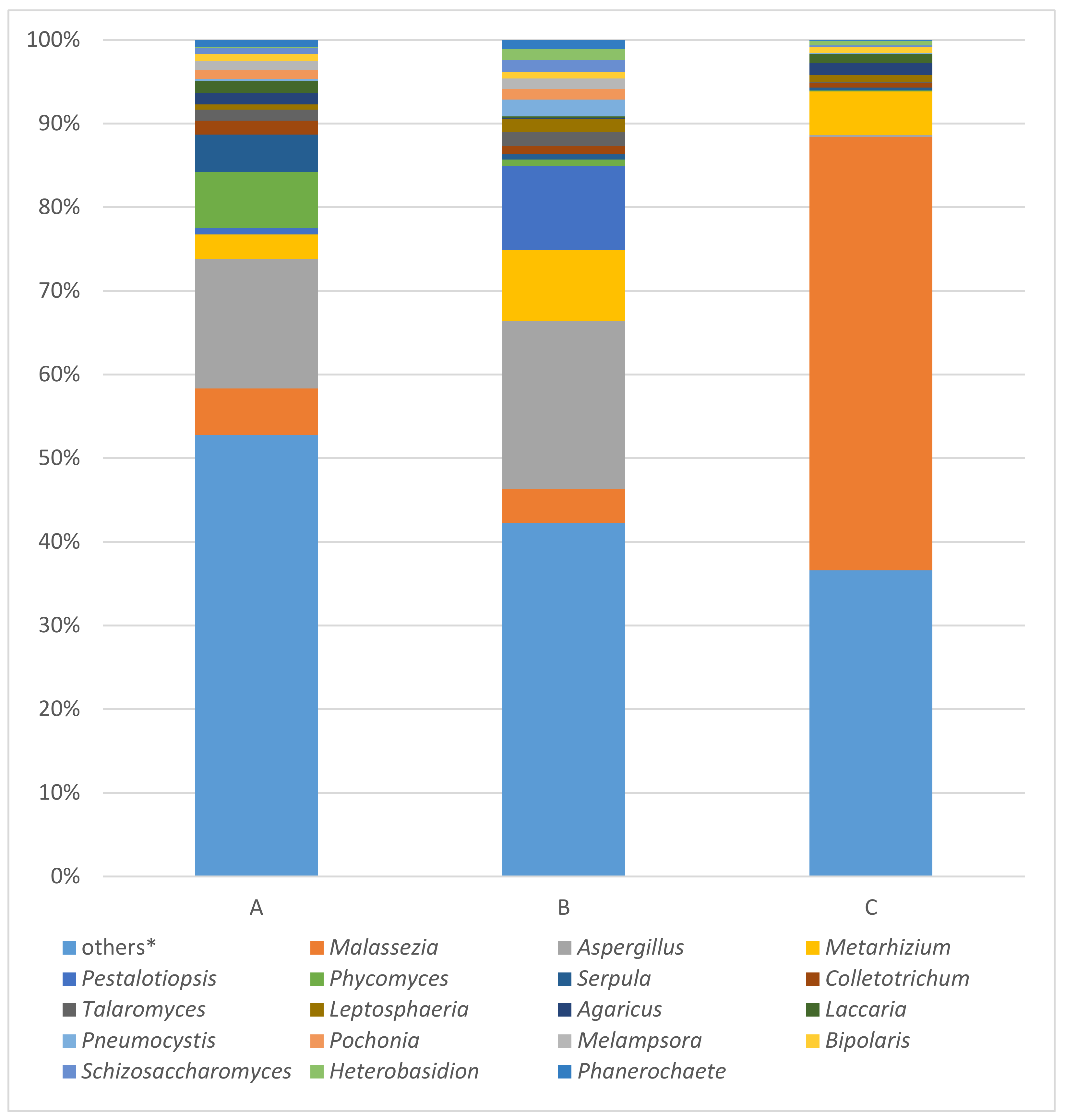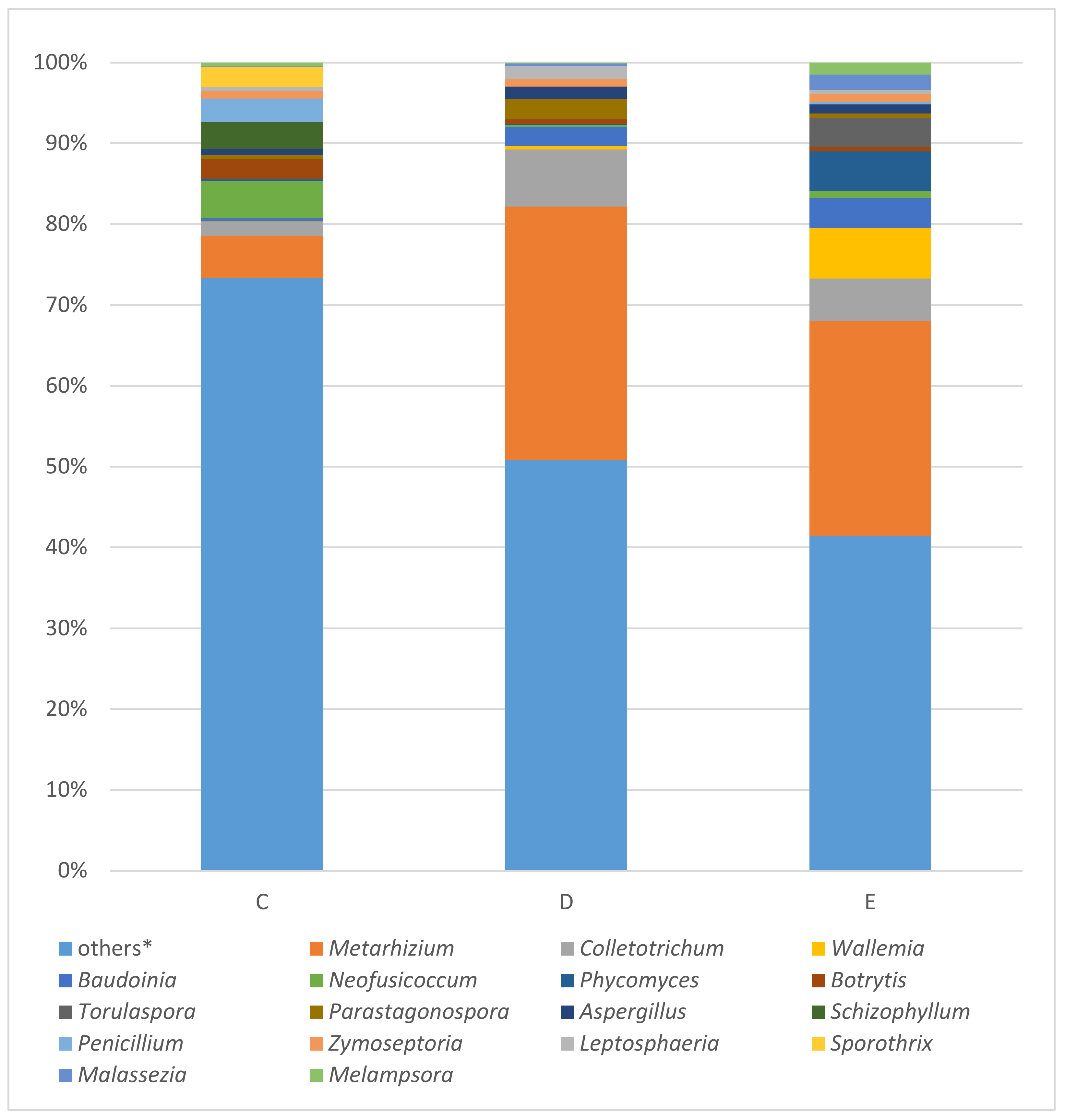Abstract
A historical crypt offers us a particular view of the conditions of some buried materials (in this case textiles) and the various biogenic phenomena to which they were subjected over the centuries. In addition, significant knowledge can come by studying the DNA of buried objects which allows the recognition of materials, but also to reveal some practice of the funeral ceremony. In this study, the deteriorating microbial communities colonizing various funeral textile items were identified and characterized using microscopic observation, cultivation, polymerase chain reaction (PCR) and sequencing, hydrolytic tests; and culture-independent analysis (high-throughput sequencing, MinION platform). Different PCR assays and consequent sequencing of amplicons were employed to recognize the animal origin of bodice reinforcements and the type of plant used to embellish the young girl. The analysis of ancient DNA (aDNA from animal and plant) was also completed by the application of high-throughput sequencing through Illumina platform. The combination of all these techniques permitted the identification of a complex microbiota composed by dangerous degradative microorganisms able to hydrolyze various organic substrates such as fibroin, keratin, and cellulose. Bacteria responsible for metal corrosion and bio-mineralization, and entomopathogenic and phytopathogenic fungi. The analysis of aDNA identified the animal component used in bodice manufacturing, the plant utilized as ornament and probably the season of this fatal event.
