A Methodology for Open Information Extraction and Representation from Large Scientific Corpora: The CORD-19 Data Exploration Use Case
Abstract
Featured Application
Abstract
1. Introduction
2. Related Work
2.1. Advances in Coreference Resolution
2.2. Advances in Text Summarization
2.3. Advances in Open Information Extraction
2.4. Advances in Entity Linking, Enrichment and Representation
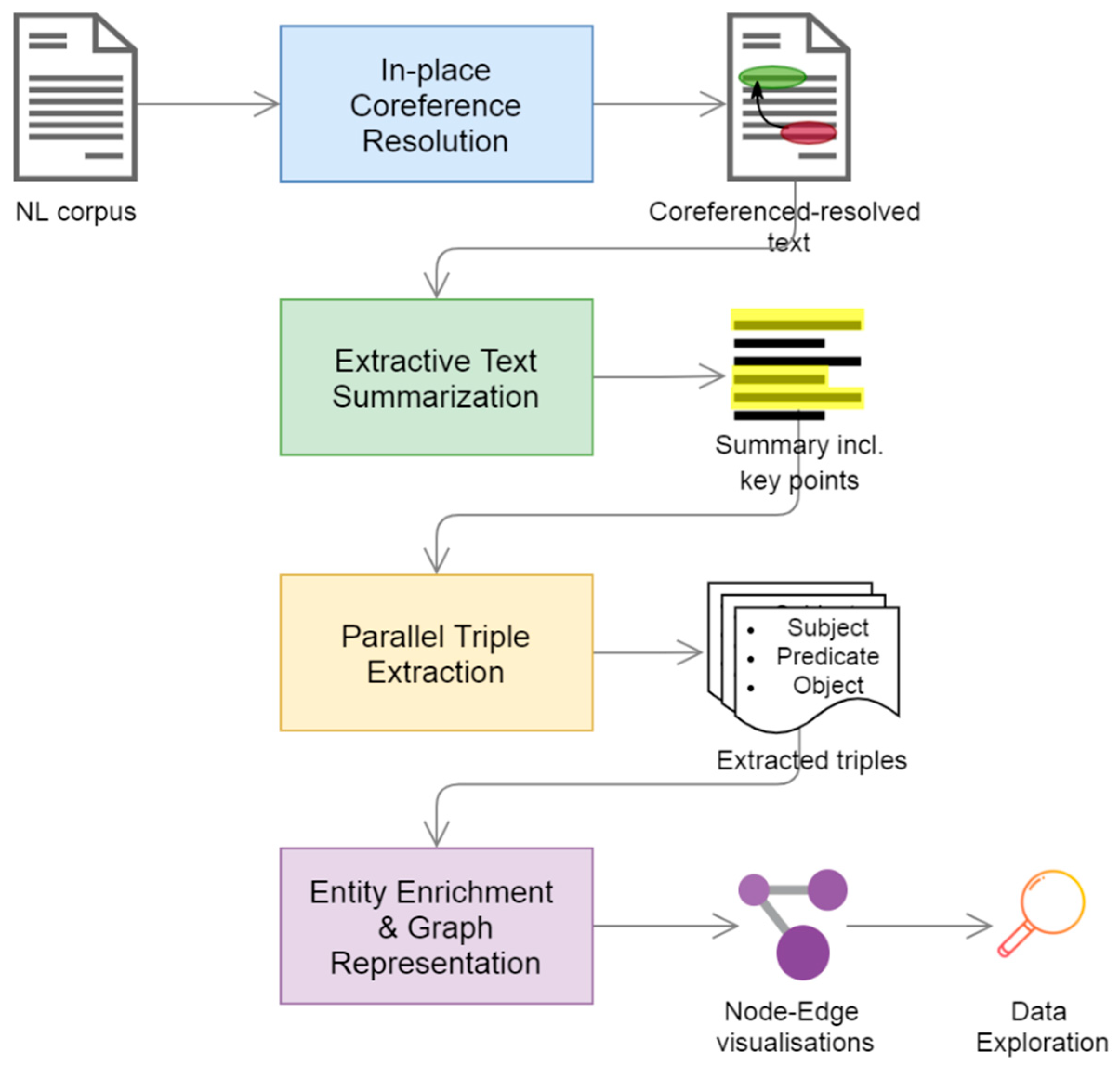
3. Materials and Methods
- an in-place neural coreference resolution component,
- an extractive text summarizer that isolates the key points of the ingested text,
- a parallel triple extraction component as our core information extraction method, and
- a toolkit of entity enrichment and representation techniques built around a graph engine.
3.1. Data
3.2. Implementation
3.2.1. In-Place Coreference Resolution
3.2.2. Extractive Text Summarization
3.2.3. Parallel Triple Extraction
- Open IE 5.1 from UW and IIT Delhi is a successor to the Ollie learning-based information extraction system [74]. The latest version is based on the combination of four different rule-based and learning-based OIE tools; namely CALMIE (specializing in triple extraction from conjunctive sentences) [75], RelNoun (for noun relations) [76], BONIE (for numerical sentences) [77], and SRLIE (based on semantic role labeling) [78].
- ClausIE from MPI follows a clause-based approach, first identifying the clause type of each sentence and then applying specific proposition extraction based on the corresponding grammatical function of the clause’s constituents. It also considers nested clauses as independent sentences. Because ClausIE detects useful pieces of information expressed in a sentence before representing them in terms of one or more extractions, it is especially useful in splitting complex sentences into many individual triples [41].
- AllenNLP OIE system formulates the triple extraction problem as a sequence BIO tagging problem and applies a bi-LSTM transducer to produce OIE tuples, which are grouped by each sentence’s predicate [72]. Given that it relies on supervised learning and contextualized word embeddings to produce independent probability distributions over possible BIO tags for each word, it has the potential of discovering richer and more complex relations. On the downside, it is not guaranteed that the neural sequence tagger will produce exactly two arguments for a given predicate (i.e., a subject and an object), thus complicating the triple extraction process.
3.2.4. Entity Enrichment & Graph Representation
- Entity linking: We leveraged the EntityLinker component from SciSpacy [79], a Python package containing models for processing biomedical, scientific, or clinical text. The component was used to perform a string overlap-based search (char-3grams) on named entities, comparing them with the concepts of the UMLS (Unified Medical Language System) knowledge base, using an approximate nearest neighbors search. The UMLS knowledge base contains over four million concepts along with additional information (e.g., definitions, hierarchies, concept–concept relations) from many health and biomedical vocabularies and standards (including CPT, ICD-10-CM, LOINC, MeSH, RxNorm, and SNOMED CT), enabling interoperability between computer systems [80]. For each triple subject or object with one or more entities linked to UMLS concepts, the coded concept name, concept description, and confidence score of the linking process was added along with the existing triple information. In order to address the ambiguity of biomedical terminology, we exploited the parametrization capabilities of the SciSpacy EntityLinker component, mainly by using the resolve_abbreviations parameter to resolve any abbreviations identified in the corpus before performing the linking and by tuning the threshold that a mention candidate must reach to be linked to a specific UMLS concept. Of course, this barely scratches the surface of biomedical terms disambiguation which remains a challenging task [81]. Overall, this entity enrichment process not only increases the contextual value of the extracted triples, but also facilitates the research for specific ontologies by mapping the existing entities with their normalized lexical variants (aliases).
- Polarity detection: By implementing polarity detection at sentence-level, we were able to classify triples that were inherently bearing a positive or a negative value. We relied on AllenNLP’s RoBERTa-based binary sentiment analyzer [63], which achieves 95.11% accuracy on the Stanford Sentiment Treebank test set.
- Triple cleaning: This process is aimed at reducing the redundant triples that resulted from the parallel triple extraction process, as described in Section 3.2.3, while also reducing the number of non-informative triples with little contextual value. To this end, we considered only “fully-linked” triples, i.e., triples whose both subject and object was linked to at least one UMLS concept. For those remaining triples, we additionally implemented a deduplication process to keep only the unique ones, based on the mentions of concepts of each sentence’s subjects and objects. In this manner, only one triple containing the same UMLS concepts was stored for each sentence.
