Enhancing U-Net with Spatial-Channel Attention Gate for Abnormal Tissue Segmentation in Medical Imaging
Abstract
:1. Introduction
- (1)
- We attempt to explore the advantages and disadvantages of the popular encoder-decoder U-Net architecture. We then propose a novel spatial-channel attention gate (scAG) while using attention mechanisms to reduce the weakness while maximizing the advantages of using skip connections.
- (2)
- The proposed scAG combines two components: the spatial attention gate (sAG) and channel attention gate (cAG). sAG automatically concentrates on ’where’ is the region of interest while cAG automatically learns ’what’ is the meaningful representation of the given feature. It significantly suppresses the drawbacks of the plain skip connections in U-Net models, thus improving the segmentation results.
- (3)
- Our proposed method achieves superior results when compared with reference state-of-the-art methods on three types of medical images.
2. Related Works
2.1. Segmentation Model
2.2. Attention and Gating Mechanism
3. Proposed Method
3.1. Overview and Motivation
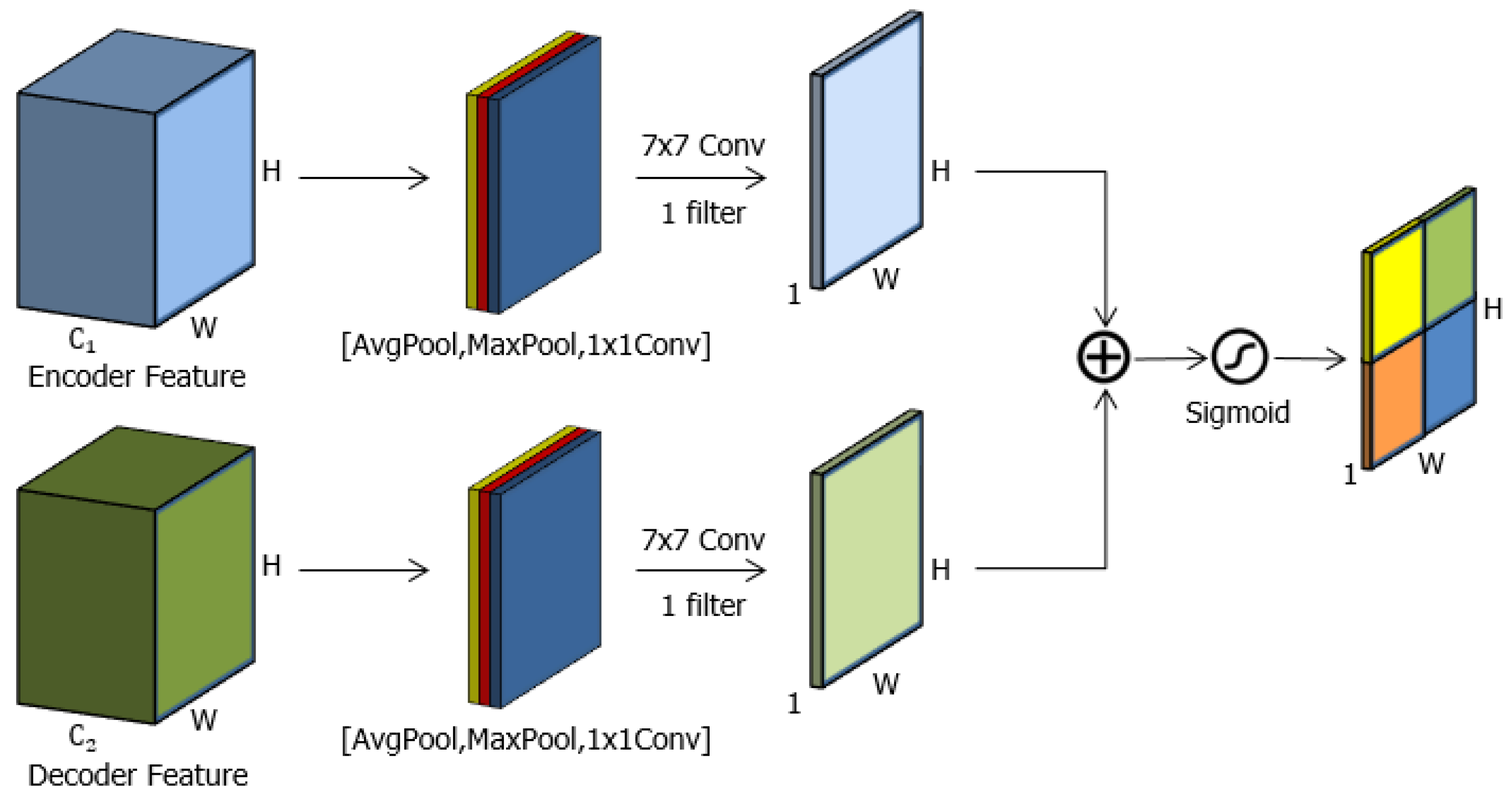
3.2. Spatial Attention Gate (sAG)
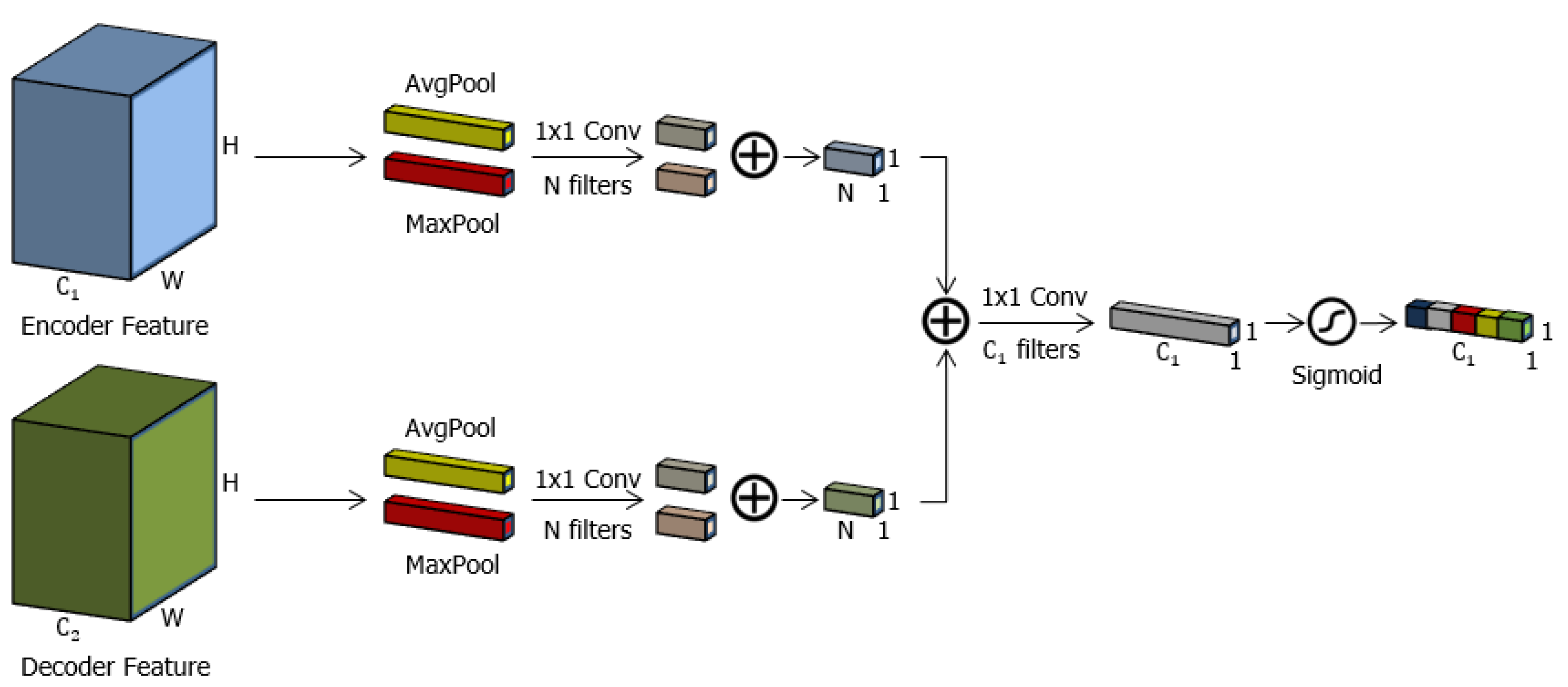
3.3. Channel Attention Gate (cAG)
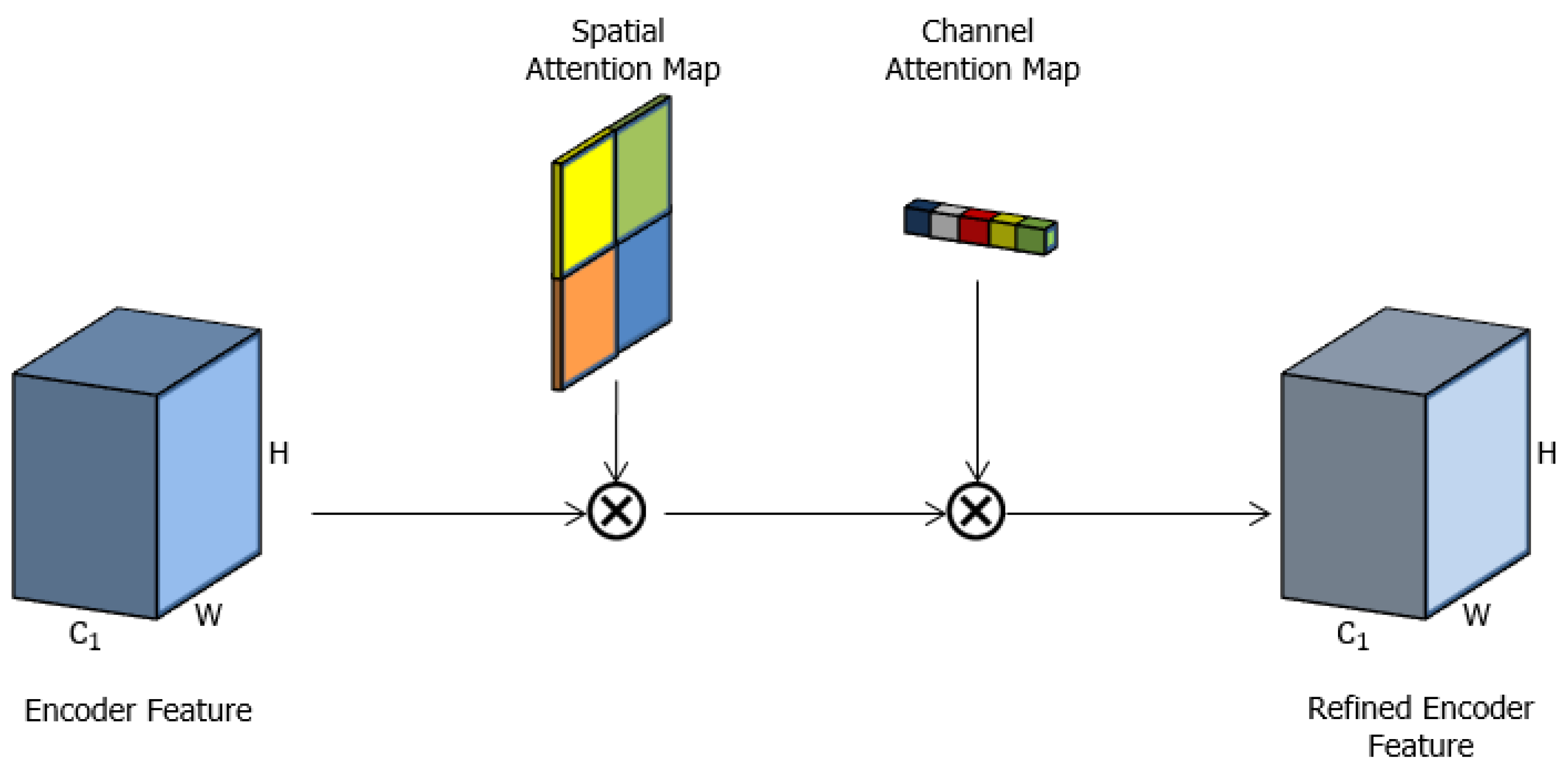
3.4. Spatial-Channel Attention Gate (scAG)
4. Experimental Results and Discussion