Keywords:
microbial community; biodeterioration; high-throughput sequencing; MinION approach; SEM; aDNA; animal; plant 1. Introduction
At the beginning of a restoration and conservation trial it is necessary, when it is possible, to analyze the target items in order to know well their characteristics and their degree of conservation. Among the various possible investigations, the microorganisms’ identification and their degradation abilities are important to perform using culture-dependent and –independent approaches [1,2]. In fact, the responsibility of microbial communities on the deterioration of cultural heritage items is largely accepted by the scientific community and operators [3,4,5]. Regarding the microbial action on fabric produced by natural fibers there is a widespread opinion that filamentous fungi are responsible for the biodeterioration of cellulosic textiles (cotton, linen, hemp, and jute), while bacteria primarily degrade animal fibers (silk and wool) [4,6]. In the past, some attempts were tried to investigate silk biodegradation in laboratory conditions [7] using bacterial strains. A deep analysis of bacterial community present on museum silk velvet was also attempted [8]. Only recently a study described the role of mold in silk deterioration, although the studied items were manuscript pages where the silking technique was applied [9]. The silk objects investigated in this manuscript are buried clothes, therefore they were subjected to particular conditions, different to those described by previous works. Moreover, to our knowledge a complete microbial investigation (including culture-dependent and culture–independent approaches) was never performed for these kinds of objects coming from this specific environment.
Various molecular biology techniques can help also to well recognize some materials, which other methods are not able to properly identify. Many cultural heritage items made by organic materials, of animal and plant origin, can be classified by the DNA analysis [10,11]. Another aim of our study was the DNA recognition of some no-well identified items.
The basilica of St. Giles is the most important monument on the principal square in Bardejov (Slovakia), which is inscribed in the UNESCO World Cultural and Natural Heritage. At the beginning of the second half of the XVII century a young girl was buried under the presbytery of this basilica. In addition to the bone remains of the girl, the archaeological study also brought to light the funeral clothes and accessories worn by the girl. Several of these girl’s items, mainly diverse textile objects, should be restored in order to set up a permanent exhibition of the finds discovered during the various archaeological studies.
We have focused our attention on those cloth objects which on their surfaces seemed to have biodeterioration phenomena and also on establishing the origin of the materials used to manufacture the stiffening material of bodice and the decorative plant relic on the girl’s head.
The microbiological survey was performed combining microscopic observation, culture-dependent, hydrolytic plate assays, and culture-independent approaches (MinION platform). The DNA analysis of bodice sticks and plant decoration was attempted using specific polymerase chain reaction (PCR) assays and high-throughput sequencing.
2. Materials and Methods
2.1. Sampling Strategy, Microbiological Cultivation, and Microscopic Observation
Several samples were taken from the three textile objects better conserved: the bodice, the cap, and the head-band. We have tried to pick up samples from parts that showed some biodeterioration phenomena under the constant supervision of restorers. The sampling campaign was carried out 4 days after the archaeological excavation. The objects were conserved in clean plastic boxes inside the sacristy of the church.
The microbiological sampling was performed using nitrocellulose membranes (5 samples; a–f) [12]. Three samples were obtained from the silk bodice; two from the front of the bodice: (a) from the ribbon; (b) from the body (Figure 1A). The third from a portion of tissue covering the stiff reinforcement (c), the sampling was performed on the backside of the bodice (Figure 1B). The other three samples were taken from the items which adorned the head of the girl: (d) from the silk bobbin lace; (e) from the linen coronet; (f) a little fragment of the decorative plant sprig (Figure 1C). A small piece of the stick used as stiff material (g) of the bodice was collected also for molecular analysis (Figure S1).

Figure 1.
(A) Front side of the bodice with the samples a and b. (B) Back side of the bodice with the sample c. (C) Decorative accessories of the head, samples d, e, and f.
The nitrocellulose membranes were cut in several small pieces, suspended in 2 mL of physiological solution and shaken overnight. This suspension was used for preparing the decimal dilution. Two-hundred microliters of each dilution were spread in diverse agar media. The bacteria were isolated on plates of Actinomycete Isolation Agar (AIA; Himedia, Mumbai, India); Reasoner’s 2A (R2A; Himedia) and LB10 agar (peptone 1 g l−1, yeast extract 0.5 g l−1, NaCl 1 g l−1, agar 15 g l−1; [13]). The agar media suitable for the cultivation of fungi were Malt Extract Agar (MEA; Himedia) and Dichloran Rose Bengal Chloramphenicol (DRBC; Himedia). The plates were incubated at room temperature (24–26 °C) and checked for the microorganisms’ growth for a minimum of two weeks. In the isolation plates a limited number of microorganisms grew. Therefore, all these microorganisms were purified performing several inoculation steps on R2A and MEA plates. The purified bacteria and fungi were maintained on plates of R2A and MEA, respectively.
Portions of the 25 mm nitrocellulose membranes, used for the microbiological sampling, were examined by scanning electron microscope (SEM; Jeol JSM 6610, Tokyo, Japan). Prior to SEM observation, the samples were sputtered with gold ions [12].
2.2. DNA Identification of Isolated Microorganisms
The DNA from bacteria was extracted using the kit Instagene (Biorad, Hercules, CA, USA) following the instruction of the manufacturer. The DNeasy Plant Mini kit (Qiagen, Hilden, Germany) was utilized to extract the fungal DNA.
The 16S rRNA gene was amplified, using the primers 27f (5’–AGA GTT TGA TCC TGG CTC AG-3’) and 685r (5’-TCT ACG CAT TTC ACC GCT AC-3’ [14]), for identifying the isolated bacteria by sequencing. The fungal isolates were identified by the amplification and consequent sequencing of the internal transcribed spacer (ITS) fragment using the primers ITS1 (5’-TCC GTA GGT GAA CCT GCG G-3’) and ITS4 (5’-TCC TCC GCT TAT TGA TAT GC-3’ [15]).
Twenty-five microliters of PCR mixture contained 50 pmol of each primer, 200 μmol l−1 of dNTP (Life Technologies, Gaithersburg, MD, USA), 1.5 U HotStar Taq plus DNA polymerase (Qiagen), 1× PCR buffer and 3 μL of the extracted bacterial or fungal DNA. The PCR program consisted of an initial denaturation at 94 °C for 5 min, followed by 30 cycles (denaturation at 94 °C for 30 s, annealing at 54 °C for 45 s, extension at 72 °C for 1 min) and a final polymerization step at 72 °C for 10 min.
PCR products from both fungal and bacterial isolates were purified using ExoSAP-IT (Affymetrix, Cleveland, OH, USA) and sequenced at a commercial facility (Eurofins Genomics, Ebersberg, Germany). The obtained sequences were directly compared with those in GenBank using BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and were subsequently deposited in GenBank under the accession numbers MT256363-MT256375 (bacterial isolates) and MT256388-MT256394 (fungal isolates).
2.3. Hydrolytic Extracellular Enzyme Assays
A loop for bacterial cells and a needle for fungal inoculation were used to transfer freshly grown cultures from the medium of maintenance to the center of Petri dishes with specific agar media, in order to assay their hydrolytic abilities. Congo red agar [13] (0.5 g l−1 KH2PO4, 0.25 g l−1 MgSO4, 2 g l−1 cellulose, 0.2 g l−1 Congo red, 2 g l−1 gelatin, 15 g l−1 agar; pH 6.8–7.2), Gelatin agar [13] (0.5 g l−1 KH2PO4, 0.25 g l−1 MgSO4, 4 g l−1 gelatin, 15 g l−1 agar), R2A supplemented with gelatin (18.12 g l−1 R2A agar and 4 g l−1 gelatin) and Fibroin agar [1] (1 mL of fibroin and 1.5 g of agar were mixed in 100 mL of distilled water) were used to assess the cellulolytic, proteolytic (gelatin-based agars) and silk deterioration properties of isolates, respectively. The keratinolytic activity was assayed through a keratin medium [2] (0.5 g l−1 KH2PO4, 0.25 g l−1 MgSO4, 4 g l−1 keratin, 15 g l−1 agar). All chemicals except hydrolysed keratin (Vipo a.s., Partizánske, Slovakia) were bought from Sigma-Aldrich. A positive reaction was manifested by a clear zone around the investigated colony.
The hydrolytic assays were performed in triplicate. All plates were incubated at room temperature (24–26 °C) for a maximum of 10 days. The positive hydrolytic reaction was represented by a clear zone around the microbial colony. The size (mm) of each hydrolytic area was measured from the edge of the colony to the edge of the zone. The microbial colony was not included in the measurement.
2.4. Total Microbial Community DNA Extraction and PCR Amplifications
The rest of the suspension (described on paragraph “Sampling strategy, microbiological cultivation and microscopic observation”), containing very small membrane pieces, was centrifuged and from the obtained pellets the DNA was extracted using DNeasy PowerSoil extraction kit (Qiagen) following the protocol provided by the producer.
The bacterial 16S rRNA gene was amplified with the primers 27f (as above) and 1492r (5’-GGT ACC TTG TTA CGA CTT-3’ [14]), while the fungal community was analyzed by the amplification of the ITS fragment (ITS1 and ITS4 primers, as above) and the 28S rRNA gene by the primers NL1 (5′-GCATAT CAATAA GCG GAG GAA AAG-3′) and NL4 (5′-GGT CCG TGT TTC AAG ACG G-3′ [16]). The PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA).
2.5. Library Preparation for Microbial Communities’ Analysis
The library for nanopore sequencing by MinION platform was made from purified amplicons using Ligation Sequencing Kit 1D (SQK-LSK108) as described previously [17]. Briefly, the process comprised the following parts: (i) the ends of amplicons were prepared by Ultra II End-prep reaction buffer and Ultra II End-prep enzyme mix from the NEBNext Ultra II End Repair/da-Tailing Module (New England Biolabs, Ipswich, MA, USA), (ii) ligation step with Barcode Adapters was performed using previous End-prep. DNA, Barcode Adapter (PCR Barcoding kit 96, Oxford Nanopore Technologies, Oxford, UK) and Blunt/TA Ligase Master Mix (NEB) and (iii) Barcoding PCR was performed using one of PCR Barcode primer (BC1-BC96) from PCR Barcoding Expansion Pack 1–96 (EXP-PBC096, ONT), LongAmp Taq 2x master mix (NEB) and Nuclease-free water (Qiagen). For the Barcoding PCR, the following steps were used: initial denaturation step (95 °C, 3 min), followed by 19 cycles (95 °C for 15 s.; 62 °C for 15 s.; 65 °C for 2 min) and final extension step (65 °C for 10 min).