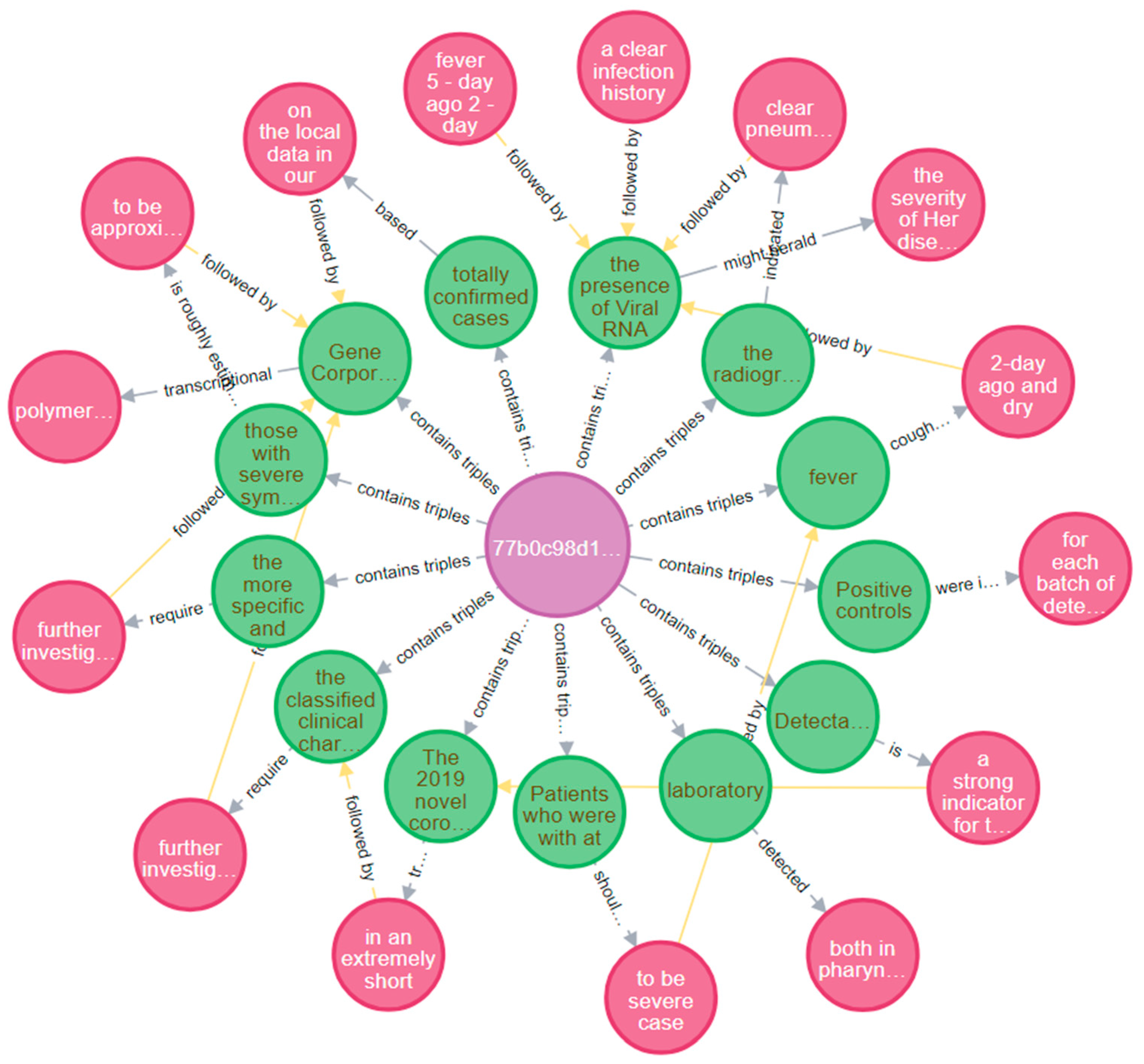
- Continuity representation: In order to enhance the readability of the extracted information, we implemented a script that connected the extracted triples with each other, based on the order of appearance in the original text. This way, the user is able to unravel the scope of the targeted content by interacting with its structured representation.
- Graph representation: It is the final step of our proposed pipeline, aiming at the practical interaction with the ingested data (e.g., visualizations and queries) that will facilitate data exploration tasks. Due to the rich internal structure that characterizes labeled property graphs, allowing each node or relationship to store more than one property (thus reducing the graph’s size), we used the Neo4J graph database based on labeled property graphs for storing, representing, and enriching the extracted relationships [82]. Neo4j is a Java-based graph database management system, described by its developers as an ACID-compliant transactional database with native graph storage and processing. It provides a web interface allowing relationships to be queried via CYPHER, a declarative graph query language that allows for expressive and efficient data querying in property graphs.
- Corpus nodes (purple): they signify the ingested text (e.g., scientific articles), are represented by the article_title and the unique article_id properties, and are connected to one or more triples via the contains triples one-directional edge,
- Subject nodes (green): they denote the subject of the extracted triple and are connected to one or more corpora (as mentioned above), as well as to one or more Objects, via predicate one-directional edges. The Subject node has the following properties: value: the natural language text of the subject; subj_entity_name: the name(s) of the entities linked to UMLS concepts; subj_entity_coded: the coded ID(s) of the entities linked to UMLS concepts; subj_entity_description: a short description of the linked entities; subj_entity_confidence: the reported confidence of the entity linking process; article_id: the unique ID of the extracted article; article_title: the title of the extracted article; sentence_text: the text of the processed sentence; sent_num: the serial number of the sentence in the whole corpus; triple_num: the serial number of the triple extracted from the given sentence; and engine: the OIE engine that from which the triple was extracted from.
- Object nodes (red): they denote the object of the extracted triple and are connected to their respective Subject through the predicate edge (as mentioned above), as well as with the “Subject” of the following triple in the corpus (if there exists one) via the followed by one-directional edge. Their properties are identical to those of the Subject nodes.
4. Results
4.1. Evaluation of the Information Extraction Process
4.2. Data Exploration Tasks
- The first task focuses on visualizing triples from the CORD-19 bibliography whose subject refers to the SARS-CoV-2 virus, the strain of coronavirus that causes the COVID-19 disease. The domain expert can either query the database for subjects containing the name of the virus or he/she can use the UMLS coded ID of the SARS-CoV-2 (C5203676), to acquire results containing all the corresponding aliases of the entity. These results are available in both tabular and graphical form (Table 5, Figure 4):
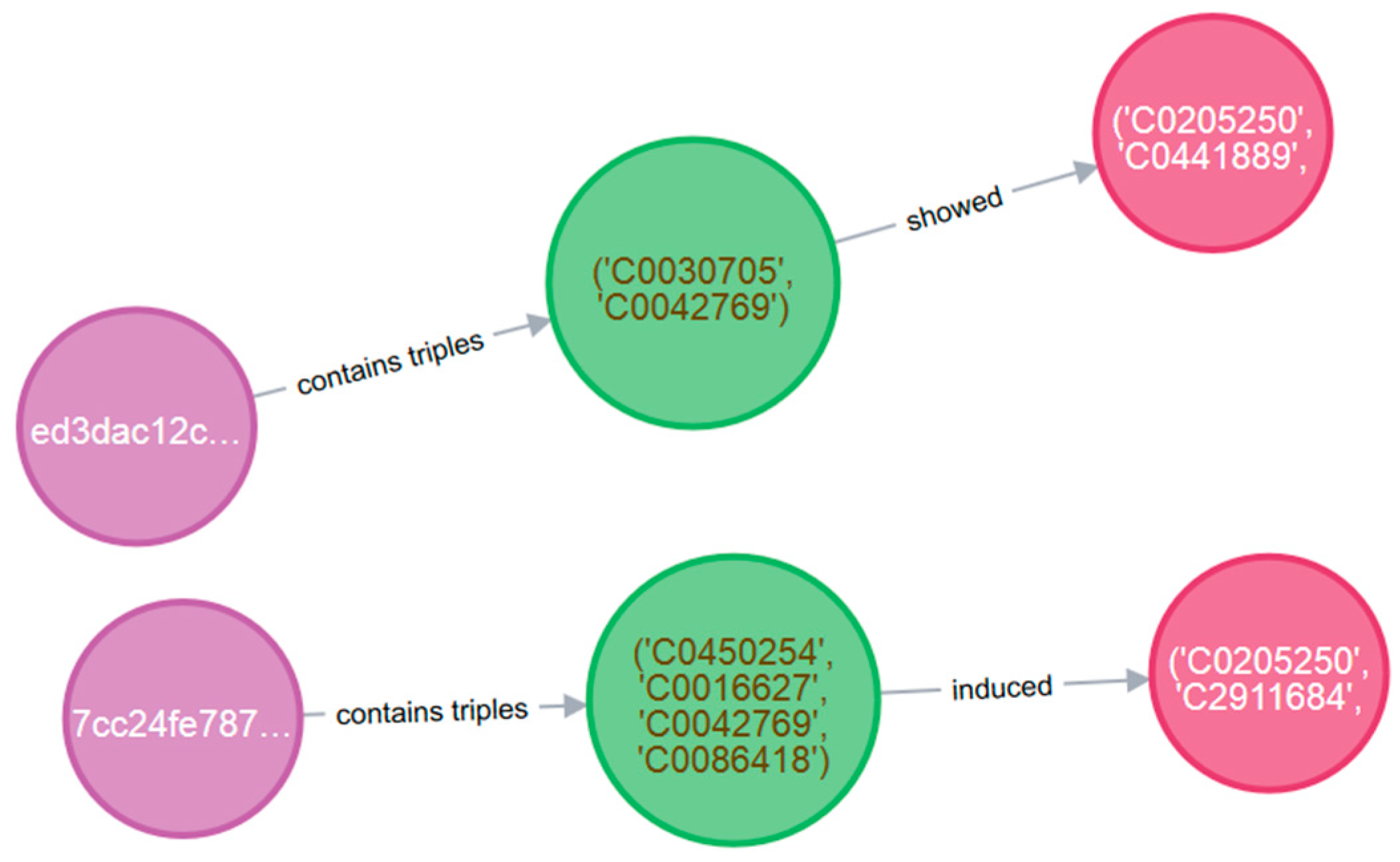
- The second task attempts to discover useful relationships regarding IL-6, a pleiotropic proinflammatory cytokine that is found in increased levels in COVID-19 patients and similar viruses. By performing a targeted search on subjects containing the “virus” entity and objects containing the “IL-6” entity, we get the results shown in Table 6 (in natural language form) and Figure 5 (as UMLS coded entities). This time, we are also interested in the title of the scientific article from where the triple was extracted.
