Disentangled Autoencoder for Cross-Stain Feature Extraction in Pathology Image Analysis
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
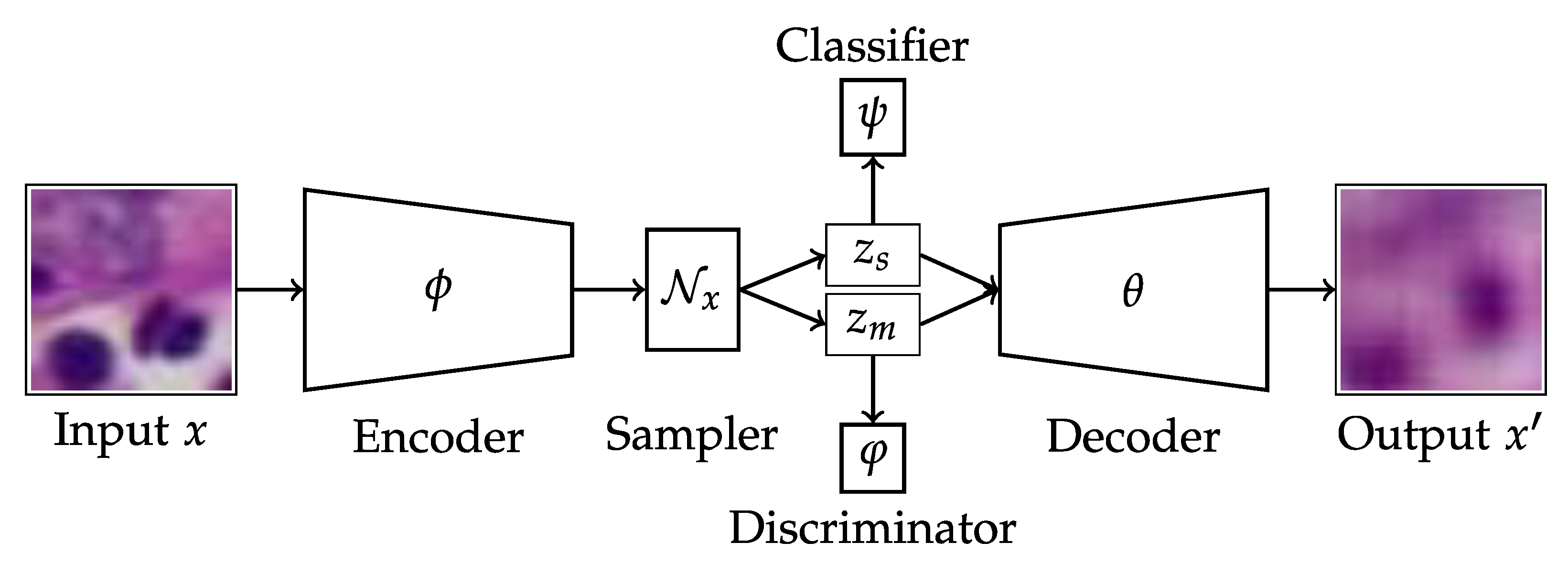
2.2.1. Disentangled Autoencoder
2.2.2. Network Architecture
2.2.3. Distances in the Latent Space
- Bhattacharyya distance (BD):
- Symmetrized Kullback–Leibler divergence (SKLD):where is Kullback–Leibler divergence as used in the latent loss from Equation (3). The Equation (11) further simplifies in the case of diagonal covariance matrices for the two distributions.
