Extended Dual-Focus Microscopy for Ratiometric-Based 3D Movement Tracking
Abstract
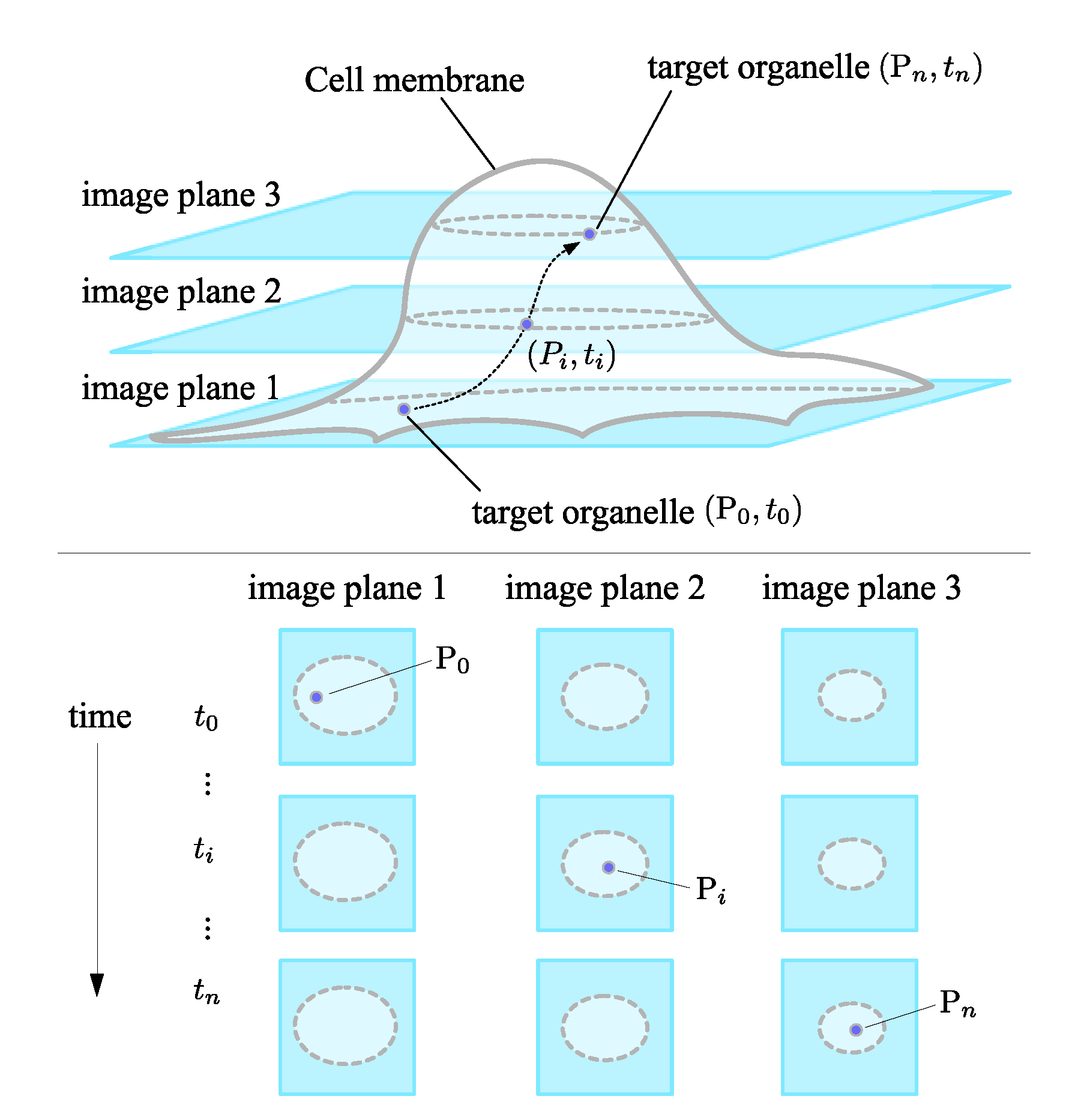
:1. Introduction
2. Methods
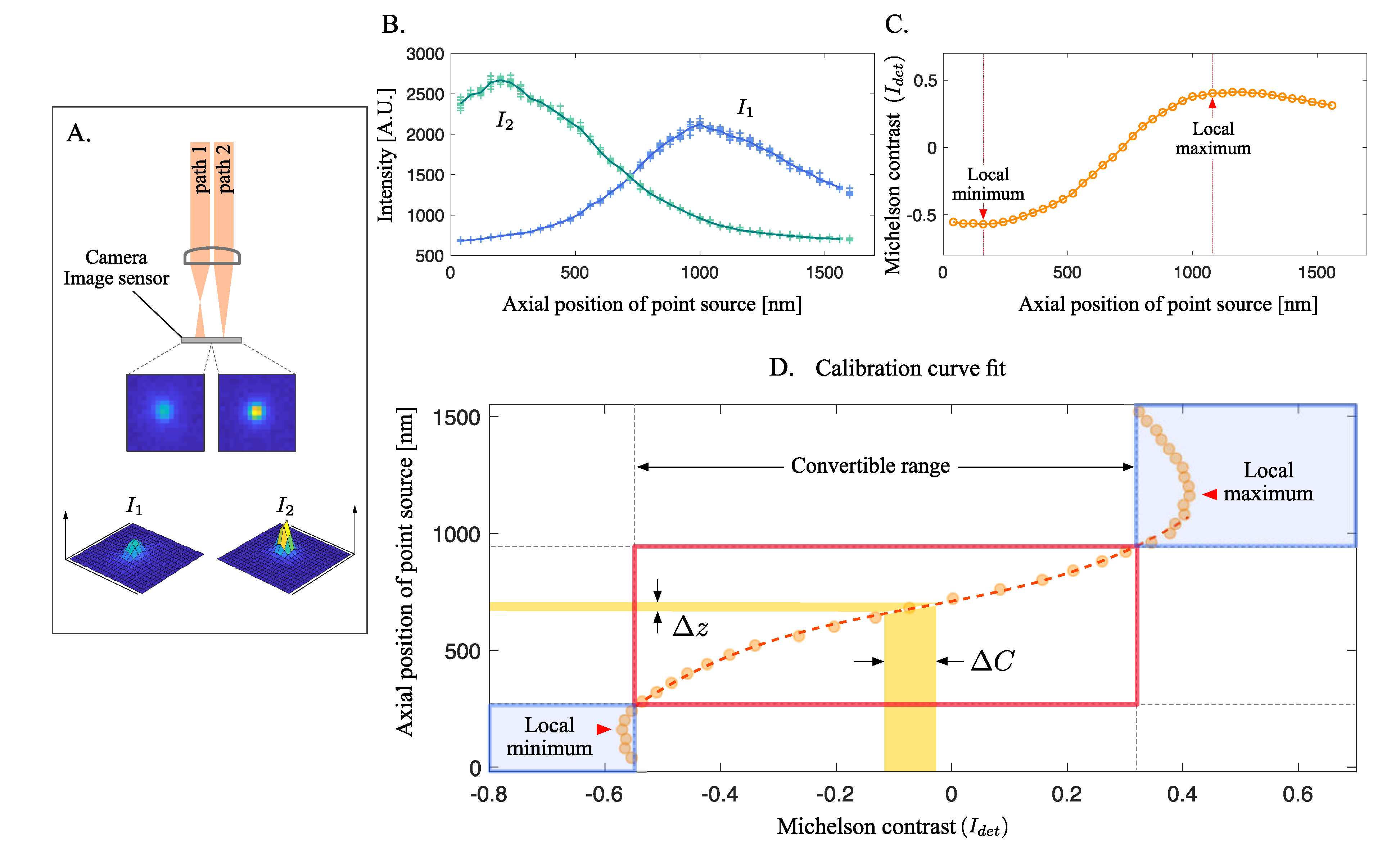
2.1. Extended Dual-Focus Optics Configuration
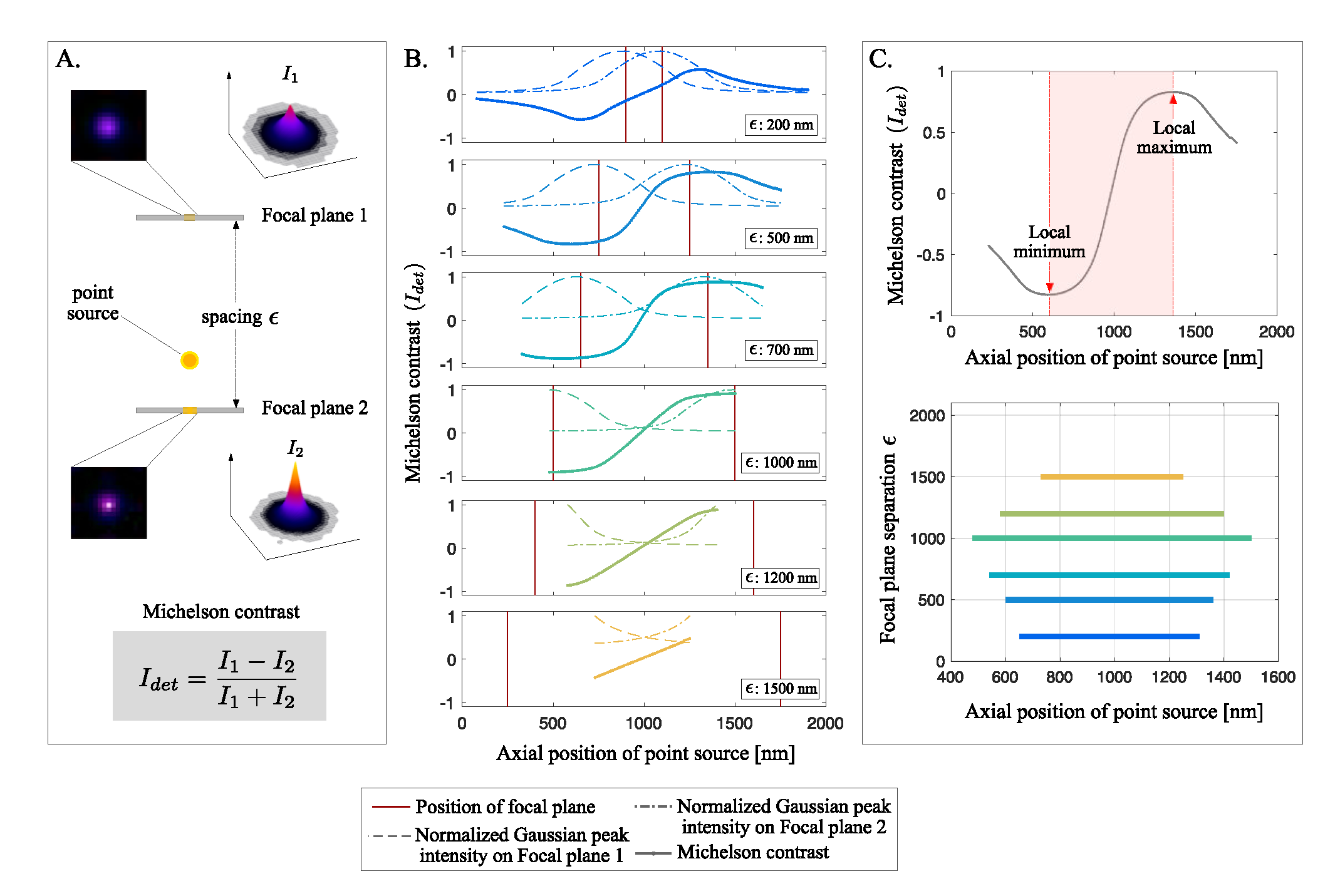
2.2. Simulation for Determining the Focal Plane Separation
2.3. Refinement on Calibration Curve
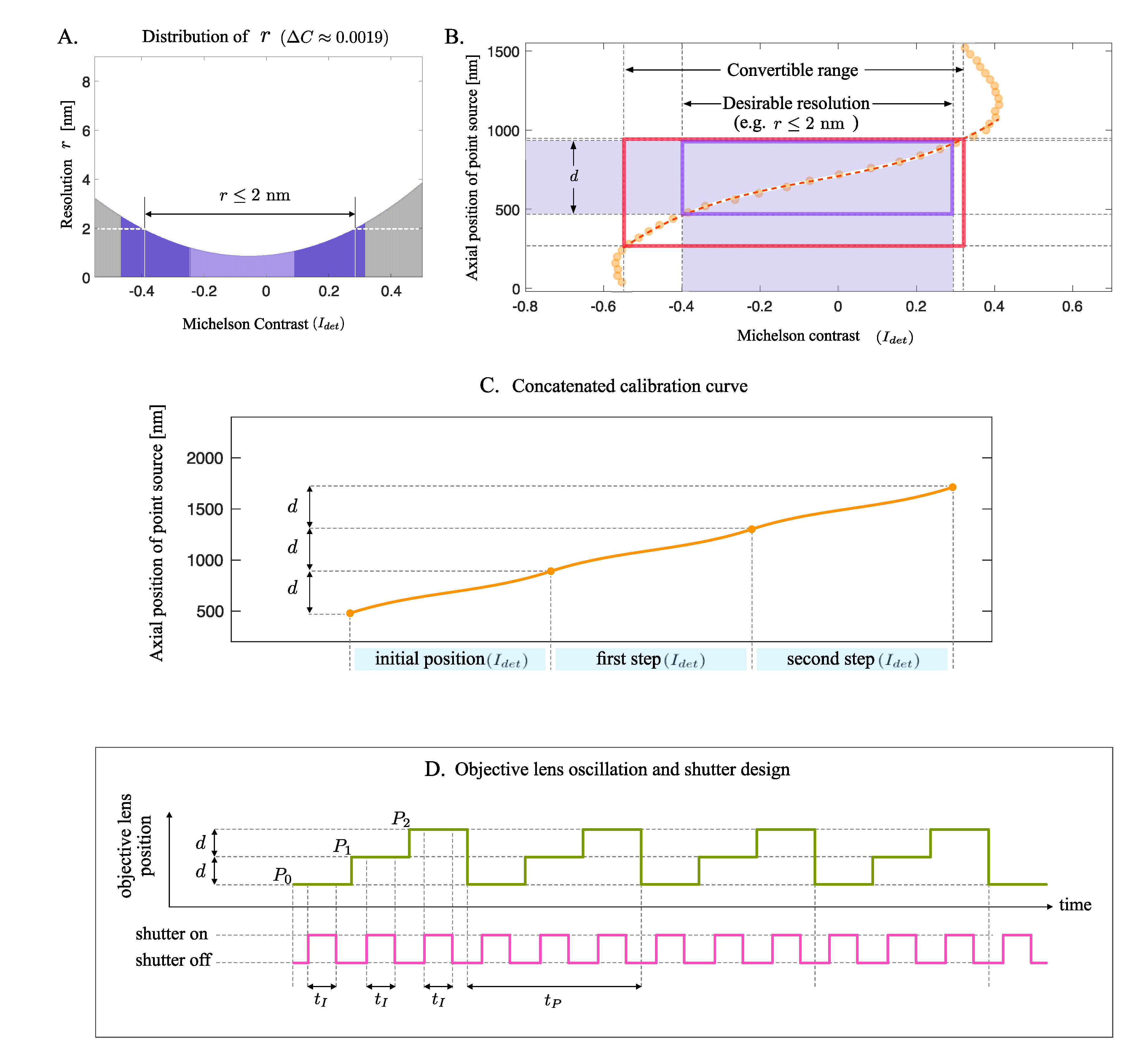
2.4. Calibration Resolution Concept
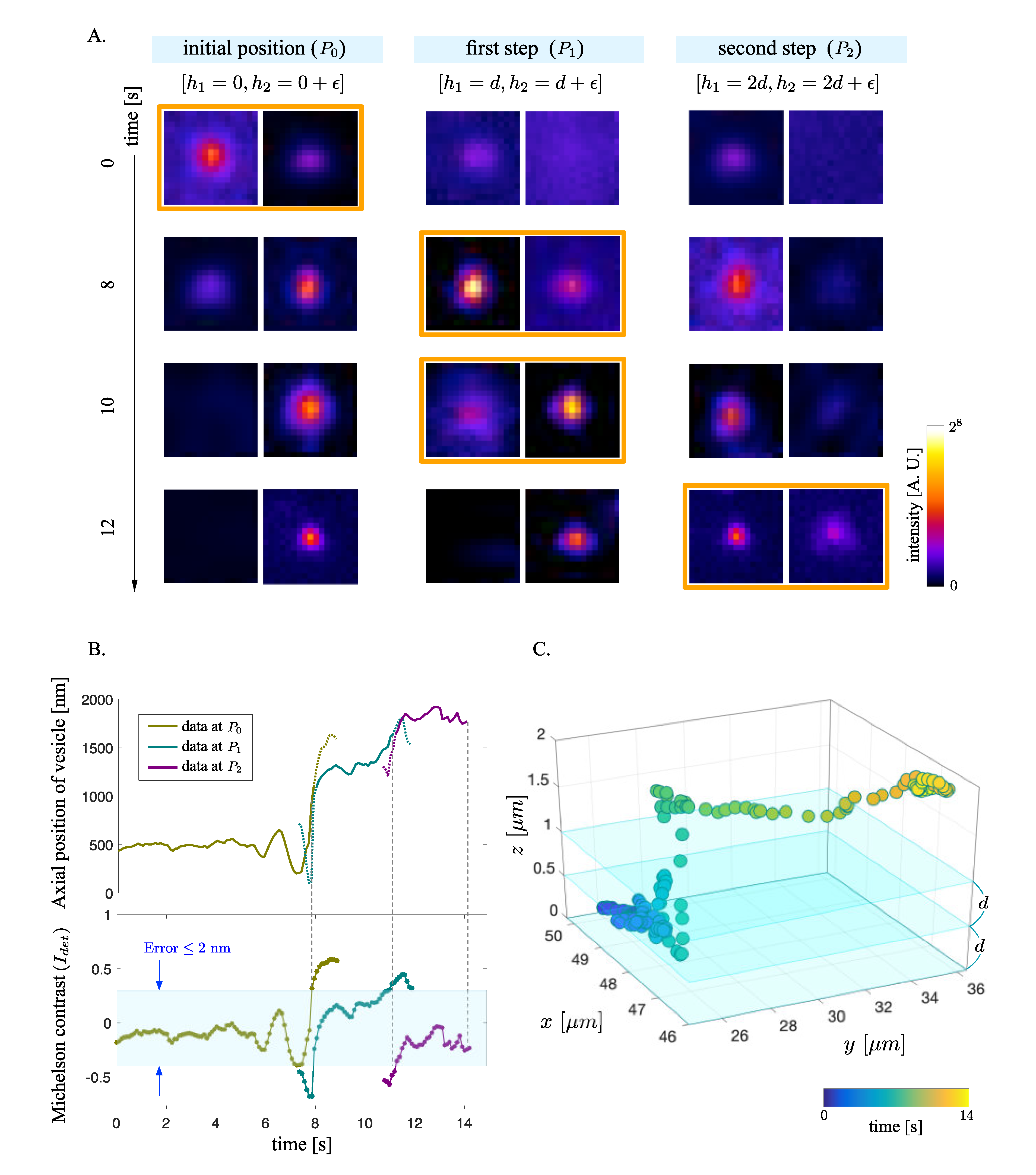
2.5. Imaging Position Step Determination
3. Results
3.1. Vesicle Tracking in Living Cell
3.2. Extension of Trackable Axial Range
3.3. Imaging of Endocytic Vesicle
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Boulaiz, H.; Alvarez, P.J.; Ramirez, A.; Marchal, J.A.; Prados, J.; Rodríguez-Serrano, F.; Perán, M.; Melguizo, C.; Aranega, A. Nanomedicine: Application areas and development prospects. Int. J. Mol. Sci. 2011, 12, 3303–3321. [Google Scholar] [CrossRef] [Green Version]
- Ray, A.; Mitra, A.K. Nanotechnology in Intracellular Trafficking, Imaging, and Delivery of Therapeutic Agents. In Emerging Nanotechnologies for Diagnostics, Drug Delivery and Medical Devices; Elsevier: Amsterdam, The Netherlands, 2017; pp. 169–188. [Google Scholar]
