Evaluating Wastewater Quality Parameters as an Alternative or Complement to Molecular Indicators for Normalization during SARS-CoV-2 Wastewater-Based Epidemiology
Abstract
1. Introduction
2. Materials and Methods
2.1. Wastewater Samples Collection
- Wastewater Treatment Plants
- University Campus Sewer Samples
2.2. Analytical Methods
2.2.1. Molecular Methods
Wastewater Treatment Plants
University Campus Sewer Samples
2.2.2. Physicochemical Characterization Methods
Wastewater Treatment Plants
University Campus Sewer Samples
2.3. Human Case Data
- Wastewater Treatment Plants
- University Campus Sewer Samples
2.4. Statistical Analysis
3. Results
3.1. Wastewater Treatment Plants
3.2. Campus University Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adhikari, S.; Halden, R.U. Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals. Environ. Int. 2022, 163, 107217. [Google Scholar] [CrossRef]
- Daughton, C.G. Wastewater surveillance for population-wide COVID-19: The present and future. Sci. Total Environ. 2020, 736, 139631. [Google Scholar] [CrossRef]
- Maal-Bared, R.; Qiu, Y.; Li, Q.; Gao, T.; Hrudey, S.E.; Bhavanam, S.; Ruecker, N.J.; Ellehoj, E.; Lee, B.E.; Pang, X. Does normalization of SARS-CoV-2 concentrations by Pepper Mild Mottle Virus improve correlations and lead time between wastewater surveillance and clinical data in Alberta (Canada): Comparing twelve SARS-CoV-2 normalization approaches. Sci. Total Environ. 2023, 856, 158964. [Google Scholar] [CrossRef]
- Mazumder, P.; Dash, S.; Honda, R.; Sonne, C.; Kumar, M. Sewage surveillance for SARS-CoV-2: Molecular detection, quantification, and normalization factors. Curr. Opin. Environ. Sci. Health 2022, 28, 100363. [Google Scholar] [CrossRef]
- Arabzadeh, R.; Grünbacher, D.M.; Insam, H.; Kreuzinger, N.; Markt, R.; Rauch, W. Data filtering methods for SARS-CoV-2 wastewater surveillance. Water Sci. Technol. 2021, 84, 1324–1339. [Google Scholar] [CrossRef]
- Wade, M.J.; Jacomo, A.L.; Armenise, E.; Brown, M.R.; Bunce, J.T.; Cameron, G.J.; Fang, Z.; Farkas, K.; Gilpin, D.F.; Graham, D.W.; et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: Lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022, 424 Pt B, 127456. [Google Scholar] [CrossRef]
- Greenwald, H.D.; Kennedy, L.C.; Hinkle, A.; Whitney, O.N.; Fan, V.B.; Crits-Christoph, A.; Harris-Lovett, S.; Flamholz, A.I.; Al-Shayeb, B.; Liao, L.D.; et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res. X 2021, 12, 100111. [Google Scholar] [CrossRef]
- Sim, W.; Park, S.; Ha, J.; Kim, D.; Oh, J.-E. Evaluation of population estimation methods for wastewater-based epidemiology in a metropolitan city. Sci. Total Environ. 2023, 857, 159154. [Google Scholar] [CrossRef]
- Lu, E.; Ai, Y.; Davis, A.; Straathof, J.; Halloran, K.; Hull, N.; Winston, R.; Weir, M.H.; Soller, J.; Bohrerova, Z.; et al. Wastewater surveillance of SARS-CoV-2 in dormitories as a part of comprehensive university campus COVID-19 monitoring. Environ. Res. 2022, 212, 113580. [Google Scholar] [CrossRef]
- Ai, Y.; Davis, A.; Jones, D.; Lemeshow, S.; Tu, H.; He, F.; Ru, P.; Pan, X.; Bohrerova, Z.; Lee, J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021, 801, 149757. [Google Scholar] [CrossRef]
- Ma, D.; Straathof, J.; Liu, Y.; Hull, N.M. Monitoring SARS-CoV-2 RNA in Wastewater with RT-qPCR and Chip-Based RT-dPCR: Sewershed-Level Trends and Relationships to COVID-19. ACS EST Water 2022, 2, 2084–2093. [Google Scholar] [CrossRef]
- Castiglioni, S.; Bijlsma, L.; Covaci, A.; Emke, E.; Hernández, F.; Reid, M.; Ort, C.; Thomas, K.V.; van Nuijs, A.L.N.; de Voogt, P.; et al. Evaluation of Uncertainties Associated with the Determination of Community Drug Use through the Measurement of Sewage Drug Biomarkers. Environ. Sci. Technol. 2013, 47, 1452–1460. [Google Scholar] [CrossRef]
- Baz-Lomba, J.A.; Di Ruscio, F.; Amador, A.; Reid, M.; Thomas, K.V. Assessing Alternative Population Size Proxies in a Wastewater Catchment Area Using Mobile Device Data. Environ. Sci. Technol. 2019, 53, 1994–2001. [Google Scholar] [CrossRef]
