Lung Field Segmentation in Chest X-rays: A Deformation-Tolerant Procedure Based on the Approximation of Rib Cage Seed Points
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Proposed Segmentation Method
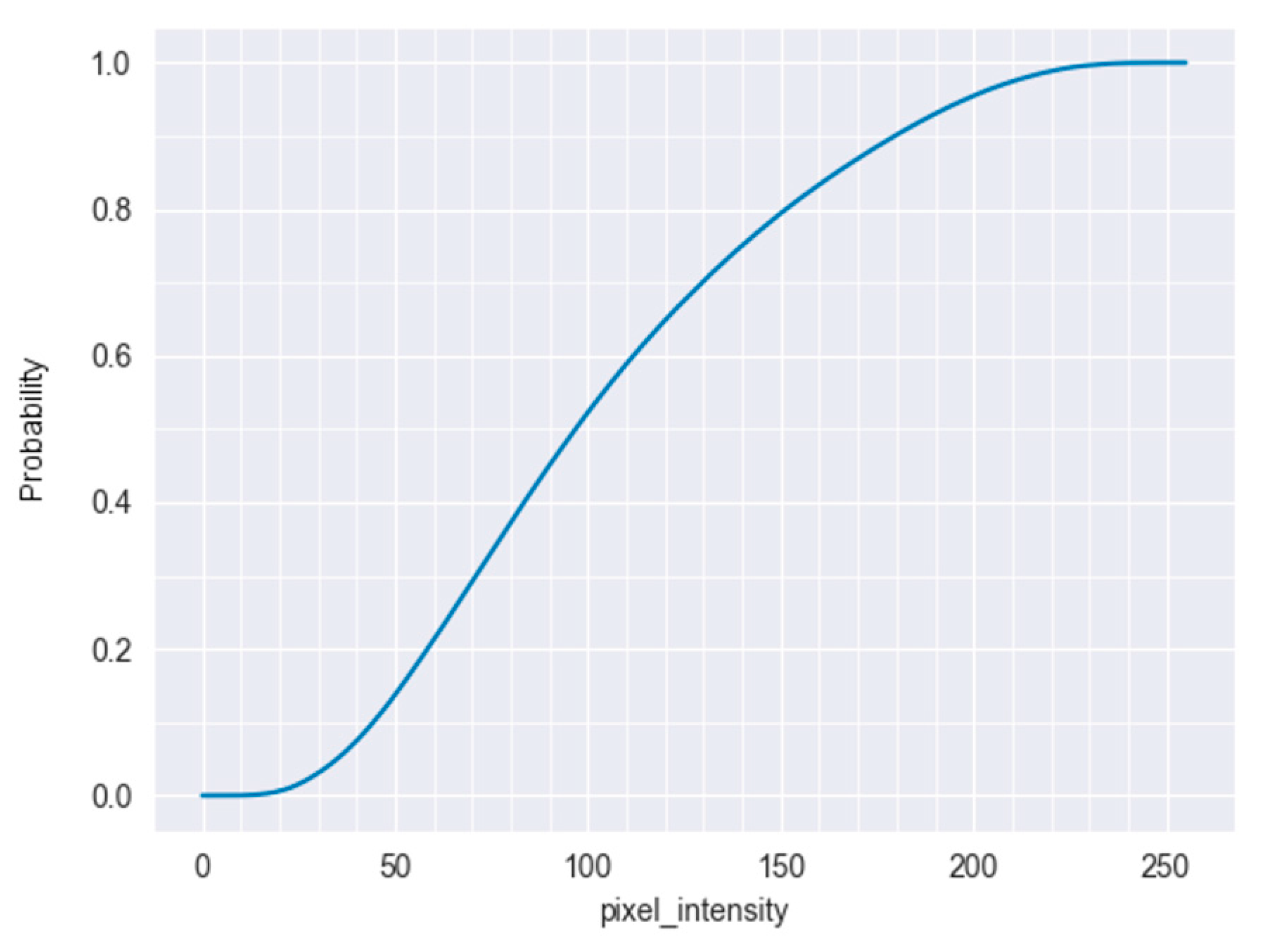
2.2. Lung Region Approximation: A Robust Method
2.3. Minimizing the Irrelevant Lung Area
2.4. Identification of Border Points
2.5. Stretching the Initial Region
2.6. Evaluation Dataset
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
- Availability of data and material: Algorithm output (images and graphs) is available at: https://drive.google.com/drive/folders/1iaq4mFhgM2Loedlj_bXBKp-ZeIPVCwEF
- The evaluation dataset used is publicly available at: https://lhncbc.nlm.nih.gov/publication/pub9931
- Code availability: https://bitbucket.org/vmposdel/cxr_image_segmentation/
References
- Raoof, S.; Feigin, D.; Sung, A.; Raoof, S.; Irugulpati, L.; Rosenow, E.C. Interpretation of Plain Chest Roentgenogram. Chest 2012, 141, 545–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurney, J.W. Why chest radiography became routine. Radiology 1995, 195, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Brady, A.P. Error and discrepancy in radiology: Inevitable or avoidable? Insights Imaging 2016, 8, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Robinson, P.J.; Wilson, D.; Coral, A.; Murphy, A.; Verow, P. Variation between experienced observers in the interpretation of accident and emergency radiographs. Br. J. Radiol. 1999, 72, 323–330. [Google Scholar] [CrossRef]
- Lodwick, G.S.; Keats, T.E.; Dorst, J.P. The coding of roentgen images for computer analysis as applied to lung cancer. Radiology 1963, 81, 185–200. [Google Scholar] [CrossRef] [PubMed]
- van Ginneken, B.; Hogeweg, L.; Prokop, M. Computer-aided diagnosis in chest radiography: Beyond nodules. Eur. J. Radiol. 2009, 72, 226–230. [Google Scholar] [CrossRef]
- Bar, Y.; Diamant, I.; Wolf, L.; Lieberman, S.; Konen, E.; Greenspan, H. Chest pathology detection using deep learning with non-medical training. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), New York, NY, USA, 16–19 April 2015; pp. 294–297. [Google Scholar]
- Anavi, Y.; Kogan, I.; Gelbart, E.; Geva, O.; Greenspan, H. Visualizing and enhancing a deep learning framework using patients age and gender for chest X-ray image retrieval. In: Medical Imaging 2016: Computer-Aided Diagnosis. Int. Soc. Opt. Photonics 2016, 9785, 978510. [Google Scholar]
- Yao, L.; Poblenz, E.; Dagunts, D.; Covington, B.; Bernard, D.; Lyman, K. Learning to diagnose from scratch by exploiting dependencies among labels. arXiv 2018, arXiv:171010501. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [Green Version]
- Candemir, S.; Antani, S. A review on lung boundary detection in chest X-rays. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 563–576. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Gao, Y.; Niu, Z.; Jiang, Y.; Li, L.; Xiao, X.; Wang, M.; Fang, E.F.; Menpes-Smith, W.; Xia, J.; et al. Weakly Supervised Deep Learning for COVID-19 Infection Detection and Classification from CT Images. IEEE Access 2020, 8, 118869–118883. [Google Scholar] [CrossRef]