4.1. Datasets
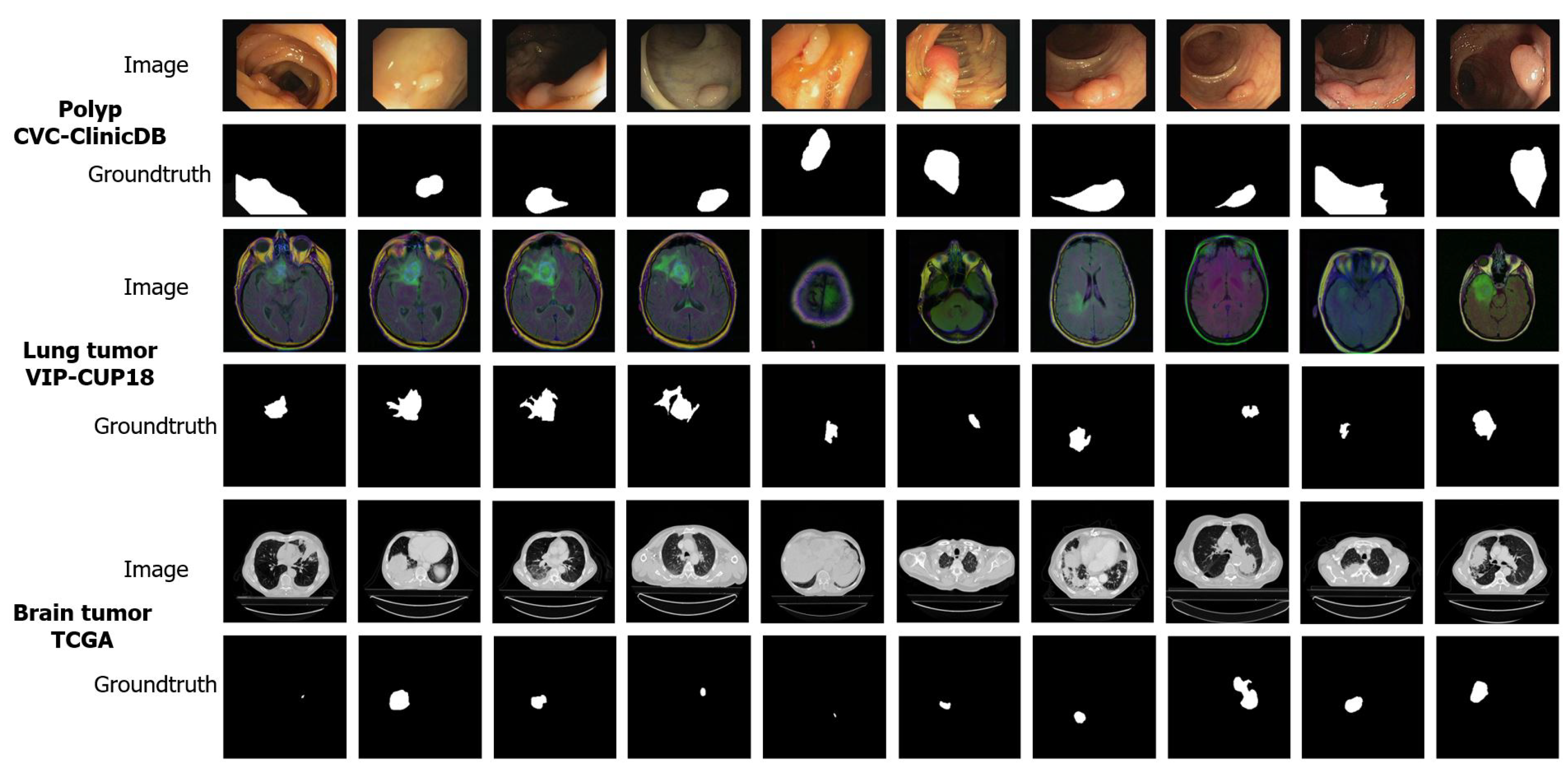
4.1.1. Colonoscopy Images (CVC-ClinicDB)
4.1.2. Lung Tumor Images (VIP-CUP18)
4.1.3. Brain Tumor Images (TCGA)
4.2. Network Architecture
4.3. Training and Implementation
4.4. Experimental Results
4.4.1. Ablation Studies
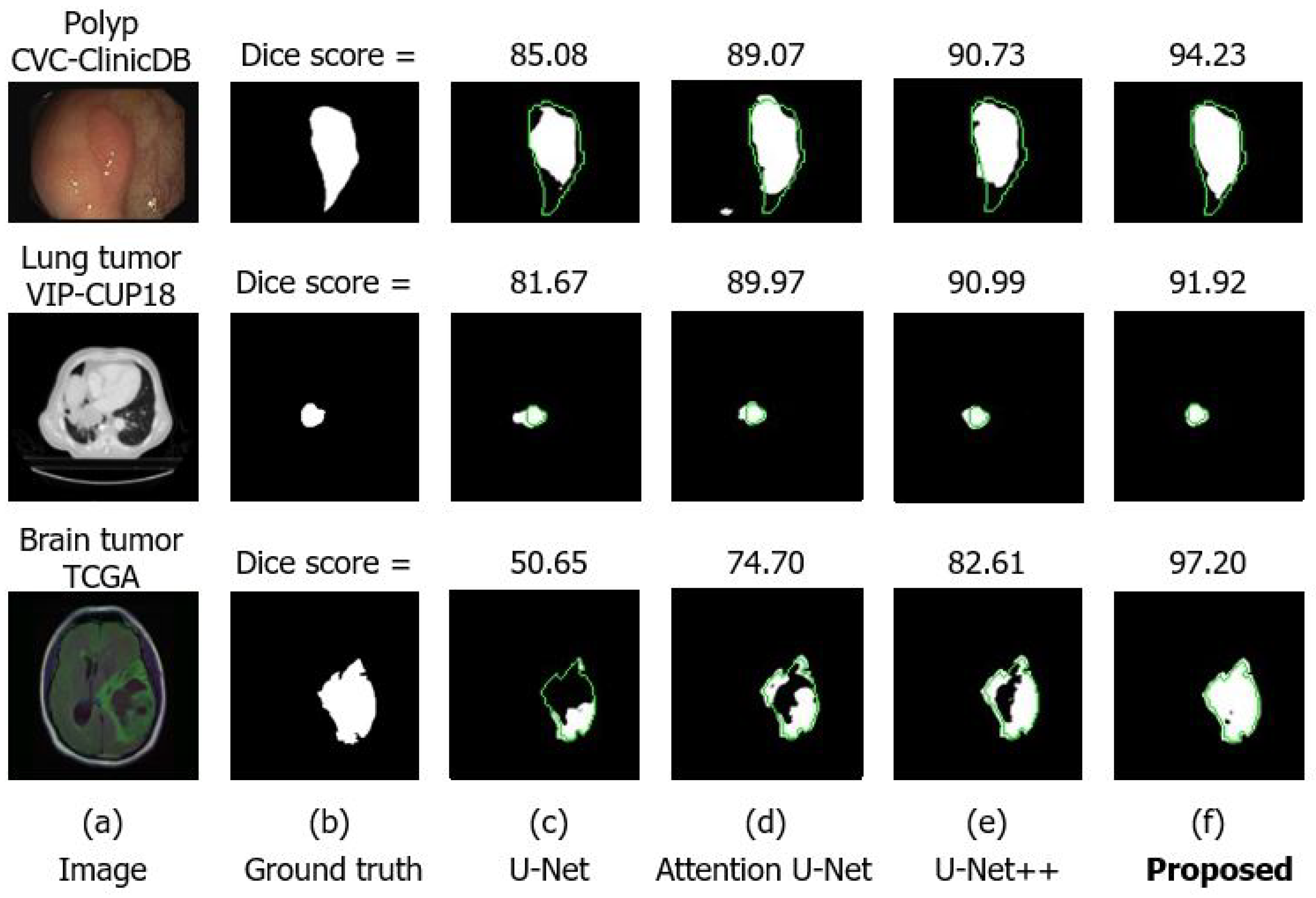
4.4.2. Comparison with Other U-Net Architectures
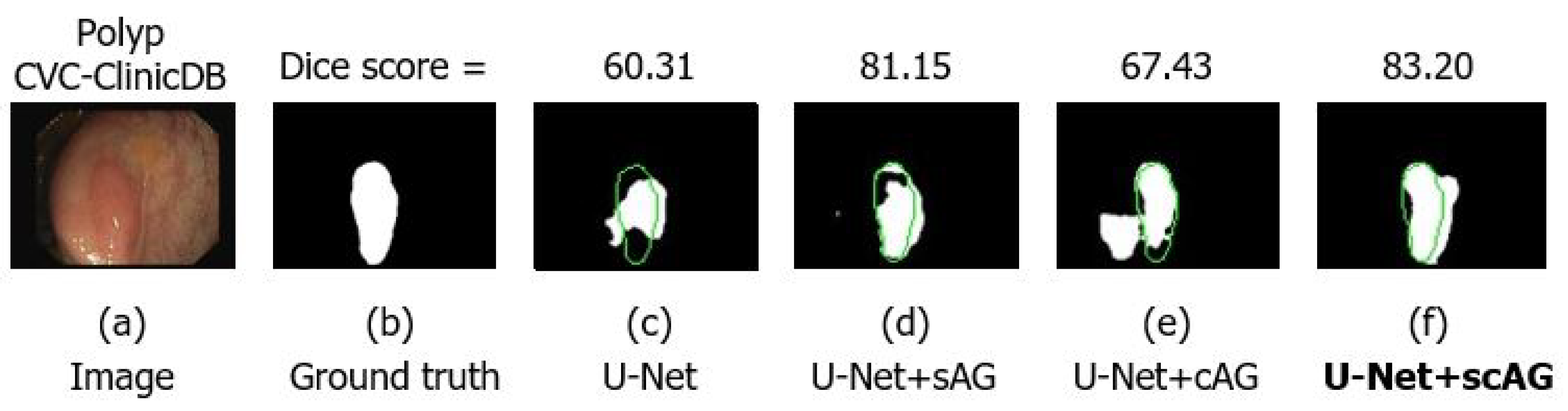
4.4.3. Combining the Spatial and Channel Attention Gate
4.4.4. Self-Attention for the Encoder Feature Map
4.4.5. Comparison with Other Attention Gate Methods
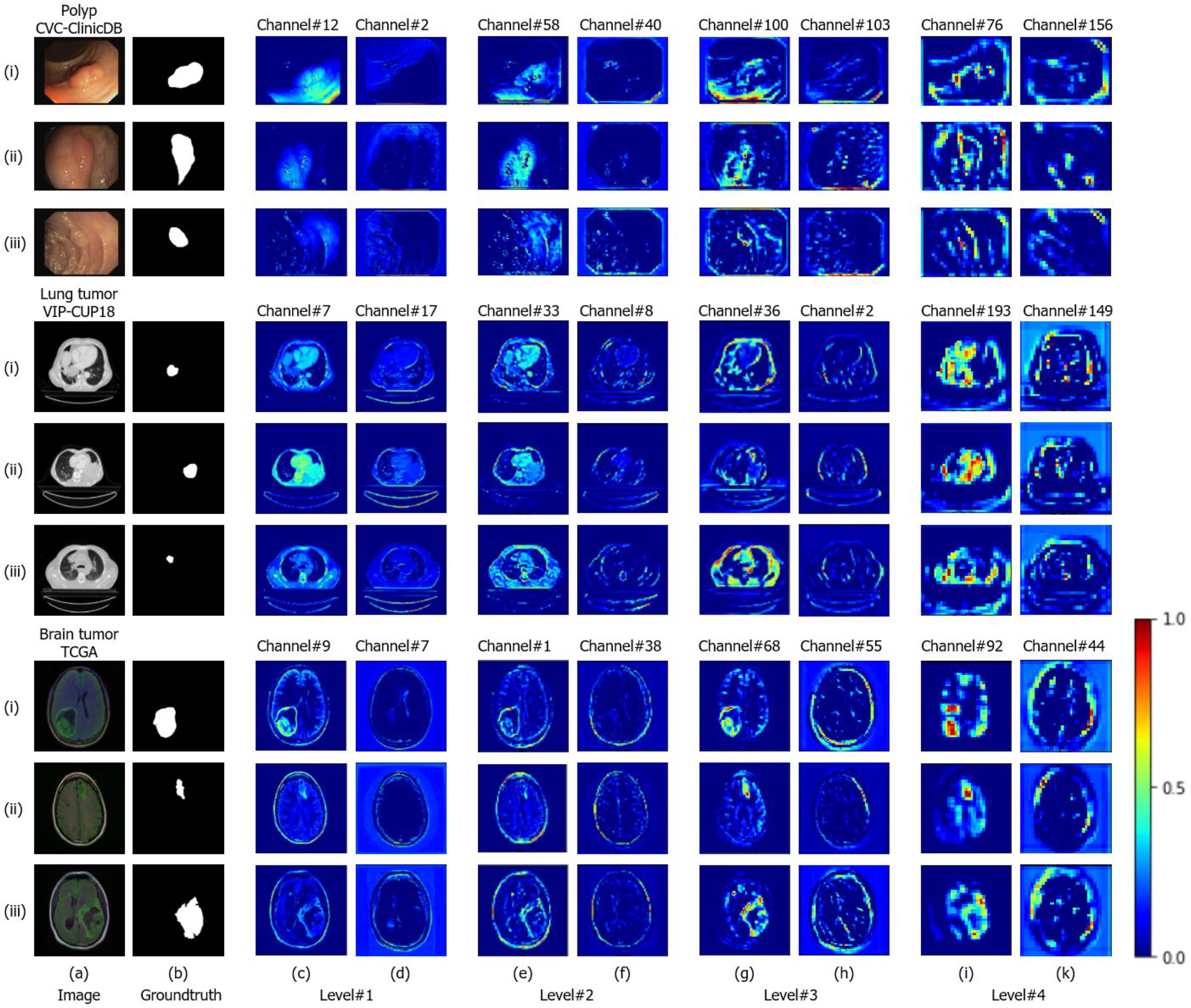
4.5. Visualization of the Attention Gate
4.6. Statistical Analysis of Variance on Dice Score, Precision and Recall Metrics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Tanzi, L.; Vezzetti, E.; Moreno, R.; Moos, S. X-ray Bone Fracture Classification Using Deep Learning: A Baseline for Designing a Reliable Approach. Appl. Sci. 2020, 10, 1507. [Google Scholar] [CrossRef] [Green Version]
- Gribaudo, M.; Moos, S.; Piazzolla, P.; Porpiglia, F.; Vezzetti, E.; Violante, M.G. Enhancing Spatial Navigation in Robot-Assisted Surgery: An Application. In International Conference on Design, Simulation, Manufacturing: The Innovation Exchange; Springer: Cham, Switzerland, 2019; pp. 95–105. [Google Scholar]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain Tumor Segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Jia, F.; Hu, Q. Automatic Segmentation of Liver Tumor in CT Images with Deep Convolutional Neural Networks. J. Comput. Commun. 2015, 3, 146–151. [Google Scholar] [CrossRef] [Green Version]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.C.; Liu, J.; Turkbey, E.B.; Summers, R.M. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. In Proceedings of the IEEE International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 556–564. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T.L. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the IEEE International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the International Conference on 3D Vision, Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.; Brox, T.; Ronneberger, O. 3d u-net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the IEEE International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Ben-Cohen, A.; Diamant, I.; Klang, E.; Amitai, M.; Greenspan, H. Fully convolutional network for liver segmentation and lesions detection. In Proceedings of the International Workshop on Deep Learning in Medical Image Analysis, Athens, Greece, 21 October 2016; pp. 77–85. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the International Workshop on Deep Learning in Medical Image Analysis, Granada, Spain, 20 September 2018; pp. 3–11. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. In Proceedings of the International Conference on Medical Imaging with Deep Learning, Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual attention network for image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6450–6458. [Google Scholar]
- Li, H.; Liu, Y.; Ouyang, W.; Wang, X. Zoom out-and-in network with map attention decision for region proposal and object detection. Int. J. Comput. Vis. 2019, 127, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Xiong, P.; An, J.; Wang, L. Pyramid Attention Network for Semantic Segmentation. In Proceedings of the British Machine Vision Conference, Northumbria, UK, 3–6 September 2018. [Google Scholar]
- Pedersoli, M.; Lucas, T.; Schmid, C.; Verbeek, J. Areas of attention for image captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, Hawaii, USA, 21–26 July 2017; pp. 1251–1259. [Google Scholar]
- Yang, Z.; He, X.; Gao, J.; Deng, L.; Smola, A. Stacked attention networks for image question answering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 21–29. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Roy, A.G.; Navab, N.; Wachinger, C. Concurrent Spatial and Channel ‘Squeeze & Excitation’ in Fully Convolutional Networks. In Proceedings of the IEEE International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 421–429. [Google Scholar]