- Feng, S.; Roguet, A.; McClary-Gutierrez, J.S.; Newton, R.J.; Kloczko, N.; Meiman, J.G.; McLellan, S.L. Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities. ACS EST Water 2021, 1, 1955–1965. [Google Scholar] [CrossRef]
- Sakarovitch, C.; Schlosser, O.; Courtois, S.; Proust-Lima, C.; Couallier, J.; Pétrau, A.; Litrico, X.; Loret, J.-F. Monitoring of SARS-CoV-2 in wastewater: What normalisation for improved understanding of epidemic trends? J. Water Health 2022, 20, 712–726. [Google Scholar] [CrossRef]
- Zhan, Q.; Babler, K.M.; Sharkey, M.E.; Amirali, A.; Beaver, C.C.; Boone, M.M.; Comerford, S.; Cooper, D.; Cortizas, E.M.; Currall, B.B.; et al. Relationships between SARS-CoV-2 in Wastewater and COVID-19 Clinical Cases and Hospitalizations, with and without Normalization against Indicators of Human Waste. ACS EST Water 2022, 2, 1992–2003. [Google Scholar] [CrossRef]
- Jarret, R.L.; Gillaspie, A.G.; Barkley, N.A.; Pinnow, D.L. The Occurrence and Control of Pepper Mild Mottle Virus (PMMoV) in the USDA/ARS Capsicum Germplasm Collection. Seed Technol. 2008, 30, 26–36. [Google Scholar]
- Peng, J.; Shi, B.; Zheng, H.; Lu, Y.; Lin, L.; Jiang, T.; Chen, J.; Yan, F. Detection of pepper mild mottle virus in pepper sauce in China. Arch. Virol. 2015, 160, 2079–2082. [Google Scholar] [CrossRef]
- Zhou, W.P.; Li, Y.Y.; Li, F.; Tan, G.L. First report of natural infection of tomato by pepper mild mottle virus in China. J. Plant Pathol. 2021, 103, 363. [Google Scholar] [CrossRef]
- D’Aoust, P.M.; Graber, T.E.; Mercier, E.; Montpetit, D.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021, 770, 145319. [Google Scholar] [CrossRef]
- Nagarkar, M.; Keely, S.; Jahne, M.; Wheaton, E.; Hart, C.; Smith, B.; Garland, J.; Varughese, E.; Braam, A.; Wiechman, B.; et al. SARS-CoV-2 monitoring at three sewersheds of different scales and complexity demonstrates distinctive relationships between wastewater measurements and COVID-19 case data. Sci. Total Environ. 2022, 816, 151534. [Google Scholar] [CrossRef]
- Greaves, J.; Stone, D.; Wu, Z.; Bibby, K. Persistence of emerging viral fecal indicators in large-scale freshwater mesocosms. Water Res. X 2020, 9, 100067. [Google Scholar] [CrossRef]
- LaTurner, Z.W.; Zong, D.M.; Kalvapalle, P.; Gamas, K.R.; Terwilliger, A.; Crosby, T.; Ali, P.; Avadhanula, V.; Santos, H.H.; Weesner, K.; et al. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology-PMC. Water Res. 2021, 197, 117043. [Google Scholar] [CrossRef]
- Glassmeyer, S.T.; Furlong, E.T.; Kolpin, D.W.; Cahill, J.D.; Zaugg, S.D.; Werner, S.L.; Meyer, M.T.; Kryak, D.D. Transport of Chemical and Microbial Compounds from Known Wastewater Discharges: Potential for Use as Indicators of Human Fecal Contamination. Environ. Sci. Technol. 2005, 39, 5157–5169. [Google Scholar] [CrossRef]
- Sankararamakrishnan, N.; Guo, Q. Chemical tracers as indicator of human fecal coliforms at storm water outfalls. Environ. Int. 2005, 31, 1133–1140. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bibby, K.; Haramoto, E.; Hewitt, J.; Huygens, F.; Gyawali, P.; Korajkic, A.; Riddell, S.; Sherchan, S.P.; et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. [Google Scholar] [CrossRef]
- Yang, S.; Dong, Q.; Li, S.; Cheng, Z.; Kang, X.; Ren, D.; Xu, C.; Zhou, X.; Liang, P.; Sun, L.; et al. Persistence of SARS-CoV-2 RNA in wastewater after the end of the COVID-19 epidemics. J. Hazard. Mater. 2022, 429, 128358. [Google Scholar] [CrossRef]
- Achak, M.; Bakri, S.A.; Chhiti, Y.; Alaoui, F.E.M.; Barka, N.; Boumya, W. SARS-CoV-2 in hospital wastewater during outbreak of COVID-19: A review on detection, survival and disinfection technologies. Sci. Total Environ. 2021, 761, 143192. [Google Scholar] [CrossRef]
- Hamouda, M.; Mustafa, F.; Maraqa, M.; Rizvi, T.; Hassan, A.A. Wastewater surveillance for SARS-CoV-2: Lessons learnt from recent studies to define future applications. Sci. Total Environ. 2021, 759, 143493. [Google Scholar] [CrossRef]
- Casanova, L.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Survival of surrogate coronaviruses in water. Water Res. 2009, 43, 1893–1898. [Google Scholar] [CrossRef]
