RRM Prediction of Erythrocyte Band3 Protein as Alternative Receptor for SARS-CoV-2 Virus
Abstract
:Featured Application
Abstract
1. Introduction
2. Methods and Materials
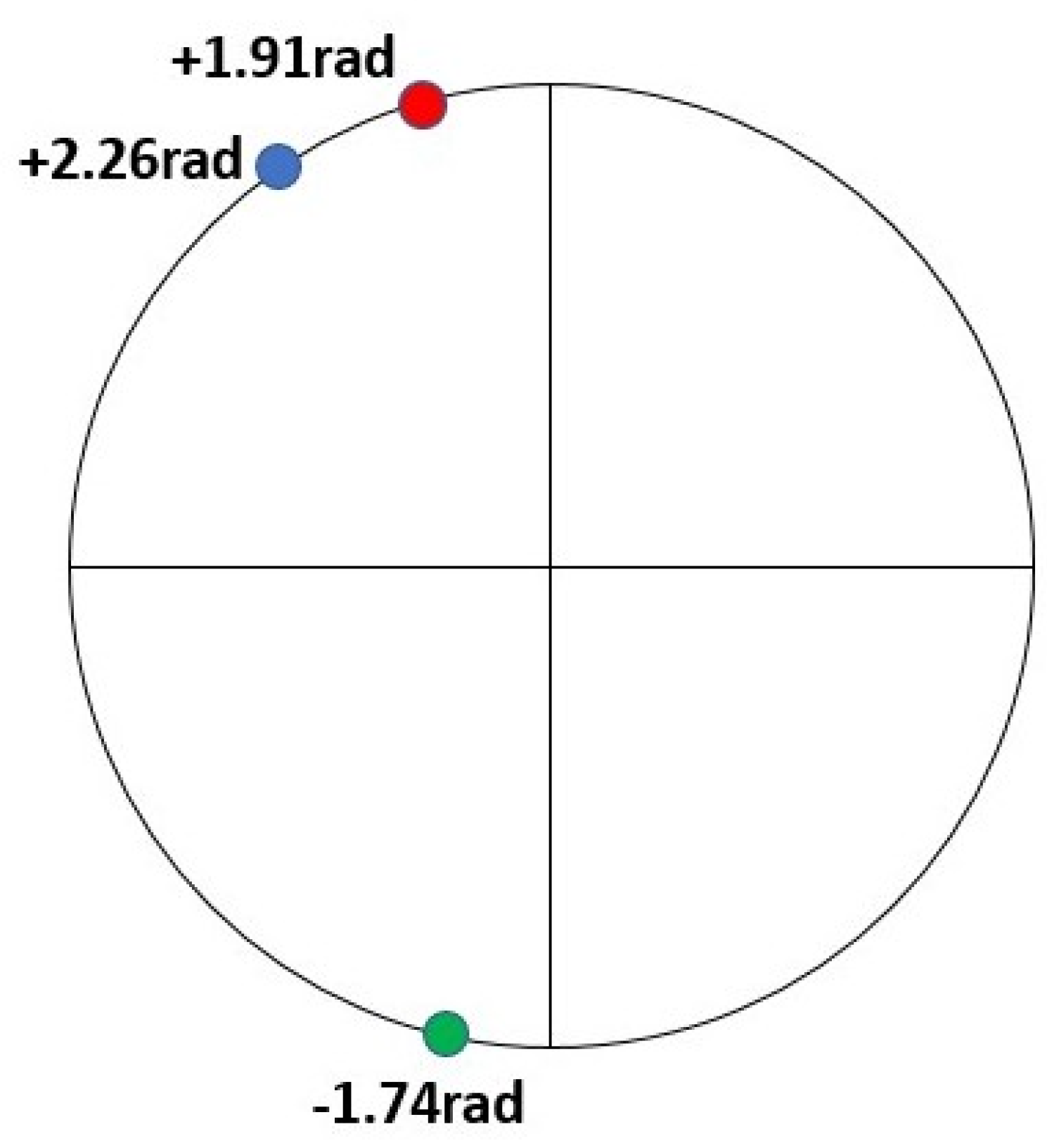
2.1. Methods-Resonant Recognition Model (RRM)
2.2. Materials-Protein Sequences Analysed by RRM
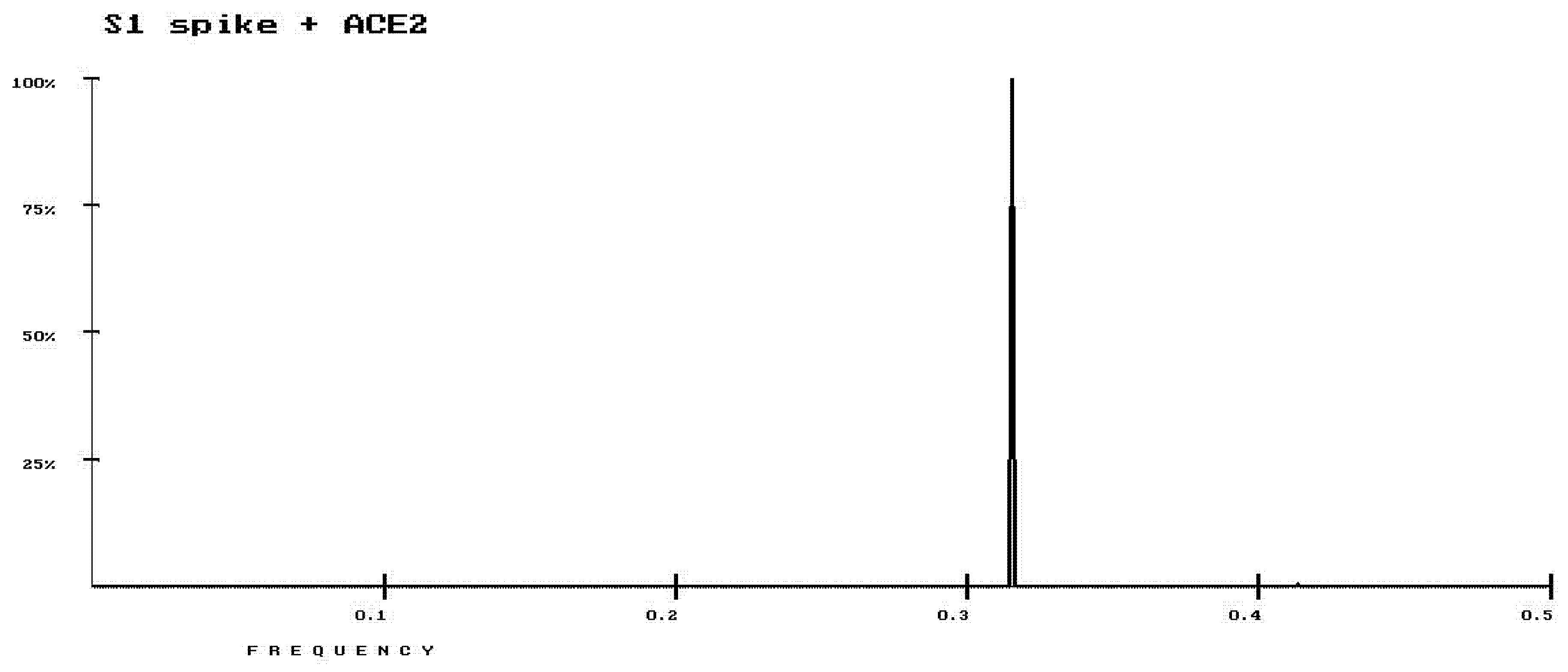
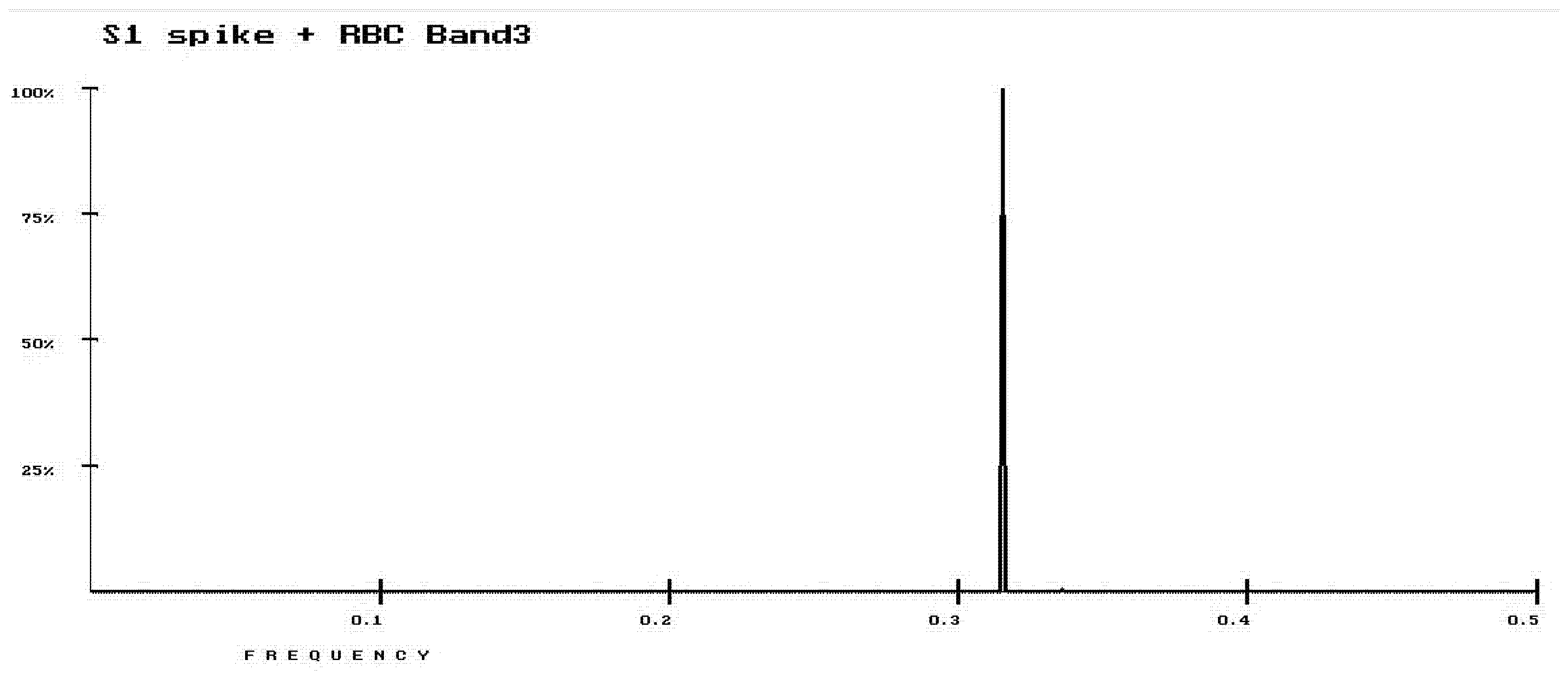
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Makin, S. How Coronaviruses Cause Infection—from Colds to Deadly Pneumonia. Available online: https://www.scientificamerican.com/article/how-coronaviruses-cause-infection-from-colds-to-deadly-pneumonia1 (accessed on 5 February 2020).
- Couzin-Frankel, J. Why Don’t Some Coronavirus Patients Sense Their Alarmingly Low Oxygen Levels? Available online: https://www.sciencemag.org/news/2020/04/why-don-t-some-coronavirus-patients-sense-their-alarmingly-low-oxygen-levels (accessed on 28 April 2020).
- Cosic, I. Macromolecular Bioactivity: Is it Resonant Interaction between Macromolecules? -Theory and Applications. IEEE Trans Biomed. Eng. 1994, 41, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I. Virtual spectroscopy for fun and profit. Biotechnology 1995, 13, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I. The Resonant Recognition Model of Macromolecular Bioactivity: Theory and Applications; Birkhäuser Verlag: Basel, Switzerland, 2012. [Google Scholar]