The intermediate purification steps by Agencourt AMPure XP magnetic beads (Beckman Coulter) and the DNA fluorometric quantification procedures with Qubit v2 Fluorometer (Invitrogen, Waltham, MA, USA) using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) were performed between each of the three abovementioned parts.
2.6. Pooled Library Preparation and Flow-Cell Processing
Each sample labeled by unique barcode was equimolar pooled to final 1 µg of DNA in 45 µL Nuclease free water (Qiagen). The procedure of second end- preparation using NEBNext Ultra II End Repair/da-Tailing Module (New England Biolabs) chemistry and control DNA CS (DCS; Ligation Sequencing kit 1D, Oxford Nanopore Technologies), and also the second adapter ligation procedure of thus end-prepared DNA using Adapter Mix 1D (AMX1D; Ligation Sequencing kit 1D, ONT) and Blunt/TA Ligase Master Mix (NEB) were done following the instructions in 1D PCR barcoding (96) genomic DNA (SQK-LSK108) protocol.
Also the final cleaning procedure using Agencourt AMPure XP beads (Beckman Coulter), Adapter Bead Binding buffer (ABB; Ligation Sequencing kit 1D, Oxford Nanopore Technologies) and Elution Buffer (ELB; Ligation Sequencing kit 1D, Oxford Nanopore Technologies) was performed following the recommendations of the manual supplied by the manufacturers (1D PCR barcoding (96) genomic DNA SQK-LSK108).
The prepared library was stored on ice during the priming of the SpotON flow cell (FLO-MIN 106D R9 Version, Oxford Nanopore Technologies). Seventy five microliters of sample composed from Running Buffer (RBF, Oxford Nanopore Technologies), Loading Beads (LLB; Oxford Nanopore Technologies), Nuclease-free water and DNA library was loaded to the flow cell via the SpotON sample port according manufacturer’s recommendations.
2.7. Sequencing Performance and Evaluation
Sequencing run was performed for 48 h on MinION sequencer (Oxford Nanopore Technologies) using R9 flow cell, connected to the appropriate personal computer meeting the recommended criteria. The MinKNOW™ software was used to check the number of active pores and used with the default settings. After sequencing, the Fast5.tmp files were converted to Fast5 files via command prompt. Reads were then base called using Albacore (Oxford Nanopore Technologies) and split by barcodes with EPI2me Desktop Agent (Oxford Nanopore Technologies Metrichor). Taxonomic classification and quantitative analysis of reads derived from 28S rRNA amplicons were performed using “What’s in my pot” tool (WIMP, Oxford Nanopore Technologies). In the case of bacterial taxa classification and quantification, the workflow for 16S rRNA gene encoded amplicon analysis in EPI2ME Agent was chosen. The minimal quality score in both cases was set to 7.
2.8. DNA Analysis of Plant Sprig and Stiff Material
The DNA from the samples f (decorative plant sprig) and g (stiff material of the bodice) was extracted by the kit ChargeSwitch Forensic DNA Purification Kit (Invitrogen) following the recommendations and instructions of manufacturer.
The obtained DNA of plant sprig was amplified using different combinations of primers oriented on the chloroplast trnL (UAA) intron (trnL_c-trnL_d, size of PCR product about 577 bp; trnL_c-trnL_h, product size about 187 bp; trnL_g-trnL_h, product size about 79 bp; trnL_g-trnL_d, product size about 474 bp [18]).
The DNA of stiff material was amplified with the primers and PCR protocols developed by [19] and oriented on the mitochondrial DNA D-loop region. We have tried the PCR assays comprising the pair of primers Pair 1, Pair 2, and Pair 3, but, also, we have combined the primers in this way: Pair1_Fw–Pair3_Rv; Pair2_Fw–Pair3_Rv.
The PCR products were separated on 1.5% agarose gel in TAE buffer. Gels were stained with ethidium bromide and visualized under UV light. The expected bands were excised from the gel and purified by QIAquick Gel Extraction kit (Qiagen). Then the purified bands were sequenced on both strands by a commercial facility (Eurofins Genomics). The resulting sequences were directly compared with those in GenBank using BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.9. Illumina Sequencing of Plant Sprig and Stiff Material DNA
The DNA extracted from the samples f (decorative plant sprig) and g (stiff material of the bodice) was sequenced by the MiSeq system (Illumina, San Diego, CA, USA). Totally 20 ng of isolated double-stranded DNA was used for transposon-based sample fragmentation with Nextera XT DNA Library Prep Kit (Illumina, CA, USA). Samples were directly amplified and indexed using Nextera XT Index Kit (Illumina, CA, USA) according to the manufacturer’s protocol. For PCR product purification 1,8x sample volume of Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA) was used. The final DNA library quantity was determined with Qubit 2.0 Fluorometer (ThermoFisher Scientific, MA, USA) while for quality (fragment length) assessment Agilent 2100 Bioanalyzer was used (Agilent Technologies, Santa Clara, CA, USA). DNA libraries were diluted to 4 nM and pooled. Paired-end sequencing 2 × 300 was performed using Illumina MiSeq platform (Illumina, San Diego, CA, USA).
The data obtained from shot-gun sequencing of both samples were processed using CLC Genomics Workbench software v 9.5.2 (Qiagen, Germany). After trimming (quality limit 0.02, min.read length 50 bp) and quality control, de novo assembly of sequences, length fraction 0.9 (animal)/0.8 (plant), similarity fraction 1.0 (animal)/0.9 (plant)) was executed. Data obtained for stiff sample were mapped against GenBank WGS database using BLAST program with megablast settings mapping against taxon Equus. Contigs obtained for the plant sample were mapped against plant part of the GenBank database also using megablast settings.
3. Results
3.1. Microbial Isolates, Hydrolytic Properties, and SEM Observation
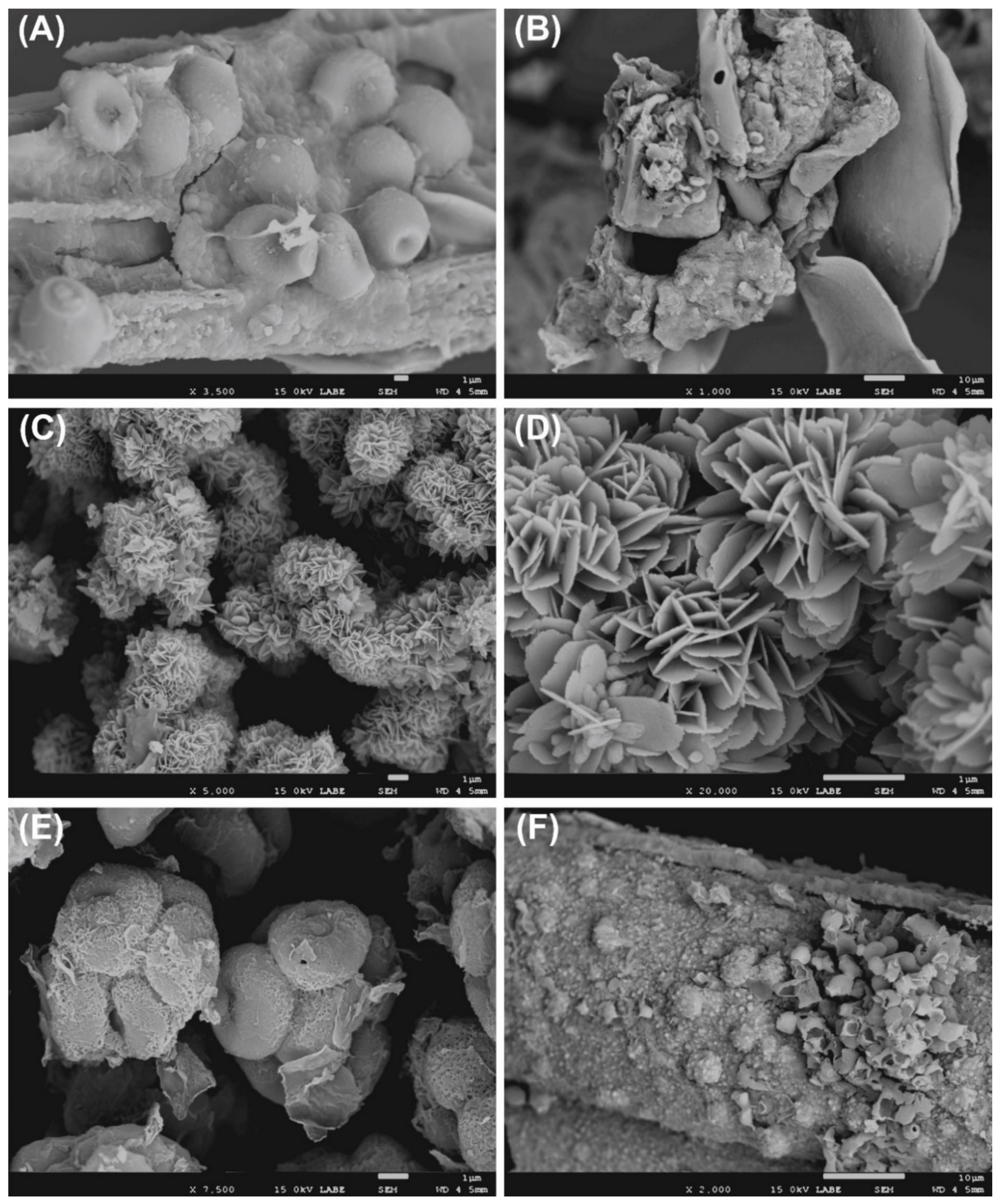
The SEM analysis displayed the presence of fungi in various samples (Figure 2), in fact it is possible to note conidia, spores, and hyphae. The occurrence of bacteria in several samples is confirmed, indirectly, by the observation of biogenic crystals (Figure 2C,D).