- Amoah, I.D.; Abunama, T.; Awolusi, O.O.; Pillay, L.; Pillay, K.; Kumari, S.; Bux, F. Effect of selected wastewater characteristics on estimation of SARS-CoV-2 viral load in wastewater. Environ. Res. 2022, 203, 111877. [Google Scholar] [CrossRef]
- Sapula, S.A.; Whittall, J.J.; Pandopulos, A.J.; Gerber, C.; Venter, H. An optimized and robust PEG precipitation method for detection of SARS-CoV-2 in wastewater. Sci. Total Environ. 2021, 785, 147270. [Google Scholar] [CrossRef]
- Schussman, M.K.; McLellan, S.L. Effect of Time and Temperature on SARS-CoV-2 in Municipal Wastewater Conveyance Systems. Water 2022, 14, 1373. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Gao, S.; Zhou, X.; Sivakumar, M.; Jiang, G. Effects of Temperature and Water Types on the Decay of Coronavirus: A Review. Water 2023, 15, 1051. [Google Scholar] [CrossRef]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- de Oliveira, L.C.; Torres-Franco, A.F.; Lopes, B.C.; Santos, B.S.d.S.; Costa, E.A.; Costa, M.S.; Reis, M.T.P.; Melo, M.C.; Polizzi, R.B.; Teixeira, M.M.; et al. Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Res. 2021, 195, 117002. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y.; et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021, 775, 145790. [Google Scholar] [CrossRef]
- Peacock, M.; Evans, C.D.; Fenner, N.; Freeman, C.; Gough, R.; Jones, T.G.; Lebron, I. UV-visible absorbance spectroscopy as a proxy for peatland dissolved organic carbon (DOC) quantity and quality: Considerations on wavelength and absorbance degradation. Environ. Sci. Process. Impacts 2014, 16, 1445–1461. [Google Scholar] [CrossRef]
- Altmann, J.; Massa, L.; Sperlich, A.; Gnirss, R.; Jekel, M. UV254 absorbance as real-time monitoring and control parameter for micropollutant removal in advanced wastewater treatment with powdered activated carbon. Water Res. 2016, 94, 240–245. [Google Scholar] [CrossRef]
- Mesquita, D.P.; Quintelas, C.; Amaral, A.L.; Ferreira, E.C. Monitoring biological wastewater treatment processes: Recent advances in spectroscopy applications. Rev. Environ. Sci. Biotechnol. 2017, 16, 395–424. [Google Scholar] [CrossRef]
- Waltham, B.; Örmeci, B. Fluorescence intensity, conductivity, and UV–vis absorbance as surrogate parameters for real-time monitoring of anaerobic digestion of wastewater sludge. J. Water Proc. Eng. 2020, 37, 101395. [Google Scholar] [CrossRef]
- Ulliman, S.L.; Korak, J.A.; Linden, K.G.; Rosario-Ortiz, F.L. Methodology for selection of optical parameters as wastewater effluent organic matter surrogates. Water Res. 2020, 170, 115321. [Google Scholar] [CrossRef]
- Bitter, L.C.; Kibbee, R.; Jiménez, G.C.; Örmeci, B. Wastewater Surveillance of SARS-CoV-2 at a Canadian University Campus and the Impact of Wastewater Characteristics on Viral RNA Detection. ACS EST Water 2022, 2, 2034–2046. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.C.; Eyre, B.D. A low cost, easy to build, portable, and universal autosampler for liquids. Methods Oceanogr. 2013, 8, 23–32. [Google Scholar] [CrossRef]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef] [PubMed]
- Duvallet, C.; Wu, F.; McElroy, K.A.; Imakaev, M.; Endo, N.; Xiao, A.; Zhang, J.; Floyd-O’sullivan, R.; Powell, M.M.; Mendola, S.; et al. Nationwide Trends in COVID-19 Cases and SARS-CoV-2 RNA Wastewater Concentrations in the United States. ACS EST Water 2022, 2, 1899–1909. [Google Scholar] [CrossRef] [PubMed]
- Bohrerova, Z.; Brinkman, N.E.; Chakravarti, R.; Chattopadhyay, S.; Faith, S.A.; Garland, J.; Herrin, J.M.; Hull, N.; Jahne, M.; Kang, D.-W.; et al. Ohio Coronavirus Wastewater Monitoring Network: Implementation of Statewide Monitoring for Protecting Public Health. J. Public Health Manag. Pract. 2023, 29, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Ohio Department of Health. Ohio Wastewater Monitoring Network Sample Results. Available online: https://data.ohio.gov/wps/portal/gov/data/view/ohio-wastewater-monitoring-network-sample-results (accessed on 17 February 2024).