- Roy, A.G.; Navab, N.; Wachinger, C. Recalibrating Fully Convolutional Networks With Spatial and Channel “Squeeze and Excitation” Blocks. IEEE Trans. Med. Imaging 2018, 38, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 818–833. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- 2018 IEEE Signal Processing Society Video and Image Processing (VIP) Cup. Available online: https://users.encs.concordia.ca/~i-sip/2018VIP-Cup/index.html (accessed on 19 May 2020).
- Buda, M.; Saha, A.; Mazurowski, M.A. Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm. Comput. Biol. Med. 2019, 109, 218–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, R.; Sivaswamy, J. M-net: A convolutional neural network for deep brain structure segmentation. Proceedings of IEEE International Symposium on Biomedical Imaging, Melbourne, Australia, 18–21 April 2017; pp. 437–440. [Google Scholar]
- Wong, K.C.L.; Moradi, M.; Tang, H.; Syeda-Mahmood, T. 3D Segmentation with Exponential Logarithmic Loss for Highly Unbalanced Object Sizes. In Proceedings of the IEEE International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 612–619. [Google Scholar]
- Lee, C.Y.; Xie, S.; Gallagher, P.; Zhang, Z.; Tu, Z. Deeply-supervised nets. In Proceedings of the Eighteenth International Conference on Artificial Intelligence and Statistics, San Diego, CA, USA, 9–12 May 2015; pp. 562–570. [Google Scholar]