- The final data exploration task allows us to focus on one of the articles and exploit the continuity representation functionality (followed by edges) to traverse through the generated triples. The result is akin to a graphical summary of the processed article. The generated chain consists of alternating subject/object nodes, depicting the sequence of their appearance in the original text (Figure 6).
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| CORD-19 Article ID: d99dbae98cc9705d9b5674bb6eb66560b4434305 |
|---|
| The current epidemic of a new coronavirus disease (COVID-19), caused by a novel coronavirus (2019-nCoV), recently officially named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has reopened the issue of the role and importance of coronaviruses in human pathology (1) (2) (3) (4) (5). This epidemic definitively confirms that this heretofore relatively harmless family of viruses, Coronaviridae, includes major pathogens of epidemic potential. The COVID-19 epidemic has clearly demonstrated the power of infectious diseases, which have been responsible for many devastating epidemics throughout history. The epidemiological potential of emerging infectious diseases, especially zoonoses, is affected by numerous environmental, epidemiological, social, and economic factors (6, 7). Emerging zoonoses pose both epidemiological and clinical challenges to health care professionals. Since the 1960s, coronaviruses have caused a wide variety of human and animal diseases. In humans, they cause up to a third of all community-acquired upper respiratory tract infections, such as the common cold, pharyngitis, and otitis media. However, more severe forms of bronchiolitis, exacerbations of asthma, and pneumonia in children and adults have also been described, sometimes with fatal outcomes in infants, the elderly, and the immunocompromised. Some coronaviruses are associated with gastrointestinal disease in children. Sporadic infections of the central nervous system have also been reported, although the role of coronaviruses in infections outside the respiratory tract has not been completely clarified (8). Most coronaviruses are adapted to their hosts, whether animal or human, although cases of possible animal-to-hu-man transmission and adaptation have been described in the past two decades, causing two epidemics. The first such outbreak originated in Guangdong, a southern province of the People’s Republic of China, in mid-November of 2002. The disease was named severe acute respiratory syndrome (SARS). The cause was shown to be a novel coronavirus (SARS-CoV), an animal virus that had crossed the species barrier and infected humans. The most likely reservoir was bats, with evidence that the virus was transmitted to a human through an intermediate host, probably a palm civet or raccoon dog (8, 9). In less than a year, SARS-CoV infected 8098 people in 26 countries, of whom 774 died (10, 11). Approximately 25% of the patients developed organ failure, most often acute respiratory distress syndrome (ARDS), requiring admission to an intensive care unit (ICU), while the case fatality rate (CFR) was 9.6%. However, in elderly patients (>60 years), the CFR was over 40%. Poor outcomes were seen in patients with certain comorbidities (diabetes mellitus and hepatitis B virus infection), patients with atypical symptoms, and those with elevated lactic acid dehydrogenase (LDH) values on admission. Interestingly, the course of the disease was biphasic in 80% of the cases, especially those with severe clinical profiles, suggesting that immunological mechanisms, rather than only the direct action of SARS-CoV, are responsible for some of the complications and fatal outcomes (8, 9). Approximately 20% of the reported cases during this epidemic were health care workers. Therefore, in addition to persons exposed to animal sources and infected family members, health care workers were among the most heavily exposed and vulnerable individuals (9, 10). During 2004, three minor outbreaks were described among laboratory personnel engaged in coronavirus research. Although several secondary cases, owing to close personal contact with infected patients, were described, there was no further spread of the epidemic. It is not clear how the SARS-CoV eventually disappeared and if it still circulates in nature among animal reservoirs. Despite ongoing surveillance, there have been no reports of SARS in humans worldwide since mid-2004 (11). In the summer of 2012, another epidemic caused by a novel coronavirus broke out in the Middle East. The disease, often complicated with respiratory and renal failure, was called Middle East respiratory syndrome (MERS), while the novel coronavirus causing it was called Middle East respiratory syndrome coronavirus (MERS-CoV). Although a coronavirus, it is not related to the coronaviruses previously described as human pathogens. However, it is closely related to a coronavirus isolated from dromedary camels and bats, which are considered the primary reservoirs, albeit not the only ones (8, 12). From 2012 to the end of January 2020, over 2500 laboratory-confirmed MERS cases, including 866 associated deaths, were reported worldwide in 27 countries (13). The largest number of such cases has been reported among the elderly, diabetics, and patients with chronic diseases of the heart, lungs, and kidneys. Over 80% of the patients required admission to the ICU, most often due to the development of ARDS, respiratory insufficiency requiring mechanical ventilation, acute kidney injury, or shock. The CFR is around 35%, and even 75% in patients >60 years of age. However, MERS-CoV, unlike its predecessor SARS-CoV, did not disappear, but still circulates among animal and human populations, occasionally causing outbreaks, either in connection with exposure to camels or infected persons (12). Overall, 19.1% of all MERS cases have been among health care workers, and more than half of all laboratory-confirmed secondary cases were transmitted from human to human in health care settings, at least in part due to shortcomings in infection prevention and control (12, 13). Post-exposure prophylaxis with ribavirin and lopinavir/ritonavir decreased the MERS-CoV risk in health care workers by 40% (14). THe eMeRGence oF covId-19 cAused by sARs-cov-2In mid-December of 2019, a pneumonia outbreak erupted once again in China, in the city of Wuhan, the province of Hube (1). The outbreak spread during the next two months throughout the country, with currently over 80 000 cases and more than 2400 fatal outcomes (CFR 2.5%), according to official reports. Exported cases have been reported in 30 countries throughout the world, with over 2400 registered cases, of which 276 are in Europe. On February 25, the first case of COVID-19 was confirmed in Zagreb, Croatia, and was linked to the current outbreak in the Lombardy and Veneto regions of northern Italy (15). The case definition was first established on January 10 and modified over time, taking into account both the virus epidemiology and clinical presentation. The clinical criteria were expanded on February 4 to include any lower acute respiratory diseases, and the epidemiological criterion was extended to the whole of China, with the possibility of expansion to some surrounding countries (16, 17). At the early stage of the outbreak, patients’ full-length genome sequences were identified, showing that the virus shares 79.5% sequence identity with SARS-CoV. Furthermore, 96% of its whole genome is identical to bat coronavirus. It was also shown that this virus uses the same cell entry receptor, ACE2, as SARS-CoV (18). The full clinical spectrum of COVID-19 ranges from asymptomatic cases, mild cases that do not require hospitalization, to severe cases that require hospitalization and ICU treatment, and those with fatal outcomes. Most cases were classified as mild (81%), 14% as severe, and 5% as critical (ie, respiratory failure, septic shock, and/or multiple organ dysfunction or failure). The overall CFR was 2.3%, while the rate in patients with comorbidities was considerably higher −10.5% for cardiovascular disease, 7.3% for diabetes, 6.3% for chronic respiratory diseases, 6.0% for hypertension, and 5.6% for cancer. The CFR in critical patients was as high as 49.0% (4).It is still not clear which factors contribute to the risk of transmitting the infection, especially by persons who are in the incubation stage or asymptomatic, as well as which factors contribute to the severity of the disease and fatal outcome. Evidence from various types of additional studies is needed to control the epidemic (19). However, it is certain that the binding of the virus to the ACE 2 receptor can induce certain immunoreactions, and the receptor diversity between humans and animal species designated as SARS-CoV-2 reservoirs further increases the complexity of COVID-19 immunopathogenicity (20). Recently, a diagnostic RT-PCR assay for the detection of SARS-CoV-19 