- Lee, S.; Kim, H.; Higuchi, H. Focus stabilization by axial position feedback in biomedical imaging microscopy. In Proceedings of the Sensors Applications Symposium (SAS), IEEE, Seoul, Korea, 12–14 March 2018; pp. 1–6. [Google Scholar]
- Ando, D.; Korabel, N.; Huang, K.C.; Gopinathan, A. Cytoskeletal network morphology regulates intracellular transport dynamics. Biophys. J. 2015, 109, 1574–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bálint, Š.; Vilanova, I.V.; Álvarez, Á.S.; Lakadamyali, M. Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. Proc. Natl. Acad. Sci. USA 2013, 110, 3375–3380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Handley, M.; Carles, G.; Harvey, A.R. Advances in 3D single particle localization microscopy. APL Photonics 2019, 4, 060901. [Google Scholar] [CrossRef] [Green Version]
- Prabhat, P.; Ram, S.; Ward, E.S.; Ober, R.J. Simultaneous imaging of different focal planes in fluorescence microscopy for the study of cellular dynamics in three dimensions. IEEE Trans. Nanobiosci. 2004, 3, 237–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehmelt, L.; Bastiaens, P.I. Spatial organization of intracellular communication: Insights from imaging. Nat. Rev. Mol. Cell Biol. 2010, 11, 440. [Google Scholar] [CrossRef]