- Cosic, I.; Cosic, D.; Lazar, K. Analysis of Tumor Necrosis Factor Function Using the Resonant Recognition Model. Cell Biochem. Biophys. 2016, 74, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Paspaliaris, V.; Cosic, D. Analysis of Protein-Receptor on an Example of Leptin-Leptin Receptor Interaction Using the Resonant Recognition Model. Appl. Sci. 2019, 9, 5169. [Google Scholar] [CrossRef] [Green Version]
- Cosic, I.; Hearn, M.T.W. Studies on Protein-DNA Interactions Using the Resonant Recognition Model: Application to Repressors and Transforming Proteins. Eur. J. Biochem. 1992, 205, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Hodder, A.N.; Aguilar, M.I.; Hearn, M.T.W. Resonant Recognition Model and protein topography: Model studies with myoglobin, haemoglobin and lysozyme. Eur. J. Biochem. 1991, 198, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Caceres, J.L.H.; Cosic, I.; Cosic, D. Application of the Resonant Recognition Model to the Study of Plasmodium Proteins Involved in Malaria Infection. MD Med. Data 2015, 7, 7–14. [Google Scholar]
- Pirogova, E.; Istivan, T.; Gan, E.; Cosic, I. Advances in methods for therapeutic peptide discovery, design and development. Curr. Pharm. Biotechnol. 2011, 12, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Pirogova, E. Bioactive Peptide Design using the Resonant Recognition Model. Nonlinear Biomed. Phys. 2007, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Drummond, A.E.; Underwood, J.R.; Hearn, M.T.W. In vitro inhibition of the actions of basic FGF by novel 16 amino acid peptides. Mol. Cell. Biochem. 