- Kingma, D.P.; Ba, J. A method for stochastic optimization. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]


| Dataset | Application | No. of Images | Input Resolution | Modality |
|---|---|---|---|---|
| CVC-ClinicDB [29] | Polyp | 612 | Endoscopy | |
| VIP-CUP18 [30] | Lung tumor | 5204 | CT | |
| TCGA [31] | Brain tumor | 1373 | MRI |
| Model | Dice Score (%) | Precistion (%) | Recall (%) |
|---|---|---|---|
| U-Net + spatial + channel (ratio = 16) | |||
| U-Net + spatial + channel (ratio = 8) | |||
| U-Net + spatial + channel (ratio = 2) |
| Model | Dice Score (%) | Precistion (%) | Recall (%) |
|---|---|---|---|
| U-Net + spatial (avg-max) + channel | |||
| U-Net + spatial (1 × 1 conv) + channel | |||
| U-Net + spatial (avg-max-conv) + channel | |||
| U-Net + spatial (k = 7) + channel | |||
| U-Net + spatial (k = 5) + channel | |||
| U-Net + spatial (k = 3) + channel |
| Model | Deep Supervision | Dataset | Dice Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|---|
| CVC-ClinicDB | |||||
| U-Net [11] | × | VIP-CUP18 | |||
| TCGA | |||||
| CVC-ClinicDB | |||||
| Attention U-Net [17,18] | × | VIP-CUP18 | |||
| TCGA | |||||
| CVC-ClinicDB | |||||
| U-Net++ [15,16] | × | VIP-CUP18 | |||
| TCGA | |||||
