Microbial Activity in Subterranean Ecosystems: Recent Advances
Abstract
:1. Introduction
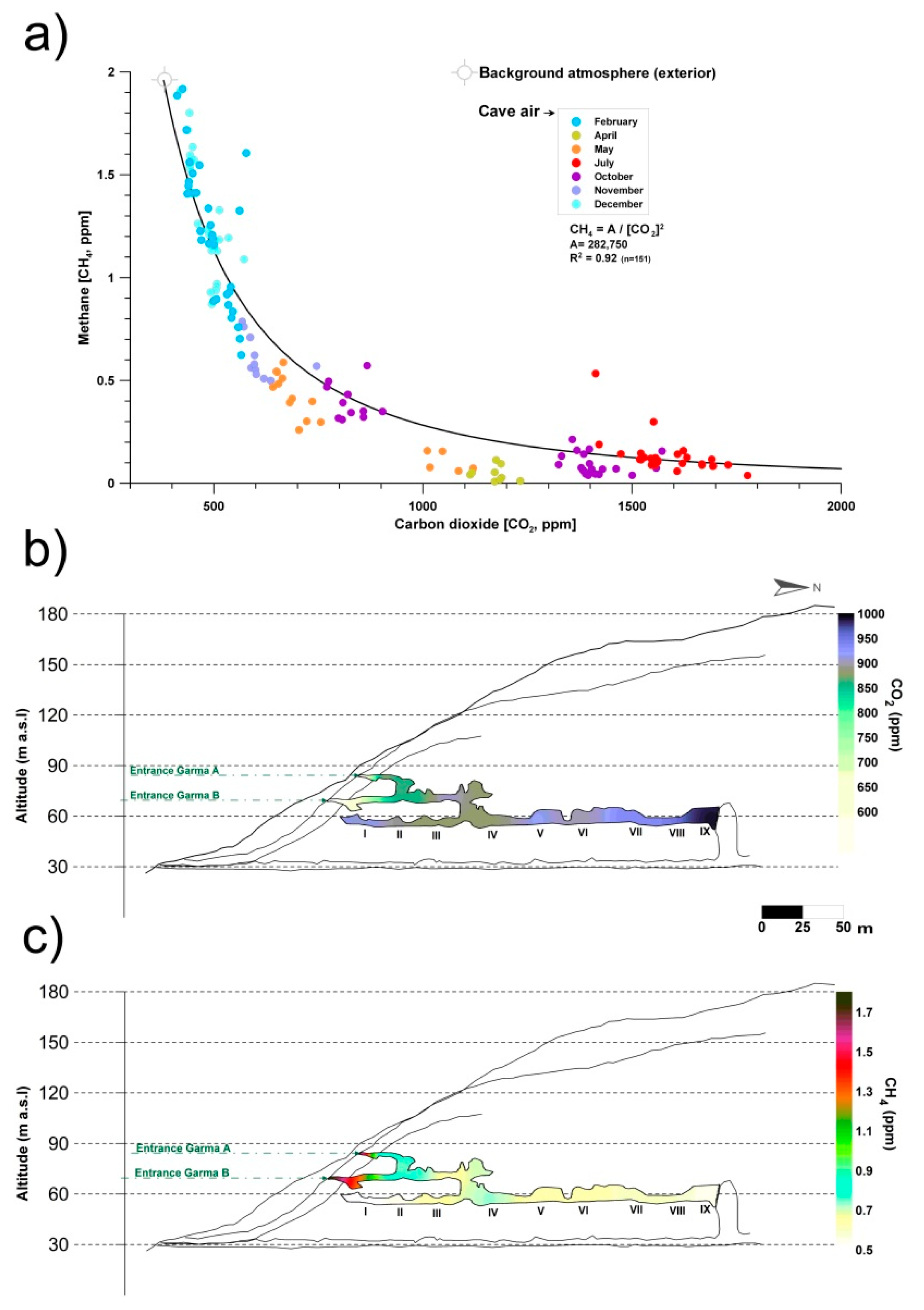
2. The Control of Greenhouse Gas Fluxes by Cave Microorganisms
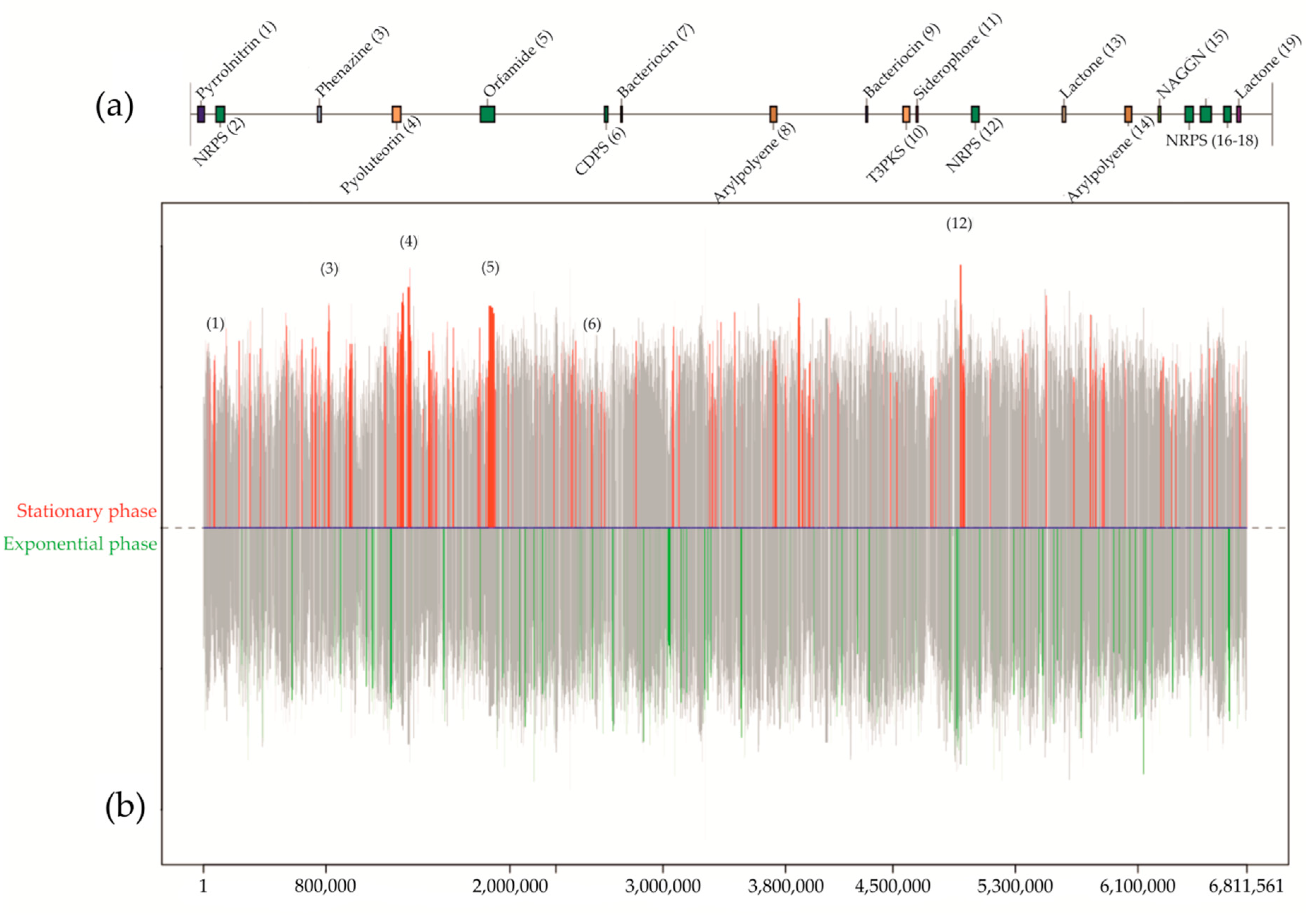
3. The Search of Antibiotics Produced by Subsurface Bacteria
3.1. Why Is There a Need of New Antibiotics?
3.2. Antibiotics from Subsurface Bacteria
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ford, D.C.; Williams, P.W. Karst Hydrogeology and Geomorphology; Wiley: Chichester, UK, 2007. [Google Scholar]
- Giardino, J.R.; Houser, C. (Eds.) Introduction to the critical zone. In Principles and Dynamics of the Critical Zone; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–13. [Google Scholar]
- Gold, T. The deep, hot biosphere. Proc. Natl. Acad. Sci. USA 1992, 89, 6045–6049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.A.; Bennett, P.C. Mineral ecology: Surface specific colonization and geochemical drivers of biofilm accumulation, composition, and phylogeny. Front. Microbiol. 2017, 8, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuezva, S.; Sanchez-Moral, S.; Saiz-Jimenez, C.; Cañaveras, J.C. Microbial communities and associated mineral fabrics in Altamira Cave, Spain. Int. J. Speleol. 2009, 38, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Northup, D.E.; Lavoie, K.H. Geomicrobiology of caves: A review. Geomicrobiol. J. 2001, 18, 199–222. [Google Scholar] [CrossRef]