1994, 130, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krsmanovic, V.; Biquard, J.M.; Sikorska-Walker, M.; Cosic, I.; Desgranges, C.; Trabaud, M.A.; Whitfield, J.F.; Durkin, J.P.; Achour, A.; Hearn, M.T. Investigation Into the Cross-reactivity of Rabbit Antibodies Raised against Nonhomologous Pairs of Synthetic Peptides Derived from HIV-1 gp120 proteins. J. Peptide. Res. 1998, 52, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Krsmanovic, V.; Cosic, I.; Biquard, J.M.; Hearn, M.T.W. Peptides immunologically related to known viral protein. U.S. Patent No. 6,294,174, 25 September 2001. [Google Scholar]
- Achour, A.; Biquard, J.M.; Krsmanovic, V.; M’Bika, J.P.; Ficheux, D.; Sikorska, M.; Cozzone, A.J. Induction of Human Immunodeficiency Virus (HIV-1) Envelope Specific Cell-Mediated Immunity by a Non-Homologus Synthetic Peptide. PLoS ONE 2007, 2, e1214. [Google Scholar] [CrossRef] [PubMed]
- Almansour, N.; Pirogova, E.; Coloe, P.; Cosic, I.; Istivan, T. Investigation of cytotoxicity of negative control peptides versus bioactive peptides on skin cancer and normal cells: A comparative study. Future Med. Chem. 2012, 4, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Istivan, T.; Pirogova, E.; Gan, E.; Almansour, N.; Coloe, P.; Cosic, I. Biological effects of a De Novo designed myxoma virus peptide analogue: Evaluation of cytotoxicity on tumor cells. PLoS ONE 2011, 6, e24809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013, 100, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millet, J.K.; Whittaker, G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 2018, 517, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, H.; Oh, S.S.; Chishti, A.H. A Presenilin-like protease associated with Plasmodium falciparum micronemes is involved in erythrocyte invasion. Mol. Biochem. Parasitol. 