O.; Sukernik, R.I. The role of natural selection in the evolution of mitochondrial haplogroups in Northeastern Eurasia. Russ. J. Genet. 2010, 46, 1105–1107. [Google Scholar] [CrossRef]
- Lancioni, H.; Di Lorenzo, P.; Ceccobelli, S.; Perego, U.A.; Miglio, A.; Landi, V.; Antognoni, M.T.; Sarti, F.M.; Lasagna, E.; Achilli, A. Phylogenetic Relationships of Three Italian Merino-Derived Sheep Breeds Evaluated through a Complete Mitogenome Analysis. PLoS ONE 2013, 8, e73712. [Google Scholar] [CrossRef]
- Colli, L.; Lancioni, H.; Cardinali, I.; Olivieri, A.; Capodiferro, M.R.; Pellecchia, M.; Rzepus, M.; Zamani, W.; Naderi, S.; Gandini, F.; et al. Whole Mitochondrial Genomes Unveil the Impact of Domestication on Goat Matrilineal Variability. BMC Genom. 2015, 16, 1115. [Google Scholar] [CrossRef]
- Lombardo, G.; Rambaldi Migliore, N.; Colombo, G.; Capodiferro, M.R.; Formenti, G.; Caprioli, M.; Moroni, E.; Caporali, L.; Lancioni, H.; Secomandi, S.; et al. The Mitogenome Relationships and Phylogeography of Barn Swallows (Hirundo rustica). Mol. Biol. Evol. 2022, 39, msac113. [Google Scholar] [CrossRef]
- Achilli, A.; Bonfiglio, S.; Olivieri, A.; Malusà, A.; Pala, M.; Kashani, B.H.; Perego, U.A.; Ajmone-Marsan, P.; Liotta, L.; Semino, O.; et al. The Multifaceted Origin of Taurine Cattle Reflected by the Mitochondrial Genome. PLoS ONE 2009, 4, e5753. [Google Scholar] [CrossRef]
- Bonfiglio, S.; Achilli, A.; Olivieri, A.; Negrini, R.; Colli, L.; Liotta, L.; Ajmone-Marsan, P.; Torroni, A.; Ferretti, L. The Enigmatic Origin of Bovine mtDNA Haplogroup R: Sporadic Interbreeding or an Independent Event of Bos primigenius Domestication in Italy? PLoS ONE 2010, 5, e15760. [Google Scholar] [CrossRef]
- Lenstra, J.A.; Ajmone-Marsan, P.; Beja-Pereira, A.; Bollongino, R.; Bradley, D.G.; Colli, L.; De Gaetano, A.; Edwards, C.J.; Felius, M.; Ferretti, L.; et al. Meta-Analysis of Mitochondrial DNA Reveals Several Population Bottlenecks during Worldwide Migrations of Cattle. Diversity 2014, 6, 178–187. [Google Scholar] [CrossRef]
- Olivieri, A.; Gandini, F.; Achilli, A.; Fichera, A.; Rizzi, E.; Bonfiglio, S.; Battaglia, V.; Brandini, S.; Gaetano, A.D.; El-Beltagi, A.; et al. Mitogenomes from Egyptian Cattle Breeds: New Clues on the Origin of Haplogroup Q and the Early Spread of Bos taurus from the Near East. PLoS ONE 2015, 10, e0141170. [Google Scholar] [CrossRef]
- Hill, E.W.; Bradley, D.G.; Al-Barody, M.; Ertugrul, O.; Splan, R.K.; Zakharov, I.; Cunningham, E.P. History and Integrity of Thoroughbred Dam Lines Revealed in Equine mtDNA Variation. Anim. Genet. 2002, 33, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Bower, M.A.; Whitten, M.; Nisbet, R.E.R.; Spencer, M.; Dominy, K.M.; Murphy, A.M.; Cassidy, R.; Barrett, E.; Hill, E.W.; Binns, M. Thoroughbred Racehorse Mitochondrial DNA Demonstrates Closer than Expected Links between Maternal Genetic History and Pedigree Records. J. Anim. Breed. Genet. 2013, 130, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Čačić, M.; Cubric-Curik, V.; Baban, M.; Barać, Z.; Curik, I. Use of Mitochondrial DNA Analyses in Verification of the Lipizzan Horse Pedigre. Agric. Conspec. Sci. 2011, 76, 365–368. [Google Scholar]
- Giontella, A.; Cardinali, I.; Lancioni, H.; Giovannini, S.; Pieramati, C.; Silvestrelli, M.; Sarti, F.M. Mitochondrial DNA Survey Reveals the Lack of Accuracy in Maremmano Horse Studbook Records. Animals 2020, 10, 839. [Google Scholar] [CrossRef]
- Engel, L.; Becker, D.; Nissen, T.; Russ, I.; Thaller, G.; Krattenmacher, N. Exploring the Origin and Relatedness of Maternal Lineages through Analysis of Mitochondrial DNA in the Holstein Horse. Front. Genet. 2021, 12, 632500. [Google Scholar] [CrossRef]