- Deschout, H.; Zanacchi, F.C.; Mlodzianoski, M.; Diaspro, A.; Bewersdorf, J.; Hess, S.T.; Braeckmans, K. Precisely and accurately localizing single emitters in fluorescence microscopy. Nat. Methods 2014, 11, 253. [Google Scholar] [CrossRef]
- Toprak, E.; Balci, H.; Blehm, B.H.; Selvin, P.R. Three-dimensional particle tracking via bifocal imaging. Nano Lett. 2007, 7, 2043–2045. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.M.; Sato, T.; Gonda, K.; Higuchi, H. Three-dimensional nanometry of vesicle transport in living cells using dual-focus imaging optics. Biochem. Biophys. Res. Commun. 2007, 359, 1–7. [Google Scholar] [CrossRef]
- Lee, S.; Kim, H.; Higuchi, H. Numerical method for vesicle movement analysis in a complex cytoskeleton network. Opt. Express 2018, 26, 16236–16249. [Google Scholar] [CrossRef]
- Lee, S.; Higuchi, H. 3D rotational motion of an endocytic vesicle on a complex microtubule network in a living cell. Biomed. Opt. Express 2019, 10, 6611–6624. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, R.; Chao, J.; Ram, S.; Ward, E.S.; Ober, R.J. Intensity-based axial localization approaches for multifocal plane microscopy. Opt. Express 2017, 25, 3394–3410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirshner, H.; Aguet, F.; Sage, D.; Unser, M. 3-D PSF fitting for fluorescence microscopy: Implementation and localization application. J. Microsc. 