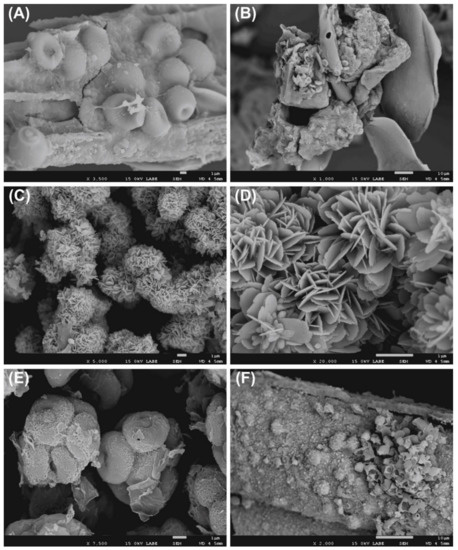
Figure 2.
Scanning electron microscope (SEM) observation of nitrocellulose membranes. (A) Conidia, (B) fungi colonizing insect remains, (C,D) biogenic crystals and their morphological details, (E) ascospores, (F) presence of fungal conidia and spores.
Bacteria were isolated from the samples a (bodice, from the ribbon), b (bodice, from the body), d (silk bobbin lace cap), and e (linen head-band). The most isolated genera were Moraxella, Kocuria, and Paracoccus. The only bacterial isolates able to hydrolyze all the tested substrates was Micrococcus yunnanensis B2. Various bacteria degrade four substrates; three isolates (the two Moraxella isolates and Paracoccus chinensis) were negative to all assays. A slight cellulolytic activity was shown only by the two Kocuria isolates and Micrococcus yunnanensis B2. The most effective keratinolytic isolate was Paenibacillus illinoisensis D1 which displayed also an extensive protease ability together with Micrococcus yunnanensis B2. Paenibacillus illinoisensis D1 evidenced also the best fibroinase activity followed by Sphingomonas sp. D2. Usually the isolates positive to proteolytic assays can degrade also the silk fibroin (Table 1).

Table 1.
DNA identification and hydrolytic properties of bacterial and fungal isolates.
From sample c (bodice, tissue covering the stiff reinforcement) only filamentous fungi were isolated belonging mainly to genera Aspergillus and Penicillium. The yeast Sporidiobolus metaroseus B2-F and Penicillium commune B1-F, recovered from sample b, were the only eukaryotic microorganisms able to produce positive results for all the hydrolytic assays. Three fungi over seven isolates displayed cellulolytic ability, on the contrary all the fungal members have significant fibroinase property.
3.2. MinION Analysis of Bacterial 16S rRNA Gene
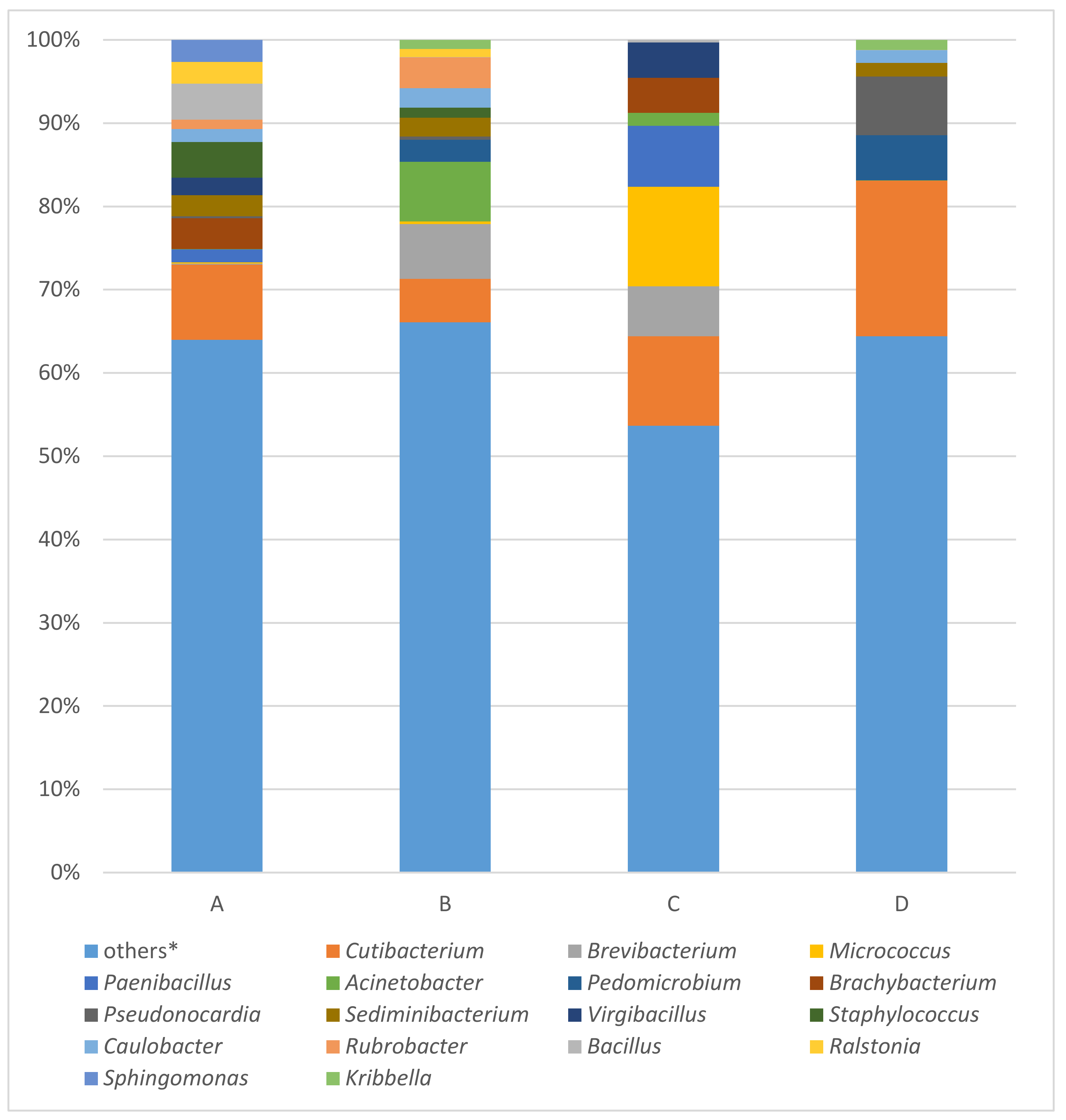
The results of all four samples (a, b, c, and d), examined by EPI2ME Desktop Agent software using “FASTQ 16S“ tool are visualized, expressing the percentage of taxa abundance on genera degree, as bar plots in Figure 3. The percentages of reads assigned to given taxa were calculated from number of classified reads. The number of classified reads in order of samples a, b, c, and d were 46,671, 378,356, 38,042, and 64,707, respectively. Because of microbial diversity, the percentage of given taxa of each samples were counted together, and the value over 2% of this sum were used as threshold. This is the reason why the number of “other“ taxa forms the biggest amount of all four analyzed samples. In the order of samples a, b, c, and d “other“ taxa comprised 63.99%, 66.07%, 53.65%, and 64.40%, respectively.
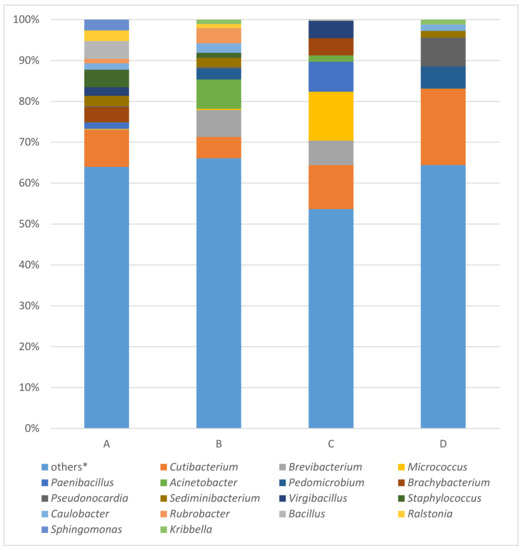
Figure 3.
Results of bacterial 16S rRNA gene analysis of samples A (the ribbon of the bodice), B (body of the bodice), C (tissue covering the stiff reinforcement of the bodice), D (silk bobbin lace cap) using MinION. * Others: the percentages of reads assigned to taxa under (below) the value of 2% in the sum of all four samples.