has been developed using synthetic nucleic acid technology, despite the lack of virus isolates and clinical samples, owing to its close relation to SARS. Additional diagnostic tests are in the pipeline, some of which are likely to become commercially available soon (21). Currently, randomized controlled trials have not shown any specific antiviral treatment to be effective for COVID-19. Therefore, treatment is based on symptomatic and supportive care, with intensive care measures for the most severe cases (22). However, many forms of specific treatment are being tried, with various results, such as with remdesivir, lopinavir/ritonavir, chloroquine phosphate, convalescent plasma from patients who have recovered from COVID-19, and others (23) (24) (25) (26). No vaccine is currently available, but researchers and vaccine manufacturers have been attempting to develop the best option for COVID-19 prevention. So far, the basic target molecule for the production of a vaccine, as well as therapeutic antibodies, is the CoV spike (S) glycoprotein (27, 28). The spread of the epidemic can only be contained and SARS-CoV-2 transmission in hospitals by strict compliance with infection prevention and control measures (contact, droplet, and airborne precautions) (22, 29). During the current epidemic, health care workers have been at an increased risk of contracting the disease and consequent fatal outcome owing to direct exposure to patients. Early reports from the beginning of the epidemic indicated that a large proportion of the patients had contracted the infection in a health care facility (as high as 41%), and that health care workers constituted a large proportion of these cases (as high as 29%). However, the largest study to date on more than 72 000 patients from China has shown that health care workers make up 3.8% of the patients. In this study, although the overall CFR was 2.3%, among health care workers it was only 0.3%. In China, the number of severe or critical cases among health care workers has declined overall, from 45.0% in early January to 8.7% in early February (4). This poses numerous psychological and ethical questions about health care workers’ role in the spread, eventual arrest, and possible consequences of epidemics. For example, during the 2014-2016 Ebola virus disease epidemic in Africa, health care workers risked their lives in order to perform life-saving invasive procedures (intravenous indwelling, hemodialysis, reanimation procedures, mechanical ventilation), and suffered high stress and fatigue levels, which may have prevented them from practicing optimal safety measures, sometimes with dire consequences (30). This third coronavirus epidemic, caused by the highly pathogenic SARS-CoV-2, underscores the need for the ongoing surveillance of infectious disease trends throughout the world. The examples of pandemic influenza, avian influenza, but also the three epidemics caused by the novel coronaviruses, indicate that respiratory infections are a major threat to humanity. Although Ebola virus disease and avian influenza are far more contagious and influenza currently has a greater epidemic potential, each of the three novel coronaviruses require urgent epidemiologic surveillance. Many infectious diseases, such as diphtheria, measles, and whooping cough, have been largely or completely eradicated or controlled through the use of vaccines. It is hoped that developments in vaccinology and antiviral treatment, as well as new preventive measures, will ultimately vanquish this and other potential threats from infectious diseases in the future. |
References
- Augenstein, I.; Padó, S.; Rudolph, S. LODifier: Generating Linked Data from Unstructured Text BT—The Semantic Web: Research and Applications; Simperl, E., Cimiano, P., Polleres, A., Corcho, O., Presutti, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 210–224. [Google Scholar]
- Clancy, R.; Ilyas, I.F.; Lin, J. Knowledge Graph Construction from Unstructured Text with Applications to Fact Verification and Beyond. In Proceedings of the Second Workshop on Fact Extraction and VERification (FEVER), Hong Kong, China, 3–7 November 2019; pp. 39–46. [Google Scholar]
- Kríž, V.; Hladká, B.; Nečaský, M.; Knap, T. Data Extraction Using NLP Techniques and Its Transformation to Linked Data BT—Human-Inspired Computing and Its Applications; Gelbukh, A., Espinoza, F.C., Galicia-Haro, S.N., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 113–124. [Google Scholar]
- Pertsas, V.; Constantopoulos, P.; Androutsopoulos, I. Ontology Driven Extraction of Research Processes BT—The Semantic Web—ISWC; Vrandečić, D., Bontcheva, K., Suárez-Figueroa, M.C., Presutti, V., Celino, I., Sabou, M., Kaffee, L.-A., Simperl, E., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 162–178. [Google Scholar]
- Exner, P.; Nugues, P. Entity Extraction: From Unstructured Text to DBpedia RDF triples. In Proceedings of the WoLE@ISWC, Boston, MA, USA, 11–15 November 2012. [Google Scholar]
- Holzinger, A.; Kieseberg, P.; Weippl, E.; Tjoa, A.M. Current advances, trends and challenges of machine learning and knowledge extraction: From machine learning to explainable AI. In International Cross-Domain Conference for Machine Learning and Knowledge Extraction; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Swizterland, 2018. [Google Scholar]
- Xiao, Z.; de Silva, T.N.; Mao, K. Evolving Knowledge Extraction from Online Resources. Int. J. Comput. Syst. Eng. 2017, 11, 746–752. [Google Scholar] [CrossRef]
- Makrynioti, N.; Grivas, A.; Sardianos, C.; Tsirakis, N.; Varlamis, I.; Vassalos, V.; Poulopoulos, V.; Tsantilas, P. PaloPro: A platform for knowledge extraction from big social data and the news. Int. J. Big Data Intell. 2017. [Google Scholar] [CrossRef]
- Wu, H.; Lei, Q.; Zhang, X.; Luo, Z. Creating A Large-Scale Financial News Corpus for Relation Extraction. In Proceedings of the 2020 3rd International Conference on Artificial Intelligence and Big Data (ICAIBD), Chengdu, China, 28–31 May 2020; pp. 259–263. [Google Scholar]
- Amith, M.; Song, H.Y.; Zhang, Y.; Xu, H.; Tao, C. Lightweight predicate extraction for patient-level cancer information and ontology development. BMC Med. Inform. Decis. Mak. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Q.; Ding, X.; Zhang, G.; Weng, L.; Ding, M. A New Method for Complex Triplet Extraction of Biomedical Texts. In International Conference on Knowledge Science, Engineering and Management; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2019. [Google Scholar]
- Haihong, E.; Xiao, S.; Song, M. A text-generated method to joint extraction of entities and relations. Appl. Sci. 2019, 9, 3795. [Google Scholar] [CrossRef]
- Kertkeidkachorn, N.; Ichise, R. T2KG: An End-to-End System for Creating Knowledge Graph from Unstructured Text. In Proceedings of the AAAI Workshops, San Francisco, CA, USA, 4–5 February 2017. [Google Scholar]
- Freitas, A.; Carvalho, D.S.; Da Silva, J.C.P.; O’Riain, S.; Curry, E. A semantic best-effort approach for extracting structured discourse graphs from wikipedia. In Proceedings of the CEUR Workshop Proceedings, Boston, MA, USA, 11–15 November 2012. [Google Scholar]
- Blomqvist, E.; Hose, K.; Paulheim, H.; Lawrynowicz, A.; Ciravegna, F.; Hartig, O. The Semantic Web: ESWC 2017 Satellite Events. In Proceedings of the ESWC 2017 Satellite Events, Portorož, Slovenia, 28 May–1 June 2017. [Google Scholar]
- Elango, P. Coreference Resolution: A Survey; Technical Report; UW-Madison: Madison, WI, USA, 2006. [Google Scholar]
- Kantor, B.; Globerson, A. Coreference resolution with entity equalization. In Proceedings of the ACL 2019—57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019. [Google Scholar]
- Hobbs, J.R. Resolving pronoun references. Lingua 1978. [Google Scholar] [CrossRef]
- Grosz, B.; Weinstein, S.; Joshi, A. Centering: A Framework for Modeling the Local Coherence of Discourse. Comput. Linguist. 1995, 21, 203–225. [Google Scholar]
- Wiseman, S.; Rush, A.M.; Shieber, S.M.; Weston, J. Learning anaphoricity and antecedent ranking features for coreference resolution. In Proceedings of the ACL-IJCNLP 2015—53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing of the Asian Federation of Natural Language Processing, Beijing, China, 26–31 July 2015. [Google Scholar]
- Clark, K.; Manning, C.D. Deep Reinforcement Learning for Mention-Ranking Coreference Models. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 2256–2262. [Google Scholar]
- Lee, K.; He, L.; Lewis, M.; Zettlemoyer, L. End-to-end Neural Coreference Resolution. In Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, Copenhagen, Denmark, 7–11 September 2017; pp. 188–197. [Google Scholar]
- Joshi, M.; Chen, D.; Liu, Y.; Weld, D.S.; Zettlemoyer, L.; Levy, O. SpanBERT: Improving Pre-training by Representing and Predicting Spans. Trans. Assoc. Comput. Linguist. 2020, 8, 64–77. [Google Scholar] [CrossRef]