- Yang, G.; Chen, J.; Gao, Z.; Li, S.; Ni, H.; Angelini, E.; Wong, T.; Mohiaddin, R.; Nyktari, E.; Wage, R.; et al. Simultaneous left atrium anatomy and scar segmentations via deep learning in multiview information with attention. Future Gener. Comput. Syst. 2020, 107, 215–228. [Google Scholar] [CrossRef]
- Li, M.; Wang, C.; Zhang, H.; Yang, G. MV-RAN: Multiview recurrent aggregation network for echocardiographic sequences segmentation and full cardiac cycle analysis. Comput. Biol. Med. 2020, 120, 103728. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, G.; Mirak, S.A.; Hosseiny, M.; Azadikhah, A.; Zhong, X.; Reiter, R.E.; Lee, Y.; Raman, S.S.; Sung, K. Automatic Prostate Zonal Segmentation Using Fully Convolutional Network with Feature Pyramid Attention. IEEE Access 2019, 7, 163626–163632. [Google Scholar] [CrossRef]
- Gordienko, Y.; Gang, P.; Hui, J.; Zeng, W.; Kochura, Y.; Alienin, O.; Rokovyi, O.; Stirenko, S. Deep Learning with Lung Segmentation and Bone Shadow Exclusion Techniques for Chest X-ray Analysis of Lung Cancer. In International Conference on Computer Science, Engineering and Education Applications; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Loog, M.; van Ginneken, B. Segmentation of the posterior ribs in chest radiographs using iterated contextual pixel classification. IEEE Trans. Med. Imaging 2006, 25, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, S.; Hu, Q. An Automatic Rib Segmentation Method on X-ray Radiographs. In Multi Media Modeling; He, X., Luo, S., Tao, D., Xu, C., Yang, J., Hasan, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 128–139. [Google Scholar]
- Wessel, J.; Heinrich, M.P.; von Berg, J.; Franz, A.; Saalbach, A. Sequential Rib Labeling and Segmentation in Chest X-ray using Mask R-CNN. arXiv 2019, arXiv:190808329. [Google Scholar]
- Cong, L.; Guo, W.; Li, Q. Segmentation of ribs in digital chest radiographs. In: Medical Imaging 2016: Biomedical Applications in Molecular, Structural, and Functional Imaging. Int. Soc. Opt. Photonics 2016, 9788, 97881T. [Google Scholar] [CrossRef]
- Wan Ahmad, W.S.H.M.; Zaki, W.M.D.W.; Ahmad Fauzi, M.F. Lung segmentation on standard and mobile chest radiographs using oriented Gaussian derivatives filter. Biomed. Eng. Online 2015, 14, 20. [Google Scholar] [CrossRef] [Green Version]
- Iakovidis, D.K.; Savelonas, M.A.; Papamichalis, G. Robust model-based detection of the lung field boundaries in portable chest radiographs supported by selective thresholding. Meas. Sci. Technol. 2009, 20, 104019. [Google Scholar] [CrossRef]
- Xu, T.; Mandal, M.; Long, R.; Basu, A. Gradient vector flow based active shape model for lung field segmentation in chest radiographs. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 3561–3564. [Google Scholar]
- Annangi, P.; Thiruvenkadam, S.; Raja, A.; Xu, H.; Sun, X.; Mao, L. A region based active contour method for X-ray lung segmentation using prior shape and low level features. In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, The Netherlands, 14–17 April 2010; pp. 892–895. [Google Scholar]
- Sundaresan, J. Dramenti/Symmetry. 2020. Available online: https://github.com/dramenti/symmetry (accessed on 20 May 2020).
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 2000. Available online: https://github.com/opencv/opencv (accessed on 20 May 2020).