- Bowling, A.T.; Del Valle, A.; Bowling, M. A Pedigree-based Study of Mitochondrial d-loop DNA Sequence Variation among Arabian Horses. Anim. Genet. 2000, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Głażewska, I.; Prusak, B.; Gralak, B. Pedigrees as a Source of Information in Mt DNA Studies of Dogs and Horses. Anim. Genet. 2013, 44, 227–230. [Google Scholar] [CrossRef]
- Perdomo-González, D.I.; Laseca, N.; Demyda-Peyrás, S.; Valera, M.; Cervantes, I.; Molina, A. Fine-Tuning Genomic and Pedigree Inbreeding Rates in Equine Population with a Deep and Reliable Stud Book: The Case of the Pura Raza Española Horse. J. Anim. Sci. Biotechnol. 2022, 13, 127. [Google Scholar] [CrossRef]
- Crisà, A.; Cardinali, I.; Giontella, A.; Silvestrelli, M.; Lancioni, H.; Buttazzoni, L. A Genetic Make Up of Italian Lipizzan Horse Through Uniparental Markers to Preserve Historical Pedigrees. Biology 2024, 13, 1087. [Google Scholar] [CrossRef]
- Brajkovic, V.; Hršak, D.; Bradić, L.; Turkalj, K.; Novosel, D.; Ristov, S.; Ajmone-Marsan, P.; Colli, L.; Cubric-Curik, V.; Sölkner, J.; et al. Mitogenome Information in Cattle Breeding and Conservation Genetics: Developments and Possibilities of the SNP Chip. Livest. Sci. 2023, 275, 105299. [Google Scholar] [CrossRef]
- Fortuna, G.M.; Zumbach, B.J.; Johnsson, M.; Pocrnic, I.; Gorjanc, G. Accounting for the Nuclear and Mito Genome in Dairy Cattle Breeding—A simulation Study. JDS Commun. 2024, 5, 572–576. [Google Scholar] [CrossRef]
- Xuan, T.P.; Georgescu, S.E.; Manea, M.A.; Hermenean, A.O.; Costache, M. Phylogenetic Relationships of Romanian Cattle to Other Cattle Populations Determined by Using Mitochondrial DNA D-Loop Sequence Variation. Rom. Biotechnol. Lett. 2010, 15, 5287–5292. [Google Scholar]
- Ivanković, A.; Paprika, S.; Ramljak, J.; Dovč, P.; Konjačić, M. Mitochondrial DNA-Based Genetic Evaluation of Autochthonous Cattle Breeds in Croatia. Czech J. Anim. Sci. 2014, 59, 519–528. [Google Scholar] [CrossRef]
- Ilie, D.E.; Cean, A.; Cziszter, L.T.; Gavojdian, D.; Ivan, A.; Kusza, S. Microsatellite and Mitochondrial DNA Study of Native Eastern European Cattle Populations: The Case of the Romanian Grey. PLoS ONE 2015, 10, e0138736. [Google Scholar] [CrossRef] [PubMed]
- Pál, H. A maremman és a magyar szarvasmarha közötti hasonlatosság vagy azonosság kérdéséről. Agrár. Szle. 1973, XV, 552–590. [Google Scholar]
- Felius, M.; Beerling, M.-L.; Buchanan, D.S.; Theunissen, B.; Koolmees, P.A.; Lenstra, J.A. On the History of Cattle Genetic Resources. Diversity 2014, 6, 705–750. [Google Scholar] [CrossRef]
- Bradley, D.G.; MacHugh, D.E.; Cunningham, P.; Loftus, R.T. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl. Acad. Sci. USA 1996, 93, 5131–5135. [Google Scholar] [CrossRef]
- Brüggerhoff, K.; Zakhartchenko, V.; Wenigerkind, H.; Reichenbach, H.D.; Prelle, K.; Schernthaner, W.; Alberio, R.; Küchenhoff, H.; Stojkovic, M.; Brem, G.; et al. Bovine somatic cell nuclear transfer using recipient oocytes recovered by ovum pick-up: Effect of maternal lineage of oocyte donors. Biol. Reprod. 2002, 66, 367–373. [Google Scholar] [CrossRef]
- Brüniche-Olsen, A.; Gravlund, P.; Lorenzen, E.D. Impacts of genetic drift and restricted gene flow in indigenous cattle breeds: Evidence from the Jutland breed. Anim. Genet. Resour. 2012, 50, 75–85. [Google Scholar] [CrossRef]
- Ginja, C.; Penedo, M.C.; Sobral, M.F.; Matos, J.; Borges, C.; Neves, D.; Rangel-Figueiredo, T.; Cravador, A. Molecular genetic analysis of a cattle population to reconstitute the extinct Algarvia breed. Genet. Sel. Evol. GSE 2010, 42, 18. [Google Scholar] [CrossRef]