- Andhale, N.; Bewoor, L.A. An overview of Text Summarization techniques. In Proceedings of the 2016 International Conference on Computing Communication Control and Automation (ICCUBEA), Pune, India, 12–13 August 2016; pp. 1–7. [Google Scholar]
- Allahyari, M.; Pouriyeh, S.A.; Assefi, M.; Safaei, S.; Trippe, E.D.; Gutierrez, J.B.; Kochut, K.J. Text Summarization Techniques: A Brief Survey. arXiv 2017, arXiv:abs/1707.02268. [Google Scholar] [CrossRef]
- Nallapati, R.; Zhou, B.; dos Santos, C.; Gulçehre, Ç.; Xiang, B. Abstractive text summarization using sequence-to-sequence RNNs and beyond. In Proceedings of the CoNLL 2016—20th SIGNLL Conference on Computational Natural Language Learning, Proceedings, Berlin, Germany, 11–12 August 2016. [Google Scholar]
- Nallapati, R.; Xiang, B.; Zhou, B.; Question, W.; Algorithms, A.; Heights, Y. Sequence-To-Sequence Rnns For Text Summarization. In Proceedings of the International Conference on Learning Representations, ICLR 2016 - Workshop Track, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Kouris, P.; Alexandridis, G.; Stafylopatis, A. Abstractive Text Summarization Based on Deep Learning and Semantic Content Generalization. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July 2019; pp. 5082–5092. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Hoang, A.; Bosselut, A.; Çelikyilmaz, A.; Choi, Y. Efficient Adaptation of Pretrained Transformers for Abstractive Summarization. arXiv 2019, arXiv:1906.00138. [Google Scholar]
- Filippova, K.; Altun, Y. Overcoming the lack of parallel data in sentence compression. In Proceedings of the EMNLP 2013—2013 Conference on Empirical Methods in Natural Language Processing, Seattle, WA, USA, 18–21 October 2013. [Google Scholar]
- Colmenares, C.A.; Litvak, M.; Mantrach, A.; Silvestri, F. HEADS: Headline generation as sequence prediction using an abstract: Feature-rich space. In Proceedings of the NAACL HLT 2015—2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Denver, CO, USA, 31 May–5 June 2015. [Google Scholar]
- Shimada, A.; Okubo, F.; Yin, C.; Ogata, H. Automatic Summarization of Lecture Slides for Enhanced Student Preview-Technical Report and User Study. IEEE Trans. Learn. Technol. 2018. [Google Scholar] [CrossRef]
- Hassel, M. Exploitation of Named Entities in Automatic Text Summarization for Swedish. In Proceedings of the NODALIDA’03–14th Nordic Conferenceon Computational Linguistics, Reykjavik, Iceland, 30–31 May 2003. [Google Scholar]
- Pal, A.R.; Saha, D. An approach to automatic text summarization using WordNet. In Proceedings of the 2014 IEEE International Advance Computing Conference (IACC), Gurgaon, India, 21–22 February 2014; pp. 1169–1173. [Google Scholar]
- Miller, D. Leveraging BERT for Extractive Text Summarization on Lectures. arXiv 2019, arXiv:1906.04165. [Google Scholar]
- Wang, Q.; Liu, P.; Zhu, Z.; Yin, H.; Zhang, Q.; Zhang, L. A Text Abstraction Summary Model Based on BERT Word Embedding and Reinforcement Learning. Appl. Sci. 2019, 9, 4701. [Google Scholar] [CrossRef]
- Zheng, H.-T.; Guo, J.-M.; Jiang, Y.; Xia, S.-T. Query-Focused Multi-document Summarization Based on Concept Importance. In Advances in Knowledge Discovery and Data Mining; Bailey, J., Khan, L., Washio, T., Dobbie, G., Huang, J.Z., Wang, R., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 443–453. [Google Scholar]
- Gupta, S.; Gupta, S.K. Abstractive summarization: An overview of the state of the art. Expert Syst. Appl. 2019, 121, 49–65. [Google Scholar] [CrossRef]
- Jurafsky, D.; Martin, J.H. Speech and Language Processing: An introduction to speech recognition, computational linguistics and natural language processing. In Language; Prentice Hall: Upper Saddle River, NJ, USA, 2007; ISBN 0131873210. [Google Scholar]
- Niklaus, C.; Cetto, M.; Freitas, A.; Handschuh, S. A Survey on Open Information Extraction. In Proceedings of the 27th International Conference on Computational Linguistics, Barcelona, Spain, 8–13 December 2018; pp. 3866–3878. [Google Scholar]
- Fader, A.; Soderland, S.; Etzioni, O. Identifying relations for Open Information Extraction. In Proceedings of the EMNLP 2011—Conference on Empirical Methods in Natural Language Processing, Edinburgh, UK, 27–31 July 2011. [Google Scholar]
- Mesquita, F.; Schmidek, J.; Barbosa, D. Effectiveness and efficiency of open relation extraction. In Proceedings of the EMNLP 2013–2013 Conference on Empirical Methods in Natural Language Processing, Seattle, WA, USA, 18–21 October 2013. [Google Scholar]
- Banko, M.; Cafarella, M.J.; Soderland, S.; Broadhead, M.; Etzioni, O. Open information extraction from the web. In Proceedings of the IJCAI International Joint Conference on Artificial Intelligence, Hyderabad, India, 6–12 January 2007. [Google Scholar]
- Wu, F.; Weld, D.S. Open Information Extraction Using Wikipedia. In Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, Uppsala, Sweden, 11–16 July 2010; pp. 118–127. [Google Scholar]
- Weld, D.S.; Hoffmann, R.; Wu, F. Using Wikipedia to bootstrap open information extraction. Acm Sigmod Rec 2009, 37, 62–68. [Google Scholar] [CrossRef]
- Del Corro, L.; Gemulla, R. ClausIE: Clause-Based Open Information Extraction. In Proceedings of the 22nd International Conference on World Wide Web; Association for Computing Machinery: New York, NY, USA, 2013; pp. 355–366. [Google Scholar]
- Angeli, G.; Premkumar, M.J.; Manning, C.D. Leveraging linguistic structure for open domain information extraction. In Proceedings of the ACL-IJCNLP 2015—53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing of the Asian Federation of Natural Language Processing, Beijing, China, 26–31 July 2015. [Google Scholar]
- Yang, Z.; Salakhutdinov, R.; Cohen, W.W. Transfer Learning for Sequence Tagging with Hierarchical Recurrent Networks. arXiv 2017, arXiv:1703.06345. [Google Scholar]
- Liu, Y.; Zhang, T.; Liang, Z.; Ji, H.; McGuinness, D.L. Seq2RDF: An End-to-end Application for Deriving Triples from Natural Language Text. arXiv 2018, arXiv:abs/1807.0. [Google Scholar]
- He, L.; Lee, K.; Lewis, M.; Zettlemoyer, L. Deep semantic role labeling: What works and what’s next. In Proceedings of the ACL 2017—55th Annual Meeting of the Association for Computational Linguistics, Vancouver, BC, Canada, 30 July–4 August 2017. [Google Scholar]
- Stanovsky, G.; Michael, J.; Zettlemoyer, L.; Dagan, I. Supervised open information extraction. In Proceedings of the NAACL HLT 2018—2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, New Orleans, LA, USA, 1–6 June 2018. [Google Scholar]
- Auer, S.; Bizer, C.; Kobilarov, G.; Lehmann, J.; Cyganiak, R.; Ives, Z. DBpedia: A nucleus for a Web of open data. In The Semantic Web; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Bollacker, K.; Evans, C.; Paritosh, P.; Sturge, T.; Taylor, J. Freebase: A collaboratively created graph database for structuring human knowledge. In Proceedings of the ACM SIGMOD International Conference on Management of Data, Vancouver, BC, Canada, 9–12 June 2008. [Google Scholar]
- Suchanek, F.M.; Kasneci, G.; Weikum, G. Yago: A core of semantic knowledge. In Proceedings of the 16th International World Wide Web Conference, Banff, AB, Canada, 8–12 May 2007. [Google Scholar]
- Mena, E.; Kashyap, V.; Illarramendi, A.; Sheth, A. Domain Specific Ontologies for Semantic Information Brokering on the Global Information Infrastructure. In Proceedings of the Formal Ontology in Information Systems: Proceedings of FOIS’98, Trento, Italy, 6–8 June 1998. [Google Scholar]
- Karadeniz, I.; Özgür, A. Linking entities through an ontology using word embeddings and syntactic re-ranking. BMC Bioinform. 2019. [Google Scholar] [CrossRef]
- Cho, H.; Choi, W.; Lee, H. A method for named entity normalization in biomedical articles: Application to diseases and plants. BMC Bioinform. 2017. [Google Scholar] [CrossRef]
- Papadakis, G.; Tsekouras, L.; Thanos, E.; Giannakopoulos, G.; Palpanas, T.; Koubarakis, M. Domain- and Structure-Agnostic End-to-End Entity Resolution with JedAI. SIGMOD Rec. 2020, 48, 30–36. [Google Scholar] [CrossRef]
- Pang, B.; Lee, L. Opinion mining and sentiment analysis. Found. Trends. Inf. Retr. 2008, 2, 1–135. [Google Scholar] [CrossRef]
- Medhat, W.; Hassan, A.; Korashy, H. Sentiment analysis algorithms and applications: A survey. Ain Shams Eng. J. 2014. [Google Scholar] [CrossRef]
- Sentiment Classification using Machine Learning Techniques. Int. J. Sci. Res. 2016. [CrossRef]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019, arXiv:abs/1907.1. [Google Scholar]
- McBride, B. The Resource Description Framework (RDF) and Its Vocabulary Description Language RDFS BT—Handbook on Ontologies; Staab, S., Studer, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 51–65. ISBN 978-3-540-24750-0. [Google Scholar]
- Angles, R. The Property Graph Database Model. In Proceedings of the AMW, Cali, Colombia, 21–25 May 2018. [Google Scholar]
- Kohlmeier, S.; Lo, K.; Wang, L.L.; Yang, J.J. COVID-19 Open Research Dataset (CORD-19). 2020. Available online: https://pages.semanticscholar.org/coronavirus-research (accessed on 18 April 2020).