- Candemir, S.; Jaeger, S.; Palaniappan, K.; Musco, J.P.; Singh, R.K.; Xue, Z.; Karargyris, A.; Antani, S.; Thoma, G.; McDonald, C.J. Lung Segmentation in Chest Radiographs Using Anatomical Atlases with Nonrigid Registration. IEEE Trans. Med. Imaging 2014, 33, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Karargyris, A.; Candemir, S.; Folio, L.; Siegelman, J.; Callaghan, F.; Xue, Z.; Palaniappan, K.; Singh, R.K.; Antani, S.; et al. Automatic Tuberculosis Screening Using Chest Radiographs. IEEE Trans. Med. Imaging 2014, 33, 233–245. [Google Scholar] [CrossRef]
- Kalinovsky, A.; Kovalev, V. Lung Image Segmentation Using Deep Learning Methods and Convolutional Neural Network. In Proceedings of the XIII International Conference on Pattern Recognition and Information Processing (PRIP-2016), Minsk, Belarus, 3–5 October 2016. [Google Scholar]
- Novikov, A.A.; Lenis, D.; Major, D.; Hladůvka, J.; Wimmer, M.; Bühler, K. Fully Convolutional Architectures for Multiclass Segmentation in Chest Radiographs. IEEE Trans. Med. Imaging 2018, 37, 1865–1876. [Google Scholar] [CrossRef] [Green Version]
- Arbabshirania, M.R.; Dallal, A.H.; Agarwal, C.; Patel, A.; Moore, G. Accurate Segmentation of Lung Fields on Chest Radiographs using Deep Convolutional Networks. In Medical Imaging 2017: Image Processing; International Society for Optics and Photonics: Orlando, Florida, USA, 12–14 February 2017; Volume 10133, p. 1013305. [Google Scholar]
- Souza, J.C.; Diniz, J.O.B.; Ferreira, J.L.; da Silva, G.L.F.; Silva, A.C.; de Paiva, A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput. Methods Prog. Biomed. 2019, 177, 285–296. [Google Scholar] [CrossRef]
- Dai, W.; Dong, N.; Wang, Z.; Liang, X.; Zhang, H.; Xing, E.P. SCAN: Structure Correcting Adversarial Network for Organ Segmentation in Chest X-rays. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Volume 11045 of Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; pp. 263–273. [Google Scholar]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR using Limited Training Data Sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Huynh, H.T.; Anh, V.N. A deep learning method for lung segmentation on large size chest X-ray image. In Proceedings of the IEEE-RIVF International Conference on Computing and Communication Technologies(RIVF), Danang, Vietnam, 20–22 March 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Chen, B.; Zheng Zhang, Z.; Lin, J.; Chen, Y.; Lu, G. Two-stream collaborative network for multi-label chest X-ray Image classification with lung segmentation. Pattern Recognit. Lett. 2020, 135, 221–227. [Google Scholar] [CrossRef]
- Chen, H.J.; Ruan, S.J.; Huang, S.W.; Peng, Y.T. Lung X-ray Segmentation using Deep Convolutional Neural Networks on Contrast-Enhanced Binarized Images. Mathematics 2020, 8, 545. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 Image Data Collection. arXiv 2020, arXiv:200311597. [Google Scholar]




| Algorithm | Main Purpose | Main Methodology | Comments |
|---|---|---|---|
| Gordienko et al. [17] | Assess how clavicles and rib shadows affect lung segmentation | UNet-based convolutional neural network | Improved accuracy by using a preprocessed version of the JSRT dataset without clavicles and rib shadows. Process is sped up by running on a GPU. |
| Loog et al. [18] | Posterior rib segmentation | Iterated contextual pixel classification | Evaluated on the JSRT dataset. Misclassifications appear in a structured way. |