2.2.4. Bag of Visual Words
2.2.5. Statistical Analyses
3. Results
3.1. Latent Space Disentanglement
3.2. Image Registration Scenario
3.3. Intra-Tumour Heterogeneity Scenario
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef]
- Viergever, M.A.; Maintz, J.B.A.; Klein, S.; Murphy, K.; Staring, M.; Pluim, J.P.W. A survey of medical image registration—Under review. Med. Image Anal. 2016, 33, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar] [PubMed]
- van der Loos, C.M. Multiple Immunoenzyme Staining: Methods and Visualizations for the Observation With Spectral Imaging. J. Histochem. Cytochem. 2008, 56, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Alsubaie, N.; Trahearn, N.; Raza, S.E.A.; Snead, D.; Rajpoot, N.M. Stain Deconvolution Using Statistical Analysis of Multi-Resolution Stain Colour Representation. PLoS ONE 2017, 12, e0169875. [Google Scholar] [CrossRef] [PubMed]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A method for normalizing histology slides for quantitative analysis. In Proceedings of the Proceedings—2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
- Gurcan, M.N.; Boucheron, L.E.; Can, A.; Madabhushi, A.; Rajpoot, N.M.; Yener, B. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2009, 2, 147–171. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Weis, C.-A.; Bianconi, F.; Melchers, S.M.; Schad, L.R.; Gaiser, T.; Marx, A.; Zöllner, F.G. Multi-class texture analysis in colorectal cancer histology. Sci. Rep. 2016, 6, 27988. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Khosla, A.; Gargeya, R.; Irshad, H.; Beck, A.H. Deep Learning for Identifying Metastatic Breast Cancer. arXiv 2016, arXiv:1606.05718. [Google Scholar]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.-A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning internal representations by error propagation. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition: Foundations; MIT Press: Cambridge, MA, USA, 1987; pp. 318–362. [Google Scholar]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Bengio, Y.; LeCun, Y. Scaling Learning Algorithms toward AI. In Large-Scale Kernel Machines; Bottou, L., Chapelle, O., DeCoste, D., Weston, J., Eds.; MIT Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Erhan, D.; Courville, A.; Bengio, Y.; Vincent, P. Why does unsupervised pre-training help deep learning? J. Mach. Learn. Res. 2010, 9, 201–208. [Google Scholar]
- Xu, J.; Xiang, L.; Liu, Q.; Gilmore, H.; Wu, J.; Tang, J.; Madabhushi, A. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans. Med Imaging 2016, 35, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Nguyen, V.; Kanevsky, A.B.; Samaras, D.; Kurc, T.M.; Zhao, T.; Gupta, R.R.; Gao, Y.; Chen, W.; Foran, D.; et al. Sparse autoencoder for unsupervised nucleus detection and representation in histopathology images. Pattern Recognit. 2019, 86, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Basavanhally, A.; Madabhushi, A. Stain Normalization using Sparse AutoEncoders (StaNoSA): Application to digital pathology. Comput. Med Imaging Graph. 2017, 57, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Awan, R.; Rajpoot, N. Deep Autoencoder Features for Registration of Histology Images. In Proceedings of the Medical Image Understanding and Analysis, Houston, TX, USA, 10 February 2018; Springer: Cham, Switzerland, 2018; pp. 371–378. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. In Proceedings of the International Conference on Learning Representations, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]
- Vincent, P.; Larochelle, H.; Bengio, Y.; Manzagol, P.-A. Extracting and composing robust features with denoising autoencoders. In Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 5–9 July 2008. [Google Scholar]
- Coates, A.; Ng, A.Y. Learning Feature Representations with K-Means. In Neural Networks: Tricks of the Trade, 2nd ed.; Springer: Berlin, Germany, 2012; Volume 7700, pp. 561–580. [Google Scholar]