The most abundant taxa identified in all four samples were Cutibacterium (a-9.03%; b-5.24%; c-10.74%; d-18.72%), Brevibacterium (0.09%; 6.57%; 6.00%; 0.00%), Micrococcus (0.17%; 0.31%; 11.95%; 0.00%), Paenibacillus (1.57%; 0.00%; 7.36%; 0.00%) and Acinetobacter (0.01%; 7.15%; 1.53%; 0.01%). The top 10 most abundant taxa enclosed genera Pedomicrobium (0.00%; 2.67%; 0.00%; 5.42%), Brachybacterium (3.73%; 0.00%; 4.19%; 0.00%), Pseudonocardia (0.20%; 0.37%; 0.01%; 7.06%), Sediminibacterium (2.53%; 2.27%; 0.00%; 1.63%), and Virgibacillus (2.12%; 0.00%; 4.25%; 0.00%).
3.3. MinION Analysis of Fungal 28S rRNA Gene
Fungal 28S rRNA gene analysis produced results only with samples a, b, and c. The results of this analysis are summarized in Figure 4. In the order of samples a, b, and c, the most occurring fungal taxa were Malassezia (5.59%; 4.08%; 51.77%), Aspergillus (15.46%; 20.08%; 0.25%), Metarhizium (2.93%; 8.41%; 5.22%), Pestalotiopsis (0.75%; 10.11%; 0.02%) and Phycomyces (6.75%; 0.73%; 0.10%) followed by Serpula (4.44%; 0.62%; 0.36%), Colletotrichum (1.69%; 1.02%; 0.52%), Talaromyces (1.30%; 1.68%; 0.12%), Leptosphaeria (0.64%; 1.50%; 0.83%), and Agaricus (1.37%; 0.11%; 1.41%), which completed the top 10 most abundant fungal taxa.
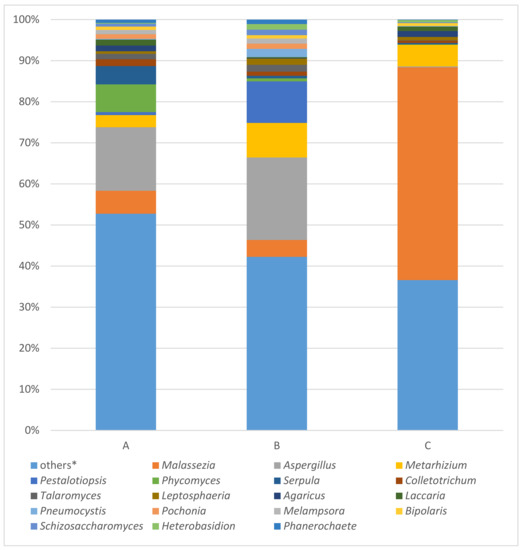
Figure 4.
Results of fungal 28S rRNA analysis of samples A (the ribbon of the bodice), B (body of the bodice), C (tissue covering the stiff reinforcement of the bodice) using MinION. * Others: the percentages of reads assigned to taxa under (below) the value of 2% in the sum of all four samples.
3.4. MinION Analysis of Fungal ITS Fragment
The fungal ITS marker amplified with the samples c, d, and e. In this order of samples, Metarhizium (5.23%; 31.36%; 26.56%), Colletotrichum (1.75%; 7.05%; 5.23%), Wallemia (0.04%; 0.41%; 6.27%), Baudoinia (0.45%; 2.37%; 3.68%), followed by Neofusicoccum (4.56%; 0.21%; 0.86%), Phycomyces (0.20%; 0.18%; 4.94%), Botrytis (2.50%; 0.58%; 0.54%), Torulaspora (0.01%; 0.00%; 3.56%), Parastagonospora (0.46%; 2.51%; 0.56%), and Aspergillus (0.80%; 1.51%; 1.13%) were identified as the most abundant fungal by WIMP tool whose results are expressed as bar plots in Figure 5.
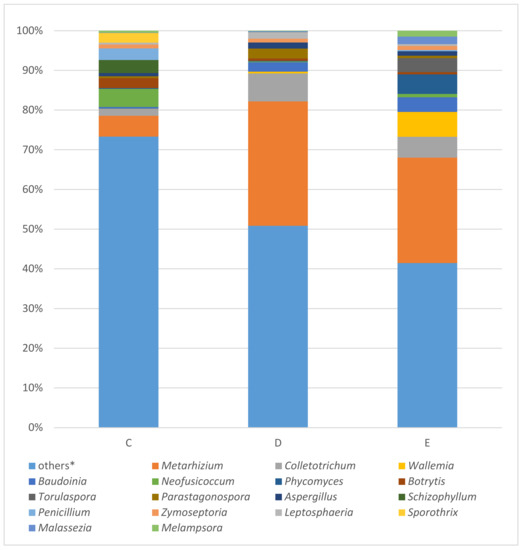
Figure 5.
Results of fungal ITS analysis of samples C (tissue covering the stiff reinforcement of the bodice), D (silk bobbin lace cap), and E (linen coronet) using MinION. * Others: the percentages of reads assigned to taxa under (below) the value of 2% in the sum of all four samples.
3.5. DNA Analysis of Animal and Plant Samples
In order to identify the animal origin of the bodice stiff material (sample g), we have tried different combination of primers, but unfortunately all PCR assays produced negative results. Using MiSeq platform (Illumina), with this sample, we obtained 1240 contigs, with a length range of 69–25, 511 bp, several of them matching Equus caballus.
Positive PCR amplifications were obtained with the plant material (sample f) using the primers combinations trnL_c - trnL_h (amplicon size about 187 bp) and trnL_g - trnL_h (about 79 bp). The consequent Sanger sequencing of the amplicons had reliable results only with the product of about 187 bp. In fact, the obtained 136 bp sequence had a 98.53% of similarity with several plant species belonging to the family of Apiaceae (MN167277, Cuminum cyminum; MN167276, Anethum graveolens; MN167272, Petroselinum crispum; MH142518, Glehnia littoralis; MK688991, Angelica sinensis). Using the DNA extracted from the plant sample, a totally of 2287 contigs with a length range of 79–1423 bp were produced by high-throughput sequencing analysis. The Poaceae family was represented by three species: Digitaria exilis, Hordeum vulgare, and Triticum aestivum. The other two detected species, Brassica juncea and Nicotiana tabacum belonged to the family Brassicaceae and Solanaceae, respectively.
4. Discussion
4.1. Bacterial Community
The two strategies of analysis (culture-dependent and culture-independent) evidenced a link with the members of the genera Micrococcus and Paenibacillus. These bacteria were detected by both approaches, but in different samples. They are frequently isolated from historical textile [1,20] and also in the past studies, as well as here, they showed significant hydrolytic properties. Members of the genus Paenibacillus produce a broad range of enzymes used in biotechnology processes [21]. In addition to Micrococcus, other isolated Actinobacteria with interesting proteolytic and fibroinolytic properties belonged to the genus Kocuria. Representatives of this genus are commune in subterranean environment [22] and were also isolated from archaeological textile [23]. Therefore, bacteria of the genera Paenibacillus, Micrococcus, and Kocuria can be considered as dangerous textile degrading microorganisms.
By high-throughput sequencing the genera Micrococcus and Paenibacillus were dominant in the sample c (tissue covering the stiff reinforcement), where M. luteus comprised 8.47% of all reads classified. Micrococcus luteus is also one of the species inhabiting the human skin [24]. Members of the genus Micrococcus were also previously identified as part of microbiota on mummy surface [2].
Regarding the MinION analysis, three species of the genus Cutibacterium (C. acnes, C. granulosum, and C. avidum), formerly known as Propionibacterium [25], were detected in all four samples (a, b, c, and d) with the remarkable prevalence of Cutibacterium acnes, which is involved in the maintenance of a healthy skin, but it can also act as an opportunistic pathogen in acne vulgaris [26] or can cause implant-associated infections [27].
Genus Acinetobacter dominated mostly in sample b (sampled from silk fabric of the bodice), whose probably touched the torso of cadaver. Hyde et al. [28] observed the highest abundance of Acinetobacter in samples of male (5–20% of relative abundance) and female (5–15% of relative abundance) cadavers studied during the time, mostly in the late stages of their decomposition.
The genus Brevibacterium dominated in the samples b and c, namely B. casei (5.63%) and B. pityocampae (4.15%), respectively. Brevibacterium was the most frequent genera identified in human archaeological remains by Philips et al. [29], also typically found in a wide range of soils and waters.
The genus Pseudonocardia was in our case the most abundant (silk bobbin lace cap) in sample d (7.06%). This genus was previously detected in the caves [30], catacombs [31], tomb walls, as part of the ancient paintings deteriorating microflora [32], or part of the so-called moonmilk [33] and also on the body of a mummy [2].
The reads identified as genus Pedomicrobium (Alphabacteria class) predominated in samples b and d (2.67% and 5.42%, respectively). Pedomicrobium is known as iron- and manganese-oxidizing and accumulating bacterium often detected in biofilms formed on different surfaces [34,35]. Probably, in these samples Pedomicrobium could be responsible for various corrosion phenomena [36] of metal materials present in the structure of the bodice, but also as decoration jewels on the head of girl. The isolates Roseomonas mucosa and Sphingomonas sp. also belonged to the class of Alphabacteria. Here they contribute to the degradation of silk fibroin and other kind of proteinaceous substrates. These bacteria have the characteristic to possess hydrolytic enzymes active also at low temperature [37,38].
The iron-oxidizing bacteria of the genus Sediminibacterium were present with approximately 2% in all samples, except of sample c. We think that their role is connect mainly with the corrosion [39] of metal materials present in these samples.
In the samples a and c, Brachybacterium was dominated by reaching 3.73% and 4.19%, respectively. Affiliates of this genus were already isolated from subterranean environment [40] and also from funeral cloths [1] demonstrating significant proteolytic property.