- Weishaar, J.L.; Aiken, G.R.; Bergamaschi, B.A.; Fram, M.S.; Fujii, R.; Mopper, K. Evaluation of specific ultraviolet absorbance as an indicator of the chemical composition and reactivity of dissolved organic carbon. Environ. Sci. Technol. 2003, 37, 4702–4708. [Google Scholar] [CrossRef] [PubMed]
- Duirk, S.E.; Valentine, R.L. Modeling dichloroacetic acid formation from the reaction of monochloramine with natural organic matter. Water Res. 2006, 40, 2667–2674. [Google Scholar] [CrossRef]
- Worrall, F.; Armstrong, A.; Holden, J. Short-term impact of peat drain-blocking on water colour, dissolved organic carbon concentration, and water table depth. J. Hydrol. 2007, 337, 315–325. [Google Scholar] [CrossRef]
- Peuravuori, J.; Pihlaja, K. Molecular size distribution and spectroscopic properties of aquatic humic substances. Anal. Chim. Acta 1997, 337, 133–149. [Google Scholar] [CrossRef]
- Graham, M.C.; Gavin, K.G.; Kirika, A.; Farmer, J.G. Processes controlling manganese distributions and associations in organic-rich freshwater aquatic systems: The example of Loch Bradan, Scotland. Sci. Total Environ. 2012, 424, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Selberg, A.; Viik, M.; Ehapalu, K.; Tenno, T. Content and composition of natural organic matter in water of Lake Pitkjärv and mire feeding Kuke River (Estonia). J. Hydrol. 2011, 400, 274–280. [Google Scholar] [CrossRef]
- Park, S.; Joe, K.S.; Han, S.H.; Kim, H.S. Characteristics of Dissolved Organic Carbon in the Leachate from Moonam Sanitary Landfill. Environ. Technol. 1999, 20, 419–424. [Google Scholar] [CrossRef]
- Moore, T.R. A Preliminary Study of the Effects of Drainage and Harvesting on Water Quality in Ombrotrophic Bogs Near Sept-Iles, Quebec. J. Am. Water Resour. Assoc. 1987, 23, 785–791. [Google Scholar] [CrossRef]
- Wilson, L.; Wilson, J.; Holden, J.; Johnstone, I.; Armstrong, A.; Morris, M. Ditch blocking, water chemistry and organic carbon flux: Evidence that blanket bog restoration reduces erosion and fluvial carbon loss. Sci. Total Environ. 2011, 409, 2010–2018. [Google Scholar] [CrossRef] [PubMed]
- Thurman, E.M. Organic Geochemistry of Natural Waters; Martinus Nijhoff/Dr W. Junk Publishers: Dordrecht, The Netherlands, 1985. [Google Scholar]
- Wallage, Z.E.; Holden, J.; McDonald, A.T. Drain blocking: An effective treatment for reducing dissolved organic carbon loss and water discolouration in a drained peatland. Sci. Total Environ. 2006, 367, 811–821. [Google Scholar] [CrossRef] [PubMed]
- The Ohio State University. COVID-19 Dashboard (Archived). Safe and Healthy Buckeyes. Available online: https://safeandhealthy.osu.edu/dashboard (accessed on 18 February 2024).