2008, 158, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallagher, P.G. Hemolytic anemias: Red blood cell membrane and metabolic defects. Available online: https://www.semanticscholar.org/paper/HEMOLYTIC-ANEMIAS-%3A-RED-BLOOD-CELL-MEMBRANE-AND-Gallagher/0f01efe030314c2fadd42d007f0683721f854156 (accessed on 5 February 2020).
- Perkins, M.E. Binding of glycophorins to Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 1984, 10, 67–78. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cosic, I.; Cosic, D.; Loncarevic, I. RRM Prediction of Erythrocyte Band3 Protein as Alternative Receptor for SARS-CoV-2 Virus. Appl. Sci. 2020, 10, 4053. https://doi.org/10.3390/app10114053
Cosic I, Cosic D, Loncarevic I. RRM Prediction of Erythrocyte Band3 Protein as Alternative Receptor for SARS-CoV-2 Virus. Applied Sciences. 2020; 10(11):4053. https://doi.org/10.3390/app10114053
Chicago/Turabian StyleCosic, Irena, Drasko Cosic, and Ivan Loncarevic. 2020. "RRM Prediction of Erythrocyte Band3 Protein as Alternative Receptor for SARS-CoV-2 Virus" Applied Sciences 10, no. 11: 4053. https://doi.org/10.3390/app10114053
APA StyleCosic, I., Cosic, D., & Loncarevic, I. (2020). RRM Prediction of Erythrocyte Band3 Protein as Alternative Receptor for SARS-CoV-2 Virus. Applied Sciences, 10(11), 4053. https://doi.org/10.3390/app10114053