2013, 249, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quammen, C.W.; Richardson, A.C.; Haase, J.; Harrison, B.D.; Taylor, R.M.; Bloom, K.S. FluoroSim: A visual problem-solving environment for fluorescence microscopy. In Eurographics Workshop on Visual Computing for Biomedicine; NIH Public Access: Bethesda, MD, USA, 2008; Volume 2008, p. 151. [Google Scholar]
- Kruth, H.S.; Jones, N.L.; Huang, W.; Zhao, B.; Ishii, I.; Chang, J.; Combs, C.A.; Malide, D.; Zhang, W.Y. Macropinocytosis is the endocytic pathway that mediates macrophage foam cell formation with native low density lipoprotein. J. Biol. Chem. 2005, 280, 2352–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franke, C.; Sauer, M.; van de Linde, S. Photometry unlocks 3D information from 2D localization microscopy data. Nat. Methods 2017, 14, 41. [Google Scholar] [CrossRef] [Green Version]
- Michelson, A.A. Studies in Optics; Courier Corporation: North Chelmsford, MA, USA, 1995. [Google Scholar]
- Kurebayashi, J.; Otsuki, T.; Tang, C.; Kurosumi, M.; Yamamoto, S.; Tanaka, K.; Mochizuki, M.; Nakamura, H.; Soono, H. Isolation and characterization of a new human breast cancer cell line, KPL-4, expressing the Erb B family receptors and interleukin-6. Br. J. Cancer 1999, 79, 707. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.W.; Monteiro-Riviere, N.A. Mechanisms of quantum dot nanoparticle cellular uptake. Toxicol. Sci. 2009, 110, 138–155. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Forry, S.P.; Gao, X.; Holbrook, R.D.; Telford, W.G.; Tona, A. Dynamics and mechanisms of quantum dot nanoparticle cellular uptake. J. Nanobiotechnol. 2010, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Ong, L.C.; Ang, L.Y.; Alonso, S.; Zhang, Y. Bacterial imaging with photostable upconversion fluorescent nanoparticles. Biomaterials 2014, 35, 2987–2998. [Google Scholar] [CrossRef]
- Park, Y.I.; Lee, K.T.; Suh, Y.D.; Hyeon, T. Upconverting nanoparticles: A versatile platform for wide-field two-photon microscopy and multi-modal in vivo imaging. Chem. Soc. Rev. 2015, 44, 1302–1317. [Google Scholar] [CrossRef]
- Bergman, J.P.; Bovyn, M.J.; Doval, F.F.; Sharma, A.; Gudheti, M.V.; Gross, S.P.; Allard, J.F.; Vershinin, M.D. Cargo navigation across 3D microtubule intersections. Proc. Natl. Acad. Sci. USA 2018, 115, 537–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdeny-Vilanova, I.; Wehnekamp, F.; Mohan, N.; Álvarez, Á.S.; Borbely, J.S.; Otterstrom, J.J.; Lamb, D.C.; Lakadamyali, M. 3D motion of vesicles along microtubules helps them to circumvent obstacles in cells. J. Cell Sci. 2017, 130, 