- von Sperling, M.; Verbyla, M.E.; Oliveira, S.M.A.C. Assessment of Treatment Plant Performance and Water Quality Data: A Guide for Students, Researchers and Practitioners; IWA Publishing: London, UK, 2020. [Google Scholar]
- McClary-Gutierrez, J.S.; Mattioli, M.C.; Marcenac, P.; Silverman, A.I.; Boehm, A.B.; Bibby, K.; Balliet, M.; Reyes, F.L.d.L.; Gerrity, D.; Griffith, J.F.; et al. SARS-CoV-2 Wastewater Surveillance for Public Health Action. Emerg. Infect. Dis. 2021, 27, e210753. [Google Scholar] [CrossRef] [PubMed]
- Been, F.; Rossi, L.; Ort, C.; Rudaz, S.; Delémont, O.; Esseiva, P. Population normalization with ammonium in wastewater-based epidemiology: Application to illicit drug monitoring. Environ. Sci. Technol. 2014, 48, 8162–8169. [Google Scholar] [CrossRef]
- Sims, N.; Kasprzyk-Hordern, B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef]
- Schill, R.; Nelson, K.L.; Harris-Lovett, S.; Kantor, R.S. The dynamic relationship between COVID-19 cases and SARS-CoV-2 wastewater concentrations across time and space: Considerations for model training data sets. Sci. Total Environ. 2023, 871, 162069. [Google Scholar] [CrossRef] [PubMed]
- Spangenberg, M.; Bryant, J.I.; Gibson, S.J.; Mousley, P.J.; Ramachers, Y.; Bell, G.R. Ultraviolet absorption of contaminants in water. Sci. Rep. 2021, 11, 3682. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, W.; Du, P.; Liu, W.; Liu, X.; Yang, C.; Jia, R.; Wang, Y.; Chen, Y.; Jia, L.; et al. Comparison of reverse-transcription qPCR and droplet digital PCR for the detection of SARS-CoV-2 in clinical specimens of hospitalized patients. Diagn. Microbiol. Infect. Dis. 2022, 103, 115677. [Google Scholar] [CrossRef] [PubMed]