Other bacteria were present exclusively in samples a (2.12%) and c (4.25%), where the members of the genus Virgibacillus. In the sample a prevailed V. marseillensis, V. carmonensis, and V. halodenitrificans. In sample c mostly V. necropolis and V. carmonensis were identified. V. carmonensis, V. necropolis, and V. picturae were isolated from deteriorated mural paintings [41]. In addition, these bacteria are able to produce different extracellular enzymes, such as proteases, cellulases, xylanases, and amylases, which can degrade several substrates [42].
4.2. Fungal Community
Malassezia, the most abundant taxa detected by 28S rRNA sequencing analysis, significantly prevailed in sample c (51.77%), from which, the M. globosa reached a percentage of 21.59%. Although Malassezia yeasts are part of the cutaneous microbiota detected mainly on healthy human skin, species of this genus are also associated with mammalian cutaneous disorders such as dandruff, seborrhoeic dermatitis, pityriasis versicolor, psoriasis, folliculitis, and otitis [43]. The ITS analysis placed Malassezia in the 16th place of the most occurring taxa (Figure 5), it dominated in sample e (c-0.09%; d-0.25%; e-1.88%).
Aspergillus and Metarhizium were detected in all samples using both fungal markers; by 28S rRNA the highest occurrence, 20.08% for Aspergillus and 8.41% for Metarhizium, was recorded in sample b. ITS analysis revealed the highest percentages of these genera in sample d, 1.51% and 31.36% for Aspergillus and Metarhizium, respectively. The presence of Metarhizium is probably due to because members of this genus are known as entomopathogens [44], which infected various insects contributing to human decomposition. In both analysis, A. fumigatus was the most dominant species among the other Aspergillus spp. This species, in previous studies, was isolated from various organs of cadavers and human remains [45,46], several cultural heritage items made by organic materials [47], and it showed also proteolytic, keratinolytic, and fibroinolytic abilities [1]. Therefore it is not surprising, that the Aspergilli and Penicilli isolated in this study possessed also interesting hydrolytic properties. They were recovered almost exclusively from a portion of silk tissue, which covered the stiff reinforcement made by animal substances. These characteristics evidenced their biodeteioration dangerousness for textile and other proteinaceous materials occurring in various cultural heritage objects.
Pestalotiopsis is another abundant taxa in sample b (10.11%) according to 28S rRNA analysis, and their species are known as plant pathogens and as the producers of antifungal active metabolites [48,49,50]. The ITS analysis did not detect this taxa.
Both taxa Phycomyces and Colletotrichum were detected by 28S rRNA (Phycomyces: a-6.75%, b-0.73%, c-0.10%; Colletotrichum: a-1.69%, b-1.02%, c-0.52%) and ITS (Phycomyces: c-0.20%, d-0.18%, e-4.94%; Colletotrichum: c-1.75%, d-7.05%, e-5.23%) analyses. These fungi are not common colonizers of historical items, in fact, to our knowledge, the first detection of Phycomyces in two canvas paintings was very recent [51].
Genus Serpula, dominated significantly only in sample a (a-4.44%, b-0.62%, c-0.36%) analyzed by 28S rRNA genes sequencing. All reads assigned to the genus Serpula belonged to only one species - S. lacrymans, a common wood-decaying dry rot causing deterioration of wood monuments [52] and buildings [53]. The ITS analysis revealed lower occurrence of Serpula taxa with highest presence in sample c (0.34%). Other phytopathogens were detected in sample c (ITS) in remarkable abundance, e.g., Neofusicoccum with one representative species of this genus N. parvum (c-4.56%), known as phytotoxins-producing grapevine canker agent [54] and Botrytis with one representative species of this genus B. cinerea (2.50%), well known pathogen causing serious losses in more than 200 crop species worldwide [55]. The genus Torulaspora represented 3.56% in sample e (ITS), with one representing species T. delbrueckii, which is also typical inhabitant of the grape surface, known as well studied non-Saccharomyces yeasts used in winemaking [56]. Another plant pathogen, Phaeosphaeria nodorum was detected mostly in sample d (2.51%). It is known as fungal pathogen of wheat (Triticum aestivum). The relevant detection of plant pathogens is due of the wood of coffin, but also of the presence of several types of plant species that usually served to adorn the body of the dead.
Wallemia ichthyophaga, as the only representatives of the taxa Wallemia, occurring in highest abundance in the sample e (6.27%), is a halophilic fungus with optimally growth on media with salinity above 15% of NaCl (w/v) [57]. Another extremophile yeast [58] was the isolate Sporidiobolus metaroseus recovered from the silk bodice (sample b). It displayed significant hydrolytic properties including fibroin degradation.
4.3. Microscopic Observation
The combination of sampling by nitrocellulose membrane and the consequent SEM screening, can be considered as a suitable method for the microscopic analysis of fragile cultural heritage items. Usually, the adhesive tape sampling is applied in combination of microscopic observation [59], but depending of the fragility of surface, the use of membranes is a valid alternative.
The SEM monitoring permitted to display the presence of an active microbial communities, mainly fungi that are able to degrade various types of substrates. In addition, the observation of ascospores [60] confirmed the results of microbial and sequencing analysis where different Aspergillus members were detected.
The presence of bacteria was evidenced by the observation of bio-mineralization phenomena, such as the occurrence of CaCO3 crystals, probably vaterite or aragonite. These CaCO3 formations are generally produced by various bacterial members belonging to the phylum Proteobacteria (vaterite forming) and Firmicutes (aragonite) [61,62]. Also, these findings are in accordance with the bacteria isolated and detected by both culture-dependent and culture-independent strategies.
4.4. Plant and Animal DNA Analysis
Usually, during the XVII century, the stiff materials used as reinforcement of the bodices were whalebone from the jaws of the whale, wood, or horn [63]. Therefore, when we have observed the reinforcement of the bodice by microscope (Figure S1), these busks were particular because they appeared as several fibers bonded, presumably, with animal glue. The restorers supposed that this structure was formed by horsehair, and the molecular analysis confirm such hypothesis. Although, the horsehair was used for different types of textile materials (horsehair interlining; [64]), in literature we did not find any previous study, where this kind of reinforcement material was described, so the analysis of the DNA help us to unambiguously identify the origin of these accessories of the bodice.
DNA of Apiaceae was identified from the small portion of plant material through PCR sequencing analysis. Unfortunately, the small sequence did not permit a specific recognition of the plant species. However, excluding Glehnia littoralis which is a typical plant growing at temperate sandy coasts around the North Pacific Ocean [65], and Angelica sinensis which is also characteristic of different world regions other than Europe [66]. One of the others species (Cuminum cyminum, Anethum graveolens and Petroselinum crispum) probably were the main ornamental plants on the head of the young girl. These species present nice inflorescence [67], which can create beautiful decorative effects. Moreover, the use of these kinds of plants in burial custom was evidenced in several archaeological studies [68,69,70].
The Illumina analysis on plant DNA did not confirm the results of PCR and sequencing approach, but other interesting outcomes were produced. In fact, the detection of several sequences is due by the presence of traces of plants, probably pollen belonged mainly to the species Hordeum vulgare (barley), Triticum aestivum (common wheat), Brassica juncea (canola), and Nicotiana tabacum (tobacco) which were commonly cultivated also in central Europe. We think that the identification of these plants perhaps indicates somehow the period of the funeral which could be in the late spring or summer. The register of deceased persons has been lost, therefore our molecular investigation provided at least some hypothesis about the seasonal period of the burial ceremony.
5. Conclusions
The combination of non-invasive sampling, classical microbiological methods (cultivation, SEM observation, and hydrolytic assays), molecular approaches, and high-throughput sequencing (culture-independent strategy and DNA analysis) permitted to investigate the deteriorating microbiota and to discover various characteristics of diverse funeral accessories. The complete microbiological analysis displayed the occurring microbiota and evidenced its dangerous hydrolytic properties and bio-precipitation abilities. The MinION approach can be optimized in order to perform the analysis in situ. In fact, this high-throughput sequencing platform is portable and in the future the analysis of samples can be done directly in the place of sampling. Moreover, MinION can sequence long sequences, which facilitates the better identification of microbial communities. By the DNA analysis, we observed important information about the use of certain materials (horse-hair reinforcement) in textile manufacturing. Knowledge about the use of herbal and flower ornaments in the 17th century burial customs in central Europe were also revealed. This study evidenced again the importance of a multi-disciplinary approach applied to cultural heritage objects. Such study showed how it is important to develop periodical and precise microbial monitoring strategies which permit prompt decisions for the safeguard of these fragile items against the microbial deterioration and also to obtain valuable data on the uses and customs of a certain historical period.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/16/5451/s1; Figure S1: Stereomicroscope visualization of reinforcement material of the bodice.
Author Contributions
Conceptualization, D.P., M.B., and A.P.; Methodology, D.P., K.Š., and M.P.; Formal Analysis, Z.K., J.P., M.K., L.K., M.B., A.P., and K.Š.; Investigation, Z.K., J.P., M.K., L.K., M.B., A.P., and M.P.; Resources, Z.K. and D.P.; Writing—Original Draft Preparation, M.P. and D.P.; Writing—Review and Editing, D.P. and Z.K.; Visualization, D.P.; Supervision, D.P.; Project Administration, M.B.; Funding Acquisition, M.B. and K.Š. All authors have read and agreed to the published version of the manuscript.
Funding
This research was mainly funded by the project APVV-19-0059 (Colored stains on historical papers: biological and chemical characterization coupled with removal solutions). This publication is also the result of implementation of the projects: Research and Development Operational Programme funded by the ERDF (grant number: ITMS 26240220086) and VEGA 1/0404/19.