1904–1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, J.L.; Ali, M.Y.; Warshaw, D.M. Cargo transport: Molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol. 2008, 20, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granger, E.; McNee, G.; Allan, V.; Woodman, P. The role of the cytoskeleton and molecular motors in endosomal dynamics. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 31, pp. 20–29. [Google Scholar]
- Zajac, A.L.; Goldman, Y.E.; Holzbaur, E.L.; Ostap, E.M. Local cytoskeletal and organelle interactions impact molecular-motor-driven early endosomal trafficking. Curr. Biol. 2013, 23, 1173–1180. [Google Scholar] [CrossRef] [Green Version]
- Masters, T.A.; Buss, F. Filopodia formation and endosome clustering induced by mutant plus-end–directed myosin VI. Proc. Natl. Acad. Sci. USA 2017, 114, 1595–1600. [Google Scholar] [CrossRef] [Green Version]
- Milo, R.; Phillips, R. Cell Biology by The Numbers; Garland Science: New York, NY, USA, 2016. [Google Scholar]
- Tumbarello, D.A.; Kendrick-Jones, J.; Buss, F. Myosin VI and its cargo adaptors–linking endocytosis and autophagy. J. Cell Sci. 2013, 126, 2561–2570. [Google Scholar] [CrossRef] [Green Version]
- von Diezmann, A.; Shechtman, Y.; Moerner, W. Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem. Rev. 2017, 117, 7244–7275. [Google Scholar] [CrossRef]