| CVC-ClinicDB | |||||
| U-Net+scAG (Proposed) | × | VIP-CUP18 | |||
| TCGA | |||||
| CVC-ClinicDB | |||||
| U-Net+scAG (Proposed) | VIP-CUP18 | ||||
| TCGA |
| Model | Deep Supervision | No. of Parameters | Inference Time(ms/img) | ||
|---|---|---|---|---|---|
| CVC-ClinicDB | VIP-CUP18 | TCGA | |||
| U-Net [11] | × | 15 | 15 | 14 | |
| Attention U-Net [17,18] | × | 22 | 21 | 21 | |
| U-Net++ [15,16] | × | 28 | 26 | 26 | |
| U-Net+scAG (Proposed) | × | 33 | 32 | 32 | |
| U-Net+scAG (Proposed) | 34 | 33 | 34 | ||
| Model | No. of Parameters | Dice Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|
| U-Net | ||||
| U-Net + sAG | () | |||
| U-Net + cAG | () | |||
| U-Net + scAG | () |
| Model | Dice Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|
| U-Net | |||
| U-Net + encoder CBAM [25] | |||
| U-Net + scAG |
| Model | Dice Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|
| U-Net | |||
| U-Net + AG [17,18] | |||
| U-Net + GAU [21] | |||
| U-Net + scAG |
| Method1:Method2 | Metric | CVC-ClinicDB | TCGA | VIP-CUP18 |
|---|---|---|---|---|
| Dice score | 0.1251 | 0.0622 | 0.1822 | |
| Deep U-Net-scAG:U-Net | Precision | 0.0646 | 0.0082 | 0.3779 |
| Recall | 0.0963 | 0.8083 | 0.3072 | |
| Dice score | 0.2243 | 0.1168 | 0.2272 | |
| Deep U-Net-scAG:Attention U-Net | Precision | 0.0728 | 0.0185 | 0.0039 |
| Recall | 0.8755 | 0.3836 | 0.3507 | |