Acknowledgments
We would like to thank the members of the Academy of Fine Arts and Design in Bratislava (Zuzana Machatová, Sylvia Birkušová, Ingrid Ondrejičková Soboslayová) who permitted us to investigate these interesting samples in the frame of their restoration project and for a photograph (Figure S1). We acknowledge the archaeologists Peter Harčar and Marián Uličný of the Monument Board of the Slovak Republic, the pioneers who started the archaeological research (Uličný, M. - Harčar, P.: Bardejov - Bazilika sv. Egídia, Výskumná dokumentácia z archeologického výskumu vo svätyni baziliky v rokoch 2008 - 2009, Pamiatkový úrad SR, február 2017). We also thank Ľubomír Orovčík (Institute of Materials and Machine Mechanics, Slovak Academy of Sciences) for the opportunity to use the SEM.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pangallo, D.; Kraková, L.; Chovanová, K.; Bučková, M.; Puškarová, A.; Šimonovičová, A. Disclosing a crypt: Microbial diversity and degradation activity of the microflora isolated from funeral clothes of Cardinal Peter Pázmány. Microbiol. Res. 2013, 168, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Kraková, L.; Šoltys, K.; Puškárová, A.; Bučková, M.; Jeszeová, L.; Kucharík, M.; Budiš, J.; Orovčík, L.U.; Szemes, T.; Pangallo, D. The microbiomes of a XVIII century mummy from the castle of Krásna Hôrka (Slovakia) and its surrounding environment. Environ. Microbiol. 2018, 20, 3294–3308. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K.; Piñar, G. Microbial deterioration of cultural heritage and works of art—tilting at windmills? Appl. Microbiol. Biotechnol. 2013, 97, 9637–9646. [Google Scholar] [CrossRef] [PubMed]
- Gutarowska, B.; Pietrzak, K.; Machnowski, W.; Milczarek, J.M. Historical textiles–A review of microbial deterioration analysis and disinfection methods. Text. Res. J. 2017, 87, 2388–2406. [Google Scholar] [CrossRef]
- Marvasi, M.; Cavalieri, D.; Mastromei, G.; Casaccia, A.; Perito, B. Omics technologies for an in-depth investigation of biodeterioration of cultural heritage. Int. Biodeterior. Biodegrad. 2019, 144, 104736. [Google Scholar] [CrossRef]
- Szostak-Kotowa, J. Biodeterioration of textiles. Int. Biodeterior. Biodegrad. 2004, 53, 165–170. [Google Scholar] [CrossRef]
- Forlani, G.; Seves, A.M.; Ciferri, O. A bacterial extracellular proteinase degrading silk fibroin. Int. Biodeterior. Biodegrad. 2000, 46, 271–275. [Google Scholar] [CrossRef]
- Brzozowska, I.; Bogdanowicz, A.; Szczęsny, P.; Zielenkiewicz, U.; Laudy, A. Evaluation of bacterial diversity on historical silk velvet textiles from the Museum of King John III’s Palace at Wilanów, Poland. Int. Biodeterior. Biodegrad. 2018, 131, 78–87. [Google Scholar] [CrossRef]
- Martins, C.; Pereira, C.S.; Plechkova, N.V.; Seddon, K.R.; Wang, J.; Whitfield, S.; Wong, W. Mycobiota of silk-faced ancient Mogao Grottoes manuscripts belonging to the Stein collection in the British library. Int. Biodeterior. Biodegrad. 2018, 134, 1–6. [Google Scholar] [CrossRef]
- MacHugh, D.E.; Larson, G.; Orlando, L. Taming the past: Ancient DNA and the study of animal domestication. Annu. Rev. Anim. Biosci. 2017, 5, 329–351. [Google Scholar] [CrossRef]
- Bieker, V.C.; Martin, M.D. Implications and future prospects for evolutionary analyses of DNA in historical herbarium collections. Bot. Lett. 2018, 165, 409–418. [Google Scholar] [CrossRef]
- Puškárová, A.; Bučková, M.; Habalová, B.; Kraková, L.; Maková, A.; Pangallo, D. Microbial communities affecting albumen photography heritage: A methodological survey. Sci. Rep. 2016, 6, 20810. [Google Scholar] [CrossRef] [PubMed]
- Grivalský, T.; Bučková, M.; Puškárová, A.; Kraková, L.; Pangallo, D. Water-related environments: A multistep procedure to assess the diversity and enzymatic properties of cultivable bacteria. World J. Microbiol. Biotechnol. 2016, 32, 42. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackenbrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–148. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–321. [Google Scholar]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Šoltys, K.; Planý, M.; Biocca, P.; Vianello, V.; Bučková, M.; Puškárová, A.; Sclocchi, M.C.; Colaizzi, P.; Bicchieri, M.; Pangallo, D.; et al. Lead soaps formation and biodiversity in a XVIII Century wax seal coloured with minium. Environ. Microbiol. 2020, 22, 1517–1534. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trn L (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef]
- Vilà, C.; Leonard, J.A.; Götherström, A.; Marklund, S.; Sandberg, K.; Lidén, K.; Wayne, R.K.; Ellegren, H. Widespread origins of domestic horse lineages. Science 2001, 291, 474–477. [Google Scholar] [CrossRef]
- Wawrzyk, A.; Gutarowska, B.; Rybitwa, D.; Pietrzak, K.; Machnowski, W.; Wrzosek, H.; Papis, A.; Walawska, A.; Otlewska, A.; Szulc, J.; et al. Vapourised hydrogen peroxide (VHP) and ethylene oxide (EtO) methods for disinfecting historical cotton textiles from the Auschwitz-Birkenau State Museum in Oświęcim, Poland. Int. Biodeterior. Biodegrad. 2008, 133, 42–51. [Google Scholar]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef]
- Piñar, G.; Ettenauer, J.; Sterflinger, K. La vie en rose: A review of rosy discoloration of subsurface monuments. Conserv. Subterr. Cult. Herit. 2014, 113–124. [Google Scholar]
- Pietrzak, K.; Puchalski, M.; Otlewska, A.; Wrzosek, H.; Guiamet, P.; Piotrowska, M.; Gutarowska, B. Microbial diversity of pre-Columbian archaeological textiles and the effect of silver nanoparticles misting disinfection. J. Cult. Herit. 2017, 23, 138–147. [Google Scholar] [CrossRef]
- Davis, C.P. Chapter 6 normal flora. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN -100-9631172-1-1. [Google Scholar]
- Scholz, C.F.; Kilian, M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Pécastaings, S.; Corvec, S.; Veraldi, S.; Khammari, A.; Roques, C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: A brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. 2018, 2, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Gharamti, A.A.; Kanafani, Z.A. Cutibacterium (formerly Propionibacterium) acnes infections associated with implantable devices. Expert Rev. Anti. Infect. Ther. 2017, 15, 1083–1094. [Google Scholar] [CrossRef]
- Hyde, E.R.; Haarmann, D.P.; Petrosino, J.F.; Lynne, A.M.; Bucheli, S.R. Initial insights into bacterial succession during human decomposition. Int. J. Legal. Med. 2014, 129, 661–671. [Google Scholar] [CrossRef]
- Philips, A.; Stolarek, I.; Kuczkowska, B.; Juras, A.; Handschuh, L.; Piontek, J.; Kozlowski, P.; Figlerowicz, M. Comprehensive analysis of microorganisms accompanying human archaeological remains. Gigascience 2017, 6, 1–13. [Google Scholar] [CrossRef]
- Nakaew, N.; Pathom-aree, W.; Lumyong, S. Generic diversity of rare actinomycetes from Thai cave soils and their possible use as new bioactive compounds. Actinomycetologica 2009, 23, 21–26. [Google Scholar] [CrossRef]
- De Leo, F.; Iero, A.; Zammit, G.; Urzı, C. Chemoorganotrophic bacteria isolated from biodeteriorated surfaces in cave and catacombs. Int. J. Speleol. 2012, 41, 125–136. [Google Scholar] [CrossRef]
- Diaz-Herraiz, M.; Jurado, V.; Cuezva, S.; Laiz, L.; Pallecchi, P.; Tiano, P.; Sanchez-Moral, S.; Saiz-Jimenez, C. The actinobacterial colonization of Etruscan paintings. Sci. Rep. 2013, 3, 1440. [Google Scholar] [CrossRef]
- Cirigliano, A.; Tomassetti, M.C.; Di Pietro, M.; Mura, F.; Maneschi, M.L.; Gentili, M.D.; Cardazzo, B.; Arrighi, C.; Mazzoni, C.; Negri, R.; et al. Calcite moonmilk of microbial origin in the Etruscan Tomba degli Scudi in Tarquinia, Italy. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Braun, B.; Richert, I.; Szewzyk, U. Detection of iron-depositing Pedomicrobium species in native biofilms from the Odertal National Park by a new, specific FISH probe. J. Microbiol. Methods 2009, 79, 37–43. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, H.; Li, X.; Hu, C.; Yang, M.; Qu, J. Characterization of biofilm and corrosion of cast iron pipes in drinking water distribution system with UV/Cl2 disinfection. Water Res. 2014, 60, 174–181. [Google Scholar] [PubMed]