| Li et al. [19] | Rib recognition | Template matching, graph theory and machine learning | Evaluated on the normal X-rays of the JSRT dataset. Overlap with clavicle introduces recognition problems. High sensitivity and specificity. |
| Wessel et al. [20] | Rib segmentation and anatomical labeling | Mask R-CNN | First approach for simultaneous rib detection and segmentation. Improved detection rate. |
| Cong et al. [21] | Eliminate the ribs | Hough transform and dynamic programming | Very high sensitivity and specificity. |
| Algorithm | Main Methodology | Datasets | DSC | Ω |
|---|---|---|---|---|
| Wan Ahmad et al. [22] 1 | Oriented Gaussian derivatives filter and Fuzzy C-Means | X-ray datasets from different machine types | - | 0.69–0.87 |
| Iakovidis et al. [23] 2 | Selective thresholding and ASM | Portable chest radiographs of patients with bacterial pulmonary infections | - | 0.91–0.92 |
| Xu et al. [24] 3 | Gradient Vector Flow-based ASM | JSRT and CXR | - | 0.84–0.9 |
| Annangi et al. [25] | Active contours; low-level features at boundary | Shanghai Pulmonary Hospital and other clinical sites in China | 0.88 | - |
| Authors (Year) | Main Method | Dataset | Jaccard | Dice | Strengths | Weaknesses |
|---|---|---|---|---|---|---|
| Kalinovsky et al. (2016) [30] | Encoder/Decoder CNN | Tuberculosis portal and JSRT | - | 0.962 | Uniform Deep Learning approach. | Hardware-demanding training. |
| Novikov et al. (2018) [31] | InvertedNet with Exponential Linear Units | JSRT | 0.95 | 0.974 | Copes with overfitting and imbalanced data. Reduces parameters. Segmentation of lungs, clavicles and heart. | Training and testing on the same dataset. Computational feasibility trade-off. |
| Arbabshirani et al. (2017) [32] | Registration-based and patch-based CNN | Geisinger and JSRT | - | 0.88–0.96 | Heterogeneous dataset. Multiscale network evaluated. | Hardware-demanding training. Coarse lung boundaries in some images. |
| Souza et al. (2019) [33] | Patch-based AlexNet, ResNet-18 with 2 deep CNNs | Montgomery County | - | 0.94 | Second CNN for more complex cases. Better segmentation of lungs with dense abnormalities. | Postprocessing required after the first network. Resizing of images required due to hardware limitations. Second network does not ensure quantitative improvement and leads to decreased performance. Many parameters. |
| Dai et al. (2018) [34] | Structure-Correcting Adversarial Network (SCAN) | JSRT, Montgomery County | - | 0.973 | Segments lung fields and heart. Limited training data. Generalizes to different patient populations and disease profiles. | Like many other methods, labeled data are a necessity. |
| Oh et al. (2020) [35] | Patch-based (FC) DenseNet103 | Mixture of public CXR datasets | 0.932–0.955 | Few trainable parameters. Provides clinically interpretable saliency maps, which are useful for COVID-19 diagnosis and patient triage. Patch training leads to smaller network complexity and augmentation of dataset. Performs classification. | ||
| Huynh et al. (2019) [36] | Hybrid Network with network individuals | Hoan My Hospital | - | 0.87 | Huge improvement compared to applying a traditional CNN to the same dataset. Addresses the challenge of segmenting large-size chest X-ray images. | Not evaluated on a standard dataset. Small testing set. Boundaries not that smooth. FPs or FNs when similar density between regions or high-curvature lung regions. |