- Allen Institute for AI Coreference Resolution Demo. Available online: https://demo.allennlp.org/coreference-resolution (accessed on 18 April 2020).
- Weischedel, R.; Palmer, M.; Marcus, M.; Hovy, E.; Pradhan, S.; Ramshaw, L.; Xue, N.; Taylor, A.; Kaufman, J.; Franchini, M.; et al. OntoNotes Release 5.0 LDC2013T19. Linguist. Data Consort. 2013. [Google Scholar] [CrossRef]
- Beltagy, I.; Lo, K.; Cohan, A. {S}ci{BERT}: A Pretrained Language Model for Scientific Text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China, 3–7 November 2019; pp. 3615–3620. [Google Scholar]
- University of Washington; Indian Institute of Technology Open IE 5.1. Available online: https://github.com/dair-iitd/OpenIE-standalone (accessed on 10 June 2020).
- Max Planck Institute for Informatics ClausIE: Clause-Based Open Information Extraction. Available online: https://www.mpi-inf.mpg.de/departments/databases-and-information-systems/software/clausie/ (accessed on 18 April 2020).
- Allen Institute for AI Open Information Extraction. Available online: https://demo.allennlp.org/open-information-extraction (accessed on 18 April 2020).
- Schmitz, M.; Bart, R.; Soderland, S.; Etzioni, O. Open language learning for information extraction. In Proceedings of the EMNLP-CoNLL 2012—2012 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning, Jeju Island, Korea, 12–14 July 2012. [Google Scholar]
- Saha, S. Mausam Open Information Extraction from Conjunctive Sentences. In Proceedings of the 27th International Conference on Computational Linguistics, Barcelona, Spain, 8–13 December 2018. [Google Scholar]
- Pal, H.-M. Demonyms and Compound Relational Nouns in Nominal Open IE. In Proceedings of the 5th ACL Workshop on Automated Knowledge Base Construction, San Diego, CA, USA, 12–17 June 2018. [Google Scholar]
- Saha, S.; Pal, H. Mausam Bootstrapping for numerical open IE. In Proceedings of the ACL 2017—55th Annual Meeting of the Association for Computational Linguistics, Vancouver, BC, Canada, 30 July–4 August 2017. [Google Scholar]
- Christensen, J.; Soderland, S.; Etzioni, O. An Analysis of Open Information Extraction Based on Semantic Role Labeling Categories and Subject Descriptors. In Proceedings of the Sixth International Conference on Knowledge Capture, Banff, AB, Canada, 25–29 July 2011; pp. 113–120. [Google Scholar]
- Neumann, M.; King, D.; Beltagy, I.; Ammar, W. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. arXiv 2019, arXiv:1902.07669. [Google Scholar]
- Bodenreider, O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004. [Google Scholar] [CrossRef]
- Kormilitzin, A.; Vaci, N.; Liu, Q.; Nevado-Holgado, A. Med7: A transferable clinical natural language processing model for electronic health records. arXiv 2020, arXiv:2003.01271. [Google Scholar]
- Neo4j Neo4j—The Leader in Graph Databases. Available online: https://neo4j.com/ (accessed on 18 April 2020).
- Gotti, F.; Langlais, P. Weakly Supervised, Data-Driven Acquisition of Rules for Open Information Extraction. In Canadian Conference on Artificial Intelligence; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2019. [Google Scholar]
- Yuan, S.; Yu, B. An Evaluation of Information Extraction Tools for Identifying Health Claims in News Headlines. In Proceedings of the Workshop Events and Stories in the News, Santa Fe, NM, USA, 21–25 August 2018. [Google Scholar]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database: Update 2019. Nucleic Acids Res. 2018, 47, D948–D954. [Google Scholar] [CrossRef]
- Lever, J.; Jones, S.J. VERSE: Event and Relation Extraction in the BioNLP 2016 Shared Task. In Proceedings of the 4th BioNLP Shared Task Workshop, Berlin, Germany, 13 August 2016. [Google Scholar]



| Article ID | Extract |
|---|---|
| 42e321eedb a25d380ae4 4d16cdf0bb deab83d665 | Two articles in the top ten cited articles discussed the emergence of New Delhi metallo-β-lactamase (NDM) gene{1} responsible for carbapenem resistance. |
| 85eb641e06 b0d6b1a0b2 02275add0c 5d27e53d71 | Official health linkages{1} have served to promote good will in some otherwise difficult relationships, as has been the case with Indonesia. |
| 4c84dbfd01 f7b2009ebe d54376da8 afcbcf1ec64 | However, one should also note that the experiment is based on labelling and quantifying proteins about 4 h post-infection. This relatively early time point{1} allows one to minimize potentially confounding influences of virion particle assembly and production on cytoplasmic levels of viral proteins, but |
| 2c5d1ebec4 04ad8061eb 81e94effbe5 2a6dbe809 | Purified ALV-A virus particles{1} were incubated with PMB for 30 min at 378C, and infectivity was measured on human 293 cells expressing the ALV receptor Tva (293-Tva){2}. The effect of PMB treatment of |
| d9eb8ffffee8 147c850b00f 613a1978c18 505580 | FCoVs and CCoVs{1} are common pathogens and readily evolve. It is necessary to pursue epidemiological surveillance of |
| Sentence | Extracted Triples (S/P/O) | Engine |
|---|---|---|
| “RA and PBD blocked the attachment of IAV and 3C-like protease (3CLP) of severe acute respiratory syndrome-associated coronaviruses plays a pivotal role in viral replication and is a promising drug target”, Source: c85ca5217f9051f839115 69eed1eb52cf992f7dd | RA and PBD/blocked/the attachment of IAV | O,C |
| 3C-like protease of severe acute respiratory syndrome-associated coronaviruses/is/3CLP | O | |
| 3C-like protease of severe acute respiratory syndrome-associated coronaviruses/plays/a pivotal role in viral replication | O | |
| 3C-like protease of severe acute respiratory syndrome-associated coronaviruses/is/a promising drug target | O | |
| PBD/blocked/the attachment of 3C-like protease (3CLP) of severe acute respiratory syndrome-associated coronaviruses | C | |
| PBD blocked the attachment of IAV/is/a promising drug target | C | |
| PBD/blocked/the attachment of IAV | C | |
| RA blocked the attachment of 3C-like protease (3CLP) of severe acute respiratory syndrome-associated coronaviruses/is/a promising drug target | C | |
| RA/blocked/the attachment of 3C-like protease (3CLP) of severe acute respiratory syndrome-associated coronaviruses | C | |
| the attachment of IAV/plays/a pivotal role in viral replication | C | |
| RA and PBD/blocked/the attachment of IAV and 3C-like protease (3CLP) of severe acute respiratory syndrome-associated coronaviruses | A | |
| “CRP is an acute phase protein that has been linked to the presence and severity of bacterial infection in numerous studies during the past 2 decades [9,34,35]”, Source: b8e9c45dda9cb8c9c4321 a55704ab2a66fb34f7d | CRP/is/an acute phase protein | O |
| an acute phase protein/has been linked/to the presence and severity of bacterial infection in numerous studies during the past 2 decades | O,C | |
| an acute phase protein/has been linked/to the presence and severity of bacterial infection in numerous studies | O | |
| CRP/is/an acute phase protein that has been linked to the severity of bacterial infection in numerous studies during the past 2 decades | C,A | |