- Hassanin, A.; Ropiquet, A. Resolving a zoological mystery: The kouprey is a real species. Proc. Biol. Sci. 2007, 274, 2849–2855. [Google Scholar] [CrossRef] [PubMed]
- Hiendleder, S.; Lewalski, H.; Janke, A. Complete mitochondrial genomes of Bos taurus and Bos indicus provide new insights into intra-species variation, taxonomy and domestication. Cytogenet. Genome Res. 2008, 120, 150–156. [Google Scholar] [CrossRef]
- Hristov, P.; Teofanova, D.; Neov, B.; Radoslavov, G. Haplotype diversity in autochthonous Balkan cattle breeds. Anim. Genet. 2015, 46, 92–94. [Google Scholar] [CrossRef]
- Kantanen, J.; Edwards, C.J.; Bradley, D.G.; Viinalass, H.; Thessler, S.; Ivanova, Z.; Kiselyova, T.; Cinkulov, M.; Popov, R.; Stojanović, S.; et al. Maternal and paternal genealogy of Eurasian taurine cattle (Bos taurus). Heredity 2009, 103, 404–415. [Google Scholar] [CrossRef]
- Pellecchia, M.; Negrini, R.; Colli, L.; Patrini, M.; Milanesi, E.; Achilli, A.; Bertorelle, G.; Cavalli-Sforza, L.L.; Piazza, A.; Torroni, A.; et al. The mystery of Etruscan origins: Novel clues from Bos taurus mitochondrial DNA. Proc. Biol. Sci. 2007, 274, 1175–1179. [Google Scholar] [CrossRef]
- Schlumbaum, A.; Turgay, M.; Schibler, J. Near East mtDNA haplotype variants in Roman cattle from Augusta Raurica, Switzerland, and in the Swiss Evolène breed. Anim. Genet. 2006, 37, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Steinborn, R.; Zakhartchenko, V.; Jelyazkov, J.; Klein, D.; Wolf, E.; Müller, M.; Brem, G. Composition of parental mitochondrial DNA in cloned bovine embryos. FEBS Lett. 1998, 426, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Steinborn, R.; Schinogl, P.; Zakhartchenko, V.; Achmann, R.; Schernthaner, W.; Stojkovic, M.; Wolf, E.; Müller, M.; Brem, G. Mitochondrial DNA heteroplasmy in cloned cattle produced by fetal and adult cell cloning. Nat. Genet. 2000, 25, 255–257. [Google Scholar] [CrossRef] [PubMed]
- Tamassia, M.; Nuttinck, F.; May-Panloup, P.; Reynier, P.; Heyman, Y.; Charpigny, G.; Stojkovic, M.; Hiendleder, S.; Renard, J.P.; Chastant-Maillard, S. In vitro embryo production efficiency in cattle and its association with oocyte adenosine triphosphate content, quantity of mitochondrial DNA, and mitochondrial DNA haplogroup. Biol. Reprod. 2004, 71, 697–704. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maróti-Agóts, Á.; Wagenhoffer, Z.; Józsa, C.; Kaltenecker, E.; Kemény, B.; Csurgay, K.; Zsigmond, B.; Cardinali, I.; Lancioni, H.; Gáspárdy, A. Phylogenetic Position of Hungarian Grey Cattle Breed Based on Total-Representation Sample. Animals 2025, 15, 1186. https://doi.org/10.3390/ani15091186
Maróti-Agóts Á, Wagenhoffer Z, Józsa C, Kaltenecker E, Kemény B, Csurgay K, Zsigmond B, Cardinali I, Lancioni H, Gáspárdy A. Phylogenetic Position of Hungarian Grey Cattle Breed Based on Total-Representation Sample. Animals. 2025; 15(9):1186. https://doi.org/10.3390/ani15091186
Chicago/Turabian StyleMaróti-Agóts, Ákos, Zsombor Wagenhoffer, Csilla Józsa, Endre Kaltenecker, Balázs Kemény, Kristóf Csurgay, Benedek Zsigmond, Irene Cardinali, Hovirag Lancioni, and András Gáspárdy. 2025. "Phylogenetic Position of Hungarian Grey Cattle Breed Based on Total-Representation Sample" Animals 15, no. 9: 1186. https://doi.org/10.3390/ani15091186
APA StyleMaróti-Agóts, Á., Wagenhoffer, Z., Józsa, C., Kaltenecker, E., Kemény, B., Csurgay, K., Zsigmond, B., Cardinali, I., Lancioni, H., & Gáspárdy, A. (2025). Phylogenetic Position of Hungarian Grey Cattle Breed Based on Total-Representation Sample. Animals, 15(9), 1186. https://doi.org/10.3390/ani15091186