- Morón-López, S.; Riveira-Muñoz, E.; Urrea, V.; Gutiérrez-Chamorro, L.; Ávila-Nieto, C.; Noguera-Julian, M.; Carrillo, J.; Mitjà, O.; Mateu, L.; Massanella, M.; et al. Comparison of Reverse Transcription (RT)-Quantitative PCR and RT-Droplet Digital PCR for Detection of Genomic and Subgenomic SARS-CoV-2 RNA. Microbiol. Spectr. 2023, 11, e0415922. [Google Scholar] [CrossRef]
- Olesen, S.W.; Imakaev, M.; Duvallet, C. Making waves: Defining the lead time of wastewater-based epidemiology for COVID-19. Water Res. 2021, 202, 117433. [Google Scholar] [CrossRef]
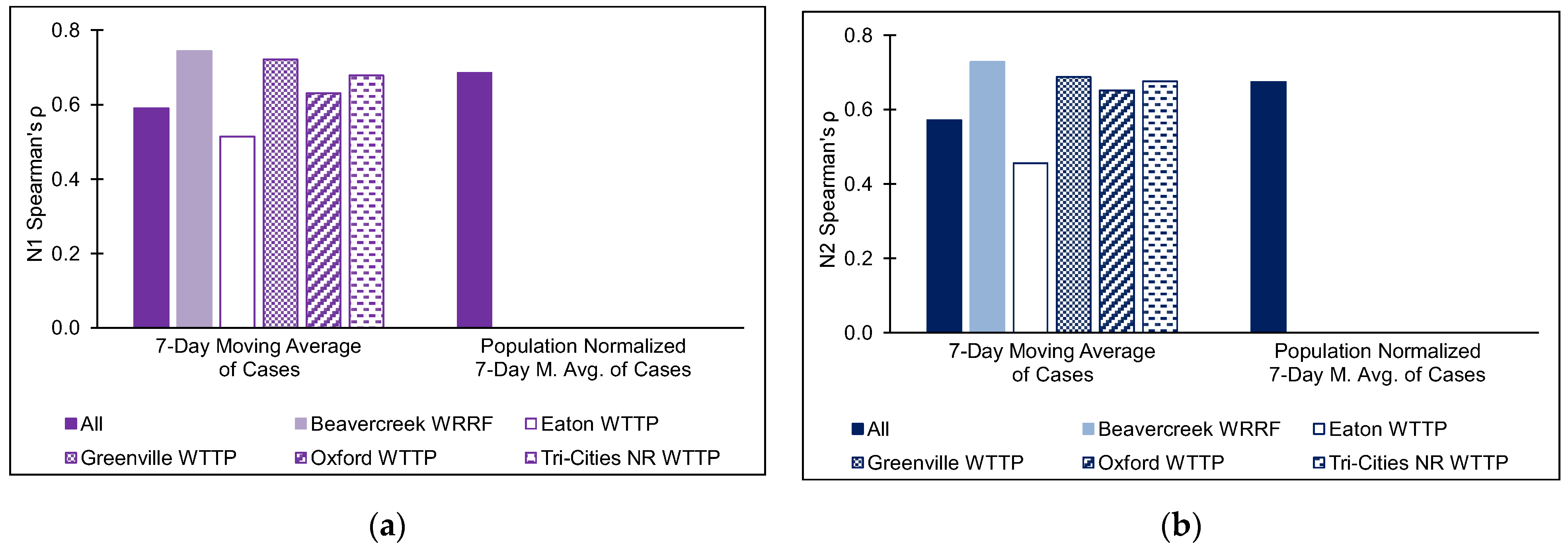

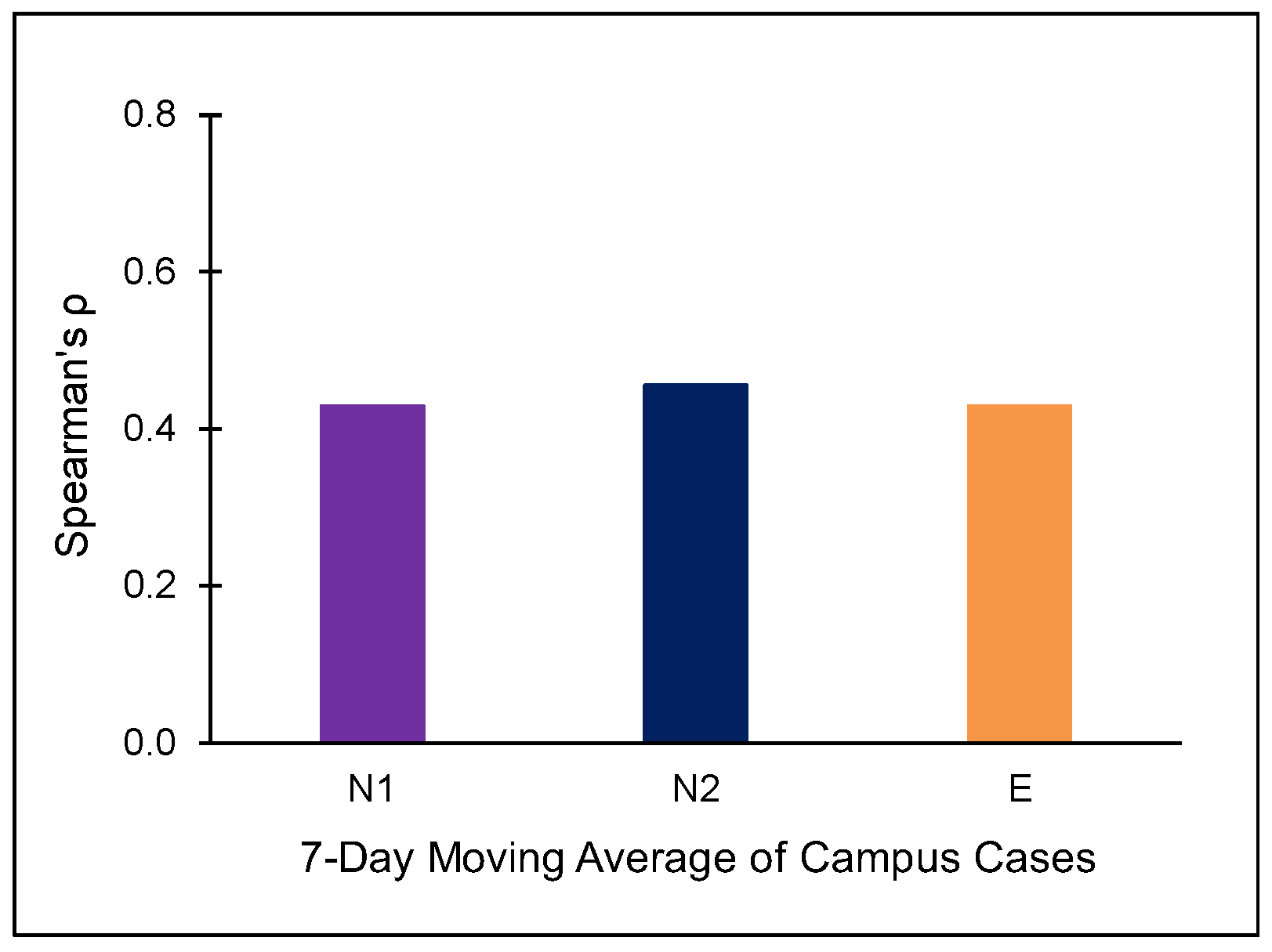
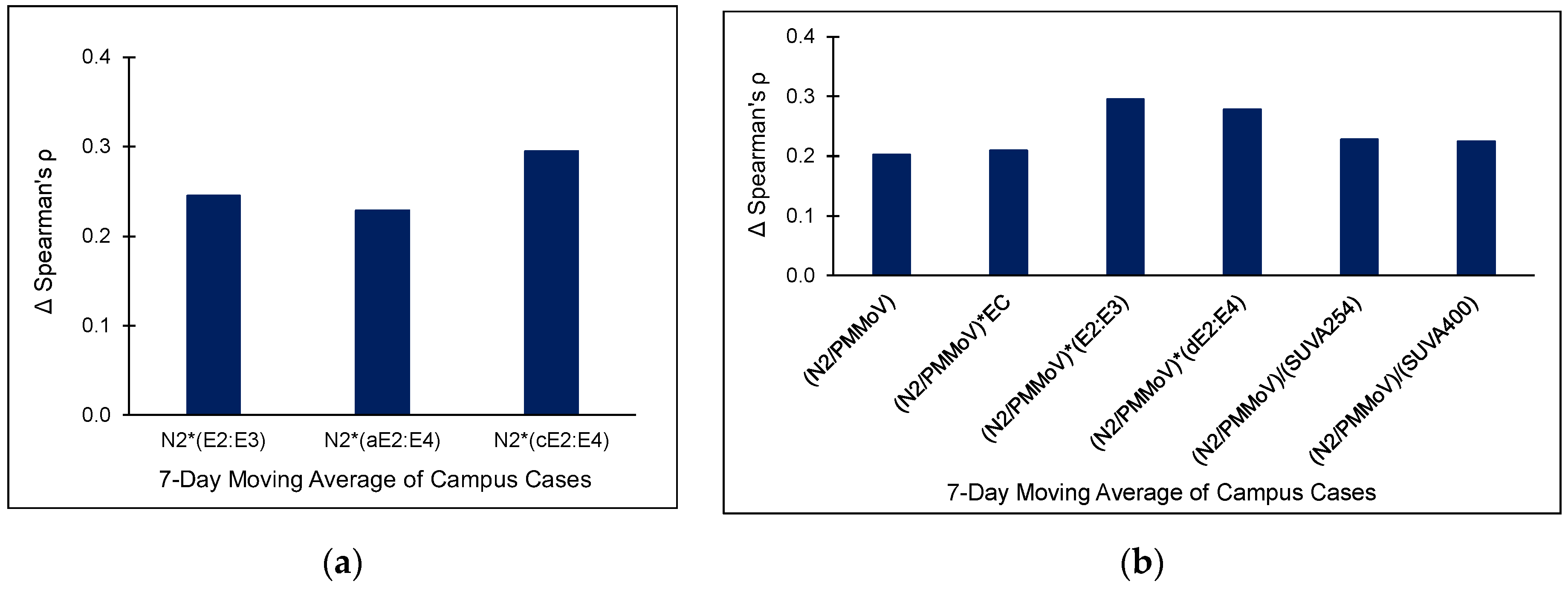
| WWTP | # of Samples (n) | Flow Rate (m3/day) | pH | Temperature (°C) | TSS (mg/L) | Population in Sewershed |
|---|---|---|---|---|---|---|
| Beavercreek WRRF | 140 | 34,990 ± 9584 | 6.32 ± 0.36 | 15.5 ± 2.75 | 152 ± 95.2 | 47,000 |
| Eaton WTTP | 48 | 5425 ± 2891 | 7.58 ± 0.54 | 17.5 ± 3.23 | 10,000 | |
| Greenville WTTP | 64 | 10,148 ± 3350 | 7.63 ± 0.12 | 14.7 ± 2.59 | 87.6 ± 35.6 | 14,000 |
| Oxford WTTP | 140 | 6507 ± 3278 | 7.49 ± 0.13 | 16.3 ± 3.55 | 276 ± 122 | 21,300 |
| Tri-Cities NR WTTP | 140 | 40,496 ± 14,935 | 7.47 ± 0.13 | 17.0 ± 3.30 | 180 ± 174 | 65,000 |