- Higgins, I.; Matthey, L.; Pal, A.; Burgess, C.; Glorot, X.; Botvinick, M.; Mohamed, S.; Lerchner, A. Β-VAE: Learning basic visual concepts with a constrained variational framework. In Proceedings of the 5th International Conference on Learning Representations, ICLR 2017—Conference Track Proceedings, Toulon, France, 24–26 April 2017. [Google Scholar]
- Burgess, C.P.; Higgins, I.; Pal, A.; Matthey, L.; Watters, N.; Desjardins, G.; Lerchner, A. Understanding disentangling in β-VAE. In Proceedings of the 2017 NIPS Workshop on Learning Disentangled Representations, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the 27th International Conference on Neural Information Processing Systems—Volume 2, Montreal, QC, Canada, 8–13 December 2014; MIT Press: Cambridge, MA, USA, 2014; pp. 2672–2680. [Google Scholar]
- Tzeng, E.; Hoffman, J.; Saenko, K.; Darrell, T. Adversarial Discriminative Domain Adaptation. Comput. Res. Repos. (CoRR) 2017, abs/1702.0, 7167–7176. [Google Scholar]
- Qin, C.; Shi, B.; Liao, R.; Mansi, T.; Rueckert, D.; Kamen, A. Unsupervised Deformable Registration for Multi-Modal Images via Disentangled Representations. Comput. Res. Repos. (CoRR) 2019, abs/1903.0, 249–261. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognit. (CVPR), Los Angeles, CA, USA, 26 June–1 July 2016; IEEE: New York, NY, USA, 2016; pp. 770–778. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. Lecture Notes in Computer Science. In Proceedings of the ECCV 2016, Amsterdam, The Netherlands, 11–14 October 2016. [Google Scholar]
- Csurka, G.; Dance, C.; Fan, L. Visual categorization with bags of keypoints. In Proceedings of the ECCV International Workshop on Statistical Learning in Computer Vision 2004, Prague, Czech Republic, 11–14 May 2004; Springer: Cham, Switzerland, 2004. [Google Scholar]
- Caicedo, J.C.; Cruz, A.; Gonzalez, F.A. Histopathology Image Classification Using Bag of Features and Kernel Functions. In Proceedings of the 12th Conference on Artificial Intelligence in Medicine, Verona, Italy, 18–22 July 2009; Combi, C., Shahar, Y., Abu-Hanna, A., Eds.; Springer: Berlin/Heidelberg, Germany; Verona, Italy, 2009; pp. 126–135. [Google Scholar]
- López-Monroy, A.P.; Montes-y-Gómez, M.; Escalante, H.J.; Cruz-Roa, A.; González, F.A. Bag-of-Visual-Ngrams for Histopathology Image Classification. In Proc SPIE 8922, IX International Seminar on Medical Information Processing and Analysis; Mexico City, Mexico, 11–14 November 2013; Brieva, J., Escalante-Ramírez, B., Eds.; SPIE: Bellingham, WA, USA, 2013. [Google Scholar]
- Jégou, H.; Perronnin, F.; Douze, M.; Sanchez, J.; Perez, P.; Schmid, C. Aggregating Local Image Descriptors into Compact Codes. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 1704–1716. [Google Scholar] [CrossRef] [PubMed]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters a density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the KDD’96: Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Simonovsky, M.; Gutiérrez-Becker, B.; Mateus, D.; Navab, N.; Komodakis, N. A Deep Metric for Multimodal Registration. arXiv 2016, arXiv:1609.05396. [Google Scholar]









| Error (Pixels) | SKLD | BD | SAD | MI |
|---|---|---|---|---|
| Average (Std. dev) | 21.8 (10.2) | 21.7 (10.2) | 24.8 (12.0) | 21.6 (11.1) |
| Median | 22.3 | 22.1 | 26.7 | 22.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hecht, H.; Sarhan, M.H.; Popovici, V. Disentangled Autoencoder for Cross-Stain Feature Extraction in Pathology Image Analysis. Appl. Sci. 2020, 10, 6427. https://doi.org/10.3390/app10186427
Hecht H, Sarhan MH, Popovici V. Disentangled Autoencoder for Cross-Stain Feature Extraction in Pathology Image Analysis. Applied Sciences. 2020; 10(18):6427. https://doi.org/10.3390/app10186427
Chicago/Turabian StyleHecht, Helge, Mhd Hasan Sarhan, and Vlad Popovici. 2020. "Disentangled Autoencoder for Cross-Stain Feature Extraction in Pathology Image Analysis" Applied Sciences 10, no. 18: 6427. https://doi.org/10.3390/app10186427
APA StyleHecht, H., Sarhan, M. H., & Popovici, V. (2020). Disentangled Autoencoder for Cross-Stain Feature Extraction in Pathology Image Analysis. Applied Sciences, 10(18), 6427. https://doi.org/10.3390/app10186427