| Chen B et al. (2020) [37] | Two-Stream Collaborative Network (TSCN) with U-net at segmentation stage | JSRT, Montgomery County and NIH | - | 0.973 | Performs classification. Few training images. Combined datasets for training and validation. | Poor performance with the Infiltration group. |
| Chen HJ et al. (2020) [38] | CNN-based architectures applied on binarized images | Montgomery County and private clinic in India | 0.842 | 0.893 | Fast training, low storage requirements. Contrast enhancement helps a lot to improve Dice score. | Contrast enhancement usefulness in terms of Jaccard measurement improvement depends on the selected Network architecture. Relatively small validation and testing set. |
| Our method | Rib cage points-driven region growing | Montgomery County | 0.862 | 0.923 | Straightforward unsupervised method. Can be used for rapid triage of patients. | Depends on at least some visibility of the rib cage and a distinguishable border curve. A few parameters still have to be selected by intuition. |
| Count of Failed Cases | Location | Reason/Diagnostic Relevance |
|---|---|---|
| 5 | Bottom part | Bright region probably due to pleural effusion |
| 4 | Bottom part | Pleural fluid causes bright areas |
| 3 | Multiple | Infiltrates due to TB—brighter regions, bone structures not sufficiently visible |
| 1 | Inner part of right lung | Increased length of cardiac silhouette, probably due to pericarditis |
| 1 | Left lung’s outer side brighter | Bright area likely because of infiltrate due to pneumonia |
| 1 | Bottom part | Bright region, localized pleural peel |
| 1 | Inner part of right lung (right hilum) | Congestive heart failure or infiltrate due to TB |
| 1 | Inner part of left lung | Extended bright region due to cardiomegaly |
| 2 | Top-left/top-right part of right lung | Inaccurate symmetry detection algorithm crop or glenohumeral joint area crop. Diagnosis irrelevant. |
| 7 | Multiple | CXRs either normal or pathological. The failure is irrelevant to the diagnosis and is probably due to poor contrast. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bosdelekidis, V.; Ioakeimidis, N.S. Lung Field Segmentation in Chest X-rays: A Deformation-Tolerant Procedure Based on the Approximation of Rib Cage Seed Points. Appl. Sci. 2020, 10, 6264. https://doi.org/10.3390/app10186264
Bosdelekidis V, Ioakeimidis NS. Lung Field Segmentation in Chest X-rays: A Deformation-Tolerant Procedure Based on the Approximation of Rib Cage Seed Points. Applied Sciences. 2020; 10(18):6264. https://doi.org/10.3390/app10186264
Chicago/Turabian StyleBosdelekidis, Vasileios, and Nikolaos S. Ioakeimidis. 2020. "Lung Field Segmentation in Chest X-rays: A Deformation-Tolerant Procedure Based on the Approximation of Rib Cage Seed Points" Applied Sciences 10, no. 18: 6264. https://doi.org/10.3390/app10186264
APA StyleBosdelekidis, V., & Ioakeimidis, N. S. (2020). Lung Field Segmentation in Chest X-rays: A Deformation-Tolerant Procedure Based on the Approximation of Rib Cage Seed Points. Applied Sciences, 10(18), 6264. https://doi.org/10.3390/app10186264