- Sánchez-Moral, S.; Portillo, M.D.C.; Janices, I.; Cuezva, S.; Cortés, Ángel, F.; Cañaveras, J.C.; Gonzalez, J.M. The role of microorganisms in the formation of calcitic moonmilk deposits and speleothems in Altamira Cave. Geomorphology 2012, 139–140, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Cuezva, S.; Fernandez-Cortes, A.; Porca, E.; Pašić, L.; Jurado, V.; Hernández, M.; Serrano-Ortiz, P.; Hermosin, B.; Cañaveras, J.C.; Sanchez-Moral, S.; et al. The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiol. Ecol. 2012, 81, 281–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, R.M.; Goordial, J.M.; Orcutt, B.N. Low energy subsurface environments as extraterrestrial analogs. Front. Microbiol. 2018, 9, 1605. [Google Scholar] [CrossRef]
- Lavoie, K.H.; Winter, A.S.; Read, K.J.H.; Hughes, E.M.; Spilde, M.N.; Northup, D.E. Comparison of bacterial communities from lava cave microbial mats to overlying surface soils from Lava Beds National Monument, USA. PLoS ONE 2017, 12, e0169339. [Google Scholar] [CrossRef]
- Miller, A.Z.; Garcia-Sanchez, A.M.; Martin-Sanchez, P.M.; Pereira, M.F.C.; Spangenberg, J.E.; Jurado, V.; Dionísio, A.; Afonso, M.J.; Chaminé, H.I.I.S.; Hermosin, B.; et al. Origin of abundant moonmilk deposits in a subsurface granitic environment. Sedimentology 2018, 65, 1482–1503. [Google Scholar] [CrossRef] [Green Version]