| Campus Site | # of Samples (n) | Turbidity (NTU) | pH | Temperature (°C) | TSS (mg/L) | Population in Dorms Served |
|---|---|---|---|---|---|---|
| 1 | 12 | 189 ± 210 | 8.47 ± 0.42 | 23.3 ± 2.76 | 493 ± 486 | 178 |
| 2 | 14 | 231 ± 299 | 8.40 ± 0.36 | 22.7 ± 2.98 | 401 ± 353 | 1685 |
| 3 | 12 | 192 ± 270 | 7.91 ± 0.57 | 22.6 ± 2.22 | 497 ± 719 | 3450 |
| 4 | 14 | 169 ± 114 | 8.48 ± 0.23 | 22.2 ± 2.98 | 511 ± 335 | 872 |
| 5 | 13 | 144 ± 47.2 | 6.63 ± 0.42 | 24.9 ± 2.89 | 218 ± 103 | 228 |
| 6 | 14 | 115 ± 27.9 | 8.44 ± 0.19 | 21.9 ± 2.63 | 313 ± 106 | 1354 |
| Site | DOC (mg/L) | SUVA 254 | SUVA 280 | SUVA 400 | E2:E3 | a E2:E4 | b E2:E4 | c E2:E4 | a E4:E6 | b E4:E6 | c E4:E6 | d E4:E6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 93.6 ± 44.8 | 0.02 ± 8 × 10−3 | 0.02 ± 6 × 10−3 | 7 × 10−3 ± 3 × 10−3 | 3.46 ± 0.51 | 5.95 ± 1.37 | 4.17 ± 1.15 | 6.55 ± 1.41 | 3.72 ± 0.34 | 4.39 ± 0.51 | 4.70 ± 0.55 | 4.86 ± 0.74 |
| 2 | 135 ± 64.8 | 0.01 ± 7 × 10−3 | 0.01 ± 5 × 10−3 | 4 × 10−3 ± 1 × 10−3 | 3.61 ± 1.38 | 6.52 ± 2.83 | 5.05 ± 2.46 | 7.37 ± 3.33 | 3.94 ± 0.73 | 4.72 ± 1.59 | 4.74 ± 1.07 | 5.13 ± 1.56 |
| 3 | 81.1 ± 41.4 | 0.02 ± 6 × 10−3 | 0.01 ± 5 × 10−3 | 6 × 10−3 ± 2 × 10−3 | 3.33 ± 0.46 | 5.50 ± 0.80 | 4.18 ± 1.22 | 5.96 ± 0.86 | 3.84 ± 0.49 | 4.65 ± 0.78 | 4.79 ± 0.63 | 4.94 ± 1.03 |
| 4 | 102 ± 28.7 | 0.01 ± 4 × 10−3 | 0.01 ± 4 × 10−3 | 5 × 10−3 ± 2 × 10−3 | 3.37 ± 0.45 | 5.64 ± 0.78 | 4.32 ± 0.95 | 6.09 ± 0.80 | 3.88 ± 0.20 | 4.82 ± 0.66 | 5.03 ± 0.73 | 5.07 ± 0.99 |