| Dice score | 0.3551 | 0.2639 | 0.5512 | |
| Deep U-Net-scAG:U-Net++ | Precision | 0.2591 | 0.0234 | 0.6049 |
| Recall | 0.8626 | 0.4368 | 0.1372 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khanh, T.L.B.; Dao, D.-P.; Ho, N.-H.; Yang, H.-J.; Baek, E.-T.; Lee, G.; Kim, S.-H.; Yoo, S.B. Enhancing U-Net with Spatial-Channel Attention Gate for Abnormal Tissue Segmentation in Medical Imaging. Appl. Sci. 2020, 10, 5729. https://doi.org/10.3390/app10175729
Khanh TLB, Dao D-P, Ho N-H, Yang H-J, Baek E-T, Lee G, Kim S-H, Yoo SB. Enhancing U-Net with Spatial-Channel Attention Gate for Abnormal Tissue Segmentation in Medical Imaging. Applied Sciences. 2020; 10(17):5729. https://doi.org/10.3390/app10175729
Chicago/Turabian StyleKhanh, Trinh Le Ba, Duy-Phuong Dao, Ngoc-Huynh Ho, Hyung-Jeong Yang, Eu-Tteum Baek, Gueesang Lee, Soo-Hyung Kim, and Seok Bong Yoo. 2020. "Enhancing U-Net with Spatial-Channel Attention Gate for Abnormal Tissue Segmentation in Medical Imaging" Applied Sciences 10, no. 17: 5729. https://doi.org/10.3390/app10175729
APA StyleKhanh, T. L. B., Dao, D.-P., Ho, N.-H., Yang, H.-J., Baek, E.-T., Lee, G., Kim, S.-H., & Yoo, S. B. (2020). Enhancing U-Net with Spatial-Channel Attention Gate for Abnormal Tissue Segmentation in Medical Imaging. Applied Sciences, 10(17), 5729. https://doi.org/10.3390/app10175729