| “Involvement of polyamines, possibly due to loss of epigenetic control of X-linked polyamine genes/is suspected in SjS since the appearance of acrolein conjugated proteins is related to the intensity of SjS and acrolein is an oxidation product of polyamines (88)”, Source: 8495f7c65f4a6cbce0e0 d53c0900f10f6740826e | Involvement of polyamines possibly due to loss of epigenetic control of X-linked polyamine genes/is suspected/in SjS since the appearance of acrolein conjugated proteins is related to the intensity of SjS | O |
| Involvement of polyamines possibly due to loss of epigenetic control of X-linked polyamine genes/is suspected/in SjS | O | |
| the appearance of acrolein conjugated proteins/is related/to the intensity of SjS | O | |
| acrolein/is/an oxidation product of polyamines | O | |
| acrolein/is/an oxidation product | O | |
| Involvement of polyamines/possibly due to loss of epigenetic control of X-linked polyamine genes/is related/to the intensity of SjS | C | |
| Involvement of polyamines, possibly due to loss of epigenetic control of X-linked polyamine/is/suspected | A | |
| “The new genome sequence was obtained by first mapping reads to a reference SARS-CoV-2 genome using BWA-MEM 0.7.5a-r405 with default parameters to generate the consensus sequence.”, Source: b5d303cbcfe6be92d733e c593118b388db77452e | The new genome sequence/obtained/by first mapping reads to a reference SARS-CoV-2 genome | O,C,A |
| a reference SARS-CoV-2 genome/be using/BWA-MEM 0.7.5a-r405 with default parameters to generate the consensus sequence/ | O | |
| BWA-MEM 0.7.5a-r405/to generate/the consensus sequence | C | |
| a reference SARS-CoV-2 genome/using/BWA-MEM 0.7.5a-r405 to generate the consensus sequence | C | |
| The new genome sequence/was/obtained | A |
| Field | Value |
|---|---|
| title | NLRP3 Inflammasome-A Key Player in Antiviral Responses |
| id | 372caa549b07492de5fd7064e9cadc62bef0f478 |
| subject | chronic intrahepatic inflammation and liver injury |
| predicate | be mediated |
| object | by the NLRP3 inflammasome |
| subj_entity_name | [‘chronic’, ‘liver’] |
| subj_entity_coded | [‘C0333383’, ‘C0160390’] |
| subj_entity_confidence | [0.7110092043876648, 1.0] |
| subj_entity_description | [‘CUI: C0333383, Name: Acute and chronic inflammation Definition: The coexistence of a chronic inflammatory process with a superimposed polymorphonuclear neutrophilic infiltration. TUI(s): T046\nAliases: (total: 8): Active chronic inflammation, Chronic Active Inflammation, Chronic active inflammation, Active Chronic Inflammation, Subacute inflammatory cell infiltrate, Subacute inflammatory cell infiltration, Subacute inflammation, Acute and chronic inflammation (morphologic abnormality)’, ‘CUI: C0160390, Name: Injury of liver Definition: Damage to liver structure or function due to trauma or toxicity. TUI(s): T037 Aliases (abbreviated, total: 12): of liver injury, Injury to liver, injuries liver, injury liver, liver injury, Injury to Liver, Hepatic injury, injury hepatic, hepatic injury, Hepatic trauma’] |
| obj_entity_name | [‘NLRP3’] |
| obj_entity_coded | [‘C1424250’] |
| obj_entity_confidence | [1.0] |
| obj_entity_description | [‘CUI: C1424250, Name: NLRP3 gene Definition: This gene plays a role in both inflammation and apoptosis. TUI(s): T028 Aliases (abbreviated, total: 29): NLRP3 gene, NLRP3 Gene, AVP, AII, MWS, CRYOPYRIN, Cryopyrin, FCAS, CIAS1 GENE, CIAS1 gene’] |
| sentiment | positive |
| sent_num | 265719 |
| sentence_text | HCV infection promotes chronic intrahepatic inflammation and liver injury mediated by the NLRP3 inflammasome (21). |
| triple_num | 1 |
| engine | 1 |
| Metric | Value |
|---|---|
| Precision | 0.78 |
| Recall | 0.76 |
| F1-score | 0.77 |
| Subject | Predicate | Object | UMLS in Subj. | UMLS in Obj. |
|---|---|---|---|---|
| the SARS-CoV-2 | might be imported | to the seafood market in a short time | (‘C5203676’) | (‘C0206208’, ‘C4526594’) |
| the mortality rate due to 2019-nCoV is comparatively lesser than the earlier outbreaks of SARS and MERS-CoVs, as well as this virus | presents | relatively mild manifestations | (‘C0026565’, ‘C5203676’, ‘C0012652’, ‘C1175743’) | (‘C1513302’) |
| the initial identification of 2019-NCoV from 7 patients | diagnosed | with unidentified viral pneumonia | (‘C0020792’, ‘C5203676’, ‘C0030705’) | (‘C0032310’) |
| 2019-nCoV cases | be detected | outside China | (‘C5203676’, ‘C0868928’) | (‘C0008115’) |
| the receptor binding domain of SARS-CoV-2 | was | capable of binding ACE2 in the context of the SARS-CoV spike protein | (‘C0597358’, ‘C5203676’) | (‘C1167622’, ‘C1422064’, ‘C1175743’) |
| In-depth understanding the underlying pathogenic mechanisms of SARS-CoV-2 | will reveal | more targets | (‘C0205125’, ‘C0450254’, ‘C0441712’, ‘C5203676’) | (‘C0085104’) |
| SARS-CoV-2 orf8 and orf10 proteins | are | other methods to transmit SARS-CoV-2 | (‘C5203676’, ‘C1710521’) | (‘C0449851’, ‘C0332289’, ‘C5203676’) |
| As of February 20 2020 the 2019 novel coronavirus now named SARS-CoV-2 causing the disease COVID-19 has caused over 75,000 | has spread | to 25 other countries World Health Organization | (‘C0205314’, ‘C0206419’, ‘C5203676’, ‘C0012634’, ‘C5203670’) | (‘C0043237’) |
| infections due to SARS-CoV-2 | have spread | to over 26 countries | (‘C3714514’, ‘C5203676’) | (‘C0454664’,) |
| 2019-nCoV S-RBD or modified S-RBD of other coronavirus | may be applied | for developing 2019-nCoV vaccines | (‘C5203676’) | (‘C0042210’) |
| DetecTable 2019-nCoV viral RNA in blood | is | a strong indicator for the further clinical severity | (‘C3830527’, ‘C5203676’, ‘C0035736’, ‘C0005767’) | (‘C0021212’, ‘C2981439’) |
| SARS-CoV-2 | is causing | the ongoing COVID-19 outbreak | (‘C5203676’) | (‘C5203670’) |
| SARS-CoV-2 | may use | integrins | (‘C5203676’) | (‘C0021701’,) |
| SARS-CoV-2 | appears | to have been transmitted during the incubation period of patients | (‘C5203676’) | (‘C1521797’, ‘C2752975’, ‘C1561960’, ‘C0030705’) |
| SARS-CoV-2 | becomes | highly transmissible | (‘C5203676’) | (‘C0162534’) |
| SARS-CoV-2 | is | an emerging new coronavirus | (‘C5203676’) | (‘C0206419’) |
| SARS-CoV-2 | might exhibit | seasonality | (‘C5203676’) | (‘C0683922’) |
| SARS-CoV-2 | was not detected | suggesting that more evidences are needed before concluding the conjunctival route as the transmission pathway of SARS-CoV-2 | (‘C5203676’) | (‘C3887511’, ‘C2959706’, ‘C0040722’, ‘C5203676’) |
| SARS-CoV-2 | is circulating | in China | (‘C5203676’) | (‘C0008115’) |
| SARS-CoV-2 | be induced | pneumonia resulting in severe respiratory distress and death | (‘C5203676’) | (‘C0032285’, ‘C1547227’, ‘C0476273’, ‘C0011065’) |
| a novel corona virus first 2019-nCov then SARS-CoV-2 | was identified | as the cause of a cluster of pneumonia cases | (‘C0205314’, ‘C0206750’, ‘C5203676’) | (‘C0015127’, ‘C1704332’, ‘C0032285’) |