- Fischer, R.S.; Wu, Y.; Kanchanawong, P.; Shroff, H.; Waterman, C.M. Microscopy in 3D: A biologist’s toolbox. Trends Cell Biol. 2011, 21, 682–691. [Google Scholar] [CrossRef] [Green Version]
- Magidson, V.; Khodjakov, A. Circumventing photodamage in live-cell microscopy. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 114, pp. 545–560. [Google Scholar]
- Lee, S.; Kim, H.; Ishikawa, M.; Higuchi, H. 3D Nanoscale Tracking Data Analysis for Intracellular Organelle Movement using Machine Learning Approach. In Proceedings of the 2019 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), IEEE, Okinawa, Japan, 11–13 February 2019; pp. 181–184. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Kim, H.; Higuchi, H. Extended Dual-Focus Microscopy for Ratiometric-Based 3D Movement Tracking. Appl. Sci. 2020, 10, 6243. https://doi.org/10.3390/app10186243
Lee S, Kim H, Higuchi H. Extended Dual-Focus Microscopy for Ratiometric-Based 3D Movement Tracking. Applied Sciences. 2020; 10(18):6243. https://doi.org/10.3390/app10186243
Chicago/Turabian StyleLee, Seohyun, Hyuno Kim, and Hideo Higuchi. 2020. "Extended Dual-Focus Microscopy for Ratiometric-Based 3D Movement Tracking" Applied Sciences 10, no. 18: 6243. https://doi.org/10.3390/app10186243