- Lee, J.S.; Little, B.J. A mechanistic approach to understanding microbiologically influenced corrosion by metal-depositing bacteria. Corrosion 2019, 75, 6–11. [Google Scholar] [CrossRef]
- Männistö, M.K.; Häggblom, M.M. Characterization of psychrotolerant heterotrophic bacteria from Finnish Lapland. Syst. Appl. Microbiol. 2006, 29, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Baik, K.S.; Park, S.C.; Rhee, M.S.; Oh, H.M.; Seong, C.N. Roseomonas frigidaquae sp. nov., isolated from a water-cooling system. Int. J. Syst. Evol. Microbiol. 2009, 59, 1630–1634. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, X.; Wang, H.; Zhang, Y.; Hu, C.; Yang, M. Characterization of the bacterial communities and iron corrosion scales in drinking groundwater distribution systems with chlorine/chloramine. Int. Biodeterior. Biodegrad. 2014, 96, 71–79. [Google Scholar] [CrossRef]
- Urzì, C.; De Leo, F.; Bruno, L.; Pangallo, D.; Krakova, L. New species description, biomineralization processes and biocleaning applications of Roman Catacombs-living bacteria. Conservation Subterr. Cult. Herit. 2014, 65–72. [Google Scholar]
- Heyrman, J.; Logan, N.A.; Busse, H.-J.; Balcaen, A.; Lebbe, L.; Rodriguez-Diaz, M.; Swings, J.; De Vos, P. Virgibacillus carmonensis sp. nov., Virgibacillus necropolis sp. nov. and Virgibacillus picturae sp. nov., three novel species isolated from deteriorated mural paintings, transfer of the species of the genus Salibacillus to Virgibacillus, as Virgibacillus marismortui comb. nov. and Virgibacillus salexigens comb. nov., and emended description of the genus Virgibacillus. Int. J. Syst. Evol. Microbiol. 2003, 53, 501–511. [Google Scholar]
- Kour, D.; Rana, K.L.; Kaur, T.; Singh, B.; Chauhan, V.S.; Kumar, A.; Rastegari, A.A.; Yadav, N.; Yadav, A.N.; Gupta, V.K. Extremophiles for hydrolytic enzymes productions: Biodiversity and potential biotechnological applications. Bioprocess. Biomol. Prod. 2019, 321–372. [Google Scholar]
- Batra, R.; Boekhout, T.; Guého, E.; Cabañes, F.J.; Dawson, T.L., Jr.; Gupta, A.K. Malassezia baillon, emerging clinical yeasts. FEMS Yeast Res. 2005, 5, 1101–1113. [Google Scholar] [CrossRef]
- Chouvenc, T.; Efstathion, C.A.; Elliott, M.L.; Su, N.-Y. Resource competition between two fungal parasites in subterranean termites. Naturwissenschaften 2012, 99, 949–958. [Google Scholar] [CrossRef]
- Martínez-Ramírez, J.A.; Strien, J.; Sanft, J.; Mall, G.; Walther, G.; Peters, F.T. Studies on drug metabolism by fungi colonizing decomposing human cadavers. Part I: DNA sequence-based identification of fungi isolated from postmortem material. Anal. Bioanal. Chem. 2013, 405, 8443–8450. [Google Scholar] [CrossRef]
- Šimonovičová, A.; Kraková, L.; Pangallo, D.; Majorošová, M.; Piecková, E.; Bodoriková, S.; Dörnhoferová, M. Fungi on mummified human remains and in the indoor air in the Kuffner family crypt in Sládkovičovo (Slovakia). Int. Biodeterior. Biodegrad. 2015, 99, 157–164. [Google Scholar] [CrossRef]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 24, 47–55. [Google Scholar] [CrossRef]
- Strobel, G.; Ford, E.; Worapong, J.; Harper, J.K.; Arif, A.M.; Grant, D.M.; Fung, P.C.W.; Chau, R.M.W. Isopestacin, an isobenzofuranone from Pestalotiopsis microspora, possessing antifungal and antioxidant activities. Phytochemistry 2002, 60, 179–183. [Google Scholar] [CrossRef]
- Harper, J.K.; Arif, A.M.; Ford, E.J.; Strobel, G.A.; Porco, J.A., Jr.; Tomer, D.P.; Oneill, K.L.; Heider, E.M.; Grant, D.M. Pestacin: A 1,3-dihydro isobenzofuran from Pestalotiopsis microspora possessing antioxidant and antimycotic activities. Tetrahedron 2003, 59, 2471–2476. [Google Scholar] [CrossRef]
- Li, E.; Jiang, L.; Guo, L.; Zhang, H.; Che, Y. Pestalachlorides A–C, antifungal metabolites from the plant endophytic fungus Pestalotiopsis adusta. Bioorg. Med. Chem. 2008, 16, 7894–7899. [Google Scholar] [CrossRef]
- Piñar, G.; Poyntner, C.; Lopandic, K.; Tafer, H.; Sterflinger, K. Rapid diagnosis of biological colonization in cultural artefacts using the MinION nanopore sequencing technology. Int. Biodeterior. Biodegrad. 2020, 148, 104908. [Google Scholar] [CrossRef]
- Blanchette, R.A. A review of microbial deterioration found in archaeological wood from different environments. Int. Biodeterior. Biodegrad. 2000, 46, 189–204. [Google Scholar] [CrossRef]
- Brebbia, C.A. Structural Studies, Repairs and Maintenance of Heritage Architecture XI; Wessex Institute of Technology: Southampton, UK, 2009; ISBN 978-1-84564-196-2. [Google Scholar]
- Evidente, A.; Punzo, B.; Andolfi, A.; Cimmino, A.; Melck, D.; Luque, J. Lipophilic phytotoxins produced by Neofusicoccum parvum, a grapevine canker agent. Phytopathol. Mediterr. 2010, 49, 74–79. [Google Scholar]
- Williamson, B.; Tudzynski, B.; Tudzynsk, P.; Van Kan, J.A.L. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef] [PubMed]
- Benito, S. The impact of Torulaspora delbrueckii yeast in winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 3081–3094. [Google Scholar] [CrossRef] [PubMed]
- Zajc, J.; Kogej, T.; Galinski, E.A.; Ramos, J.; Gunde-Cimerman, N. Osmoadaptation strategy of the most halophilic fungus, Wallemia ichthyophaga, growing optimally at salinities above 15% NaCl. Appl. Environ. Microbiol. 2014, 80, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Urbina, H.; Aime, M.C. A closer look at Sporidiobolales: Ubiquitous microbial community members of plant and food biospheres. Mycologia 2018, 110, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Urzı, C.; De Leo, F. Sampling with adhesive tape strips: An easy and rapid method to monitor microbial colonization on monument surfaces. J. Microbiol. Methods 2001, 44, 1–11. [Google Scholar] [CrossRef]
- Hubka, V.; Kolařík, M.; Kubátová, A.; Peterson, S.W. Taxonomic revision of Eurotium and transfer of species to Aspergillus. Mycologia 2013, 105, 912–937. [Google Scholar] [CrossRef]
- Silva-Castro, G.A.; Uad, I.; Gonzalez-Martinez, A.; Rivadeneyra, A.; Gonzalez-Lopez, J.; Rivadeneyra, M.A. Bioprecipitation of calcium carbonate crystals by bacteria isolated from saline environments grown in culture media amended with seawater and real brine. BioMed Res. Int. 2015, 2015, 816102. [Google Scholar] [CrossRef]
- Dhami, N.K.; Mukherjee, A.; Watkin, E.L. Microbial diversity and mineralogical-mechanical properties of calcitic cave speleothems in natural and in vitro biomineralization conditions. Front. Microbiol. 2018, 9, 40. [Google Scholar] [CrossRef]
- Waugh, N. Corsets and Crinolines; Routledge: Abingdon-on-Thames, UK, 2017. [Google Scholar]
- Arnold, J.; Tiramani, J.; Costigliolo, L.; Passot, S.; Lucas, A.; Pietsch, J. Patterns of fashion 5: The Content, Cut, Construction and Context of Bodies, Stays, Hoops and Rumps c. 1595–1795; The School of Historical Dress: London, UK, 2018. [Google Scholar]
- Yang, H.X.; Chu, J.M.; Liu, X.S. Natural persistence of the coastal plant Glehnia littoralis along temperate sandy coasts. Sci. Rep. 2017, 7, 42784. [Google Scholar] [CrossRef]
- She, M.L.; Pu, F.T.; Pan, Z.H.; Watson, M.F.; Cannon, J.F.M.; Holmes-Smith, I.; Kljuykov, E.V.; Phillippe, L.R.; Pimenov, M.G. Apiaceae. In Flora of China Editorial Committee (ED), Flora of China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2005; Volume 14, pp. 1–205. [Google Scholar]
- Sayed-Ahmad, B.; Talou, T.; Saad, Z.; Hijazi, A.; Merah, O. The apiaceae: Ethnomedicinal family as source for industrial uses. Ind. Crop. Prod. 2017, 109, 661–671. [Google Scholar] [CrossRef]
- Beneš, J.; Čulíková, V.; Kosňovská, J.; Frolík, J.; Matiášek, J. New plants at Prague Castle and Hradčany in the early modern period: A history of selected species. Interdiscip. Archaeol. 2012, 3, 103–114. [Google Scholar] [CrossRef]
- Tranberg, A. 10 burial customs in the northern Ostrobothnian region (Finland) from the late medieval period to the 20th Century. Plant remains in graves. In The Archaeology of Death in Post-Medieval Europe; De Gruyter: Berlin, Germany, 2015; pp. 189–203. [Google Scholar]
- Jarosińska, M.; Nowak, S.; Noryśkiewicz, A.M.; Badura, M. Plant identification and significance in funeral traditions exemplified by pillow filling from a child crypt burial in Byszewo (18th/19th centuries). Analecta Archaeol. Ressoviensia 2019, 14, 187–197. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).