| The accurate transmission rate of SARS-CoV-2 | is | unknown since various factors impact its transmission | (‘C0443131’, ‘C0040722’, ‘C5203676’) | (‘C1257900’, ‘C0726639’, ‘C0040722’) |
| The observed morphology of SARS-CoV-2 | is | consistent with the typical characteristics of the Coronaviridae family | (‘C1441672’, ‘C0332437’, ‘C5203676’) | (‘C0332290’, ‘C1521970’, ‘C0010076’) |
| the receptor-binding domain of SARS-CoV-2 | demonstrates | a similar structure to that of SARS-CoV | (‘C1422496’, ‘C5203676’) | (‘C0678594’, ‘C1175743’) |
| the sequence divergence between SARS-CoV-2 and RaTG13 | is | great to assign parental relationship | (‘C0004793’, ‘C0443204’, ‘C5203676’, ‘C4284300’) | (‘C0030551’, ‘C1706279’) |
| the mutation rate of SARS-CoV-2 | is | suggestive to humans | (‘C1705285’, ‘C1521828’, ‘C5203676’) | (‘C0086418’) |
| the question of whether it is a potential transmission route of SARS-CoV-2 | deserves | further evaluation | (‘C3245505’, ‘C0040722’, ‘C5203676’) | (‘C1261322’) |
| the patterns and modes of the interaction between SARS-CoV-2 and host antiviral defense | would be | similar share many features | (‘C0449774’, ‘C4054480’, ‘C0037420’, ‘C5203676’, ‘C1167395’, ‘C1880266’) | (‘C1521970’) |
| the immune response against SARS-CoV-2 | is decoupled | from viral replication | (‘C0301872’, ‘C5203676’) | (‘C0042774’) |
| the detection of SARS-CoV-2 with a high viral load in the sputum of convalescent patients | arouse | concern about prolonged shedding of the virus after recovery | (‘C1511790’, ‘C5203676’, ‘C0376705’, ‘C0038056’, ‘C0740326’, ‘C0030705’) | (‘C0439590’, ‘C0162633’, ‘C0042776’, ‘C0237820’) |
| a new coronavirus (2019-nCoV), identified through genomic sequencing | was | the culprit of the pneumonia | (‘C0206419’, ‘C5203676’, ‘C1294197’) | (‘C2068043’, ‘C0032285’) |
| SARS-CoV-2 examination of the viral genome | was | critical for identifying the pathogen | (‘C5203676’, ‘C0582103’, ‘C0042720’) | (‘C2826883’, ‘C0450254’) |
| Article | Subject | Predicate | Object | UMLS in Subj. | UMLS in Obj. |
|---|---|---|---|---|---|
| Distinct Regulation of Host Responses by ERK and JNK MAP Kinases in Swine Macrophages Infected with Pandemic (H1N1) 2009 Influenza Virus | Highly pathogenic H5N1 viral infection in human macrophages | induced | higher expression of IL-6 | (‘C0450254’, ‘C0016627’, ‘C0042769’, ‘C0086418’) | (‘C0205250’, ‘C2911684’, ‘C0021760’) |
| The Expression of IL-6, TNF-, and MCP-1 in Respiratory Viral Infection in Acute Exacerbations of Chronic Obstructive Pulmonary Disease | patients with viral infections | showed | higher levels of TNF IL-6 and MCP-1 | (‘C0030705’, ‘C0042769’) | (‘C0205250’, ‘C0441889’, ‘C1522668’, ‘C0021760’, ‘C0128897’) |
| CORD-19 Article ID: 85783a36e7e787302307f42460839435d665f4e7 | ||||||
|---|---|---|---|---|---|---|
| Article Title: SARS-CoV-2: an Emerging Coronavirus that Causes a Global Threat | ||||||
| Article body: […] Subsequently, coronaviruses with high similarity to the human SARS-CoV or civet SARS-CoV-like virus were isolated from horseshoe bats{1}, concluding | ||||||
| Extracted triples using our pipeline: | ||||||
| subject | predicate | object | subj_entity_name | obj_entity_name | ||
| coronaviruses with high similarity to the human SARS - CoV or civet SARS - CoV - like virus | were isolated | from horseshoe bats | (’coronaviruses’, ’high’, ’human’, ’SARS-CoV’, ’civet’, ’SARS-CoV-like’) | (’horseshoe’) | ||
| horseshoe bats as the potential natural reservoir of SARS-CoV | is | the natural host of SARS-CoV-2 | (’horseshoe’, ’natural’, ’SARS-CoV’) | (’natural’, ’SARS-CoV-2’) | ||
| masked palm civets | are | the intermediate host | (’palm’) | (’intermediate’, ’host’) | ||
| the receptor-binding domain of SARS-CoV-2 | demonstrates | a similar structure to that of SARS-CoV | (’receptor-binding’, ’SARS-CoV-2’) | (’structure’, ’SARS-CoV’) | ||
| a few variations in the key residues | exist | at amino acid level | (’variations’, ’residues’) | (’amino’) | ||
| Pangolin | to be | a potential intermediate host for SARS-CoV-2 | (’pangolin’) | (’potential’, ’intermediate’, ’host’, ’SARS-CoV-2’) | ||
| The EC90 of remdesivir against SARS - CoV-2 in Vero E6 cells | is | 1.76 ΒµM, which is achievable in vivo | (’EC90’, ’remdesivir’, ’SARS’, ’CoV-2’) | (’in’) | ||
| Sample of OIE triples (ClausIE) directly on article body without using the pipeline (115 triples in total): | ||||||
| subject | predicate | object | ||||
| It | is | reasonable to suspect that bat is the natural host of SARS-CoV-2 considering its similarity with SARS-CoV Subsequently | ||||
| its | has | similarity | ||||
| SARS-CoV-2 | belonged | to the Betacoronavirus genera | ||||
| SARS-CoV-2 | was | closer to SARS-like coronavirus | ||||
| Zhou and colleagues | showed | that SARS-CoV-2 had 96.2% overall genome sequence identity throughout the genome to BatCoV RaTG13 | ||||
| SARS-CoV-2 | was felled | in a basal position within the subgenus Sarbecovirus in the ORF1b tree | ||||
| This finding | implies | a possible recombination event | ||||
| SARS-CoV and MERS-CoV respectively | is | 49 | ||||
| that snake might be the intermediate host | is | 61 | ||||
| Guangxi Customs of China | found | novel coronaviruses representing two sub-lineages related to SARS-CoV-2 respectively | ||||
| RaTG13 | is | 89.20% | ||||
| Interestingly the pangolin coronavirus and SARS-CoV-2 | share | identical amino acids | ||||
| RaTG13 | possesses | one | ||||
| The effective concentration for chloroquine and remdesivir | is | EC 50 | ||||
| Chloroquine | functions | at both viral entry and post-entry stages of the SARS-CoV-2 infection in Vero E6 cells whereas remdesivir does at post-entry stage only | ||||
| potential broad-spectrum antiviral activities | is | 64 65 | ||||
| An EC90 against the SARS-CoV-2 in Vero E6 cells | is | achievable clinically | ||||
| Remdesivir | is | effective | ||||
| 1.76 µM | is | achievable | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadopoulos, D.; Papadakis, N.; Litke, A. A Methodology for Open Information Extraction and Representation from Large Scientific Corpora: The CORD-19 Data Exploration Use Case. Appl. Sci. 2020, 10, 5630. https://doi.org/10.3390/app10165630
Papadopoulos D, Papadakis N, Litke A. A Methodology for Open Information Extraction and Representation from Large Scientific Corpora: The CORD-19 Data Exploration Use Case. Applied Sciences. 2020; 10(16):5630. https://doi.org/10.3390/app10165630
Chicago/Turabian StylePapadopoulos, Dimitris, Nikolaos Papadakis, and Antonis Litke. 2020. "A Methodology for Open Information Extraction and Representation from Large Scientific Corpora: The CORD-19 Data Exploration Use Case" Applied Sciences 10, no. 16: 5630. https://doi.org/10.3390/app10165630
APA StylePapadopoulos, D., Papadakis, N., & Litke, A. (2020). A Methodology for Open Information Extraction and Representation from Large Scientific Corpora: The CORD-19 Data Exploration Use Case. Applied Sciences, 10(16), 5630. https://doi.org/10.3390/app10165630