| 5 | 213 ± 90.1 | 0.00 ± 2 × 10−3 | 9 × 10−3 ± 2 × 10−3 | 2 × 10−3 ± 9 × 10−4 | 3.74 ± 0.60 | 7.13 ± 2.41 | 5.27 ± 2.14 | 8.85 ± 5.58 | 4.18 ± 0.43 | 4.67 ± 1.10 | 4.67 ± 1.09 | 4.82 ± 0.97 |
| 6 | 91.5 ± 26.9 | 0.01 ± 6 × 10−3 | 0.01 ± 4 × 10−3 | 5 × 10−3 ± 1 × 10−3 | 3.64 ± 0.50 | 6.13 ± 0.78 | 4.69 ± 1.14 | 6.76 ± 0.92 | 3.88 ± 0.57 | 4.53 ± 0.91 | 4.61 ± 0.77 | 4.99 ± 1.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Straathof, J.; Hull, N.M. Evaluating Wastewater Quality Parameters as an Alternative or Complement to Molecular Indicators for Normalization during SARS-CoV-2 Wastewater-Based Epidemiology. Environments 2024, 11, 80. https://doi.org/10.3390/environments11040080
Straathof J, Hull NM. Evaluating Wastewater Quality Parameters as an Alternative or Complement to Molecular Indicators for Normalization during SARS-CoV-2 Wastewater-Based Epidemiology. Environments. 2024; 11(4):80. https://doi.org/10.3390/environments11040080
Chicago/Turabian StyleStraathof, Judith, and Natalie M. Hull. 2024. "Evaluating Wastewater Quality Parameters as an Alternative or Complement to Molecular Indicators for Normalization during SARS-CoV-2 Wastewater-Based Epidemiology" Environments 11, no. 4: 80. https://doi.org/10.3390/environments11040080
APA StyleStraathof, J., & Hull, N. M. (2024). Evaluating Wastewater Quality Parameters as an Alternative or Complement to Molecular Indicators for Normalization during SARS-CoV-2 Wastewater-Based Epidemiology. Environments, 11(4), 80. https://doi.org/10.3390/environments11040080