- Sasowsky, I.D.; Mylroie, J. Studies of Cave Sediments. Physical and Chemical Records of Paleoclimate; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Ghosh, S.; Kuisiene, N.; Cheeptham, N. The cave microbiome as a source for drug discovery: Reality or pipe dream? Biochem. Pharmacol. 2017, 134, 18–34. [Google Scholar] [CrossRef]
- Engel, A.S. Microbial Life of Cave Systems; De Gruiter: Berlin, Germany, 2015. [Google Scholar]
- Moldovan, O.T.; Kovac, L.; Halse, S. Cave Ecology; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Jones, B. Microbial activity in caves—A geological perspective. Geomicrobiol. J. 2001, 18, 345–357. [Google Scholar] [CrossRef]
- Barton, H.A.; Northup, D.E. Geomicrobiology in cave environments: Past, current and future perspectives. J. Cave Karst Stud. 2007, 69, 163–178. [Google Scholar]
- Engel, A.S. Microbial diversity of cave ecosystems. In Geomicrobiology: Molecular and Environmental Perspectives; Barton, L.L., Mandl, M., Loy, A., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 219–238. [Google Scholar]
- Tomczyk-Żak, K.; Zielenkiewicz, U. Microbial diversity in caves. Geomicrobiol. J. 2016, 33, 20–38. [Google Scholar] [CrossRef]
- Jones, D.S.; Macalady, J.L. The snotty and the stringy: Energy for subsurface life in caves. In Their World: A Diversity of Microbial Environments; Hurst, C.J., Ed.; Springer: Cham, Switzerland, 2016; pp. 203–224. [Google Scholar]
- De Mandal, S.; Chatterjee, R.; Kumar, N.S. Dominant bacterial phyla in caves and their predicted functional roles in C and N cycle. BMC Microbiol. 2017, 17, 90. [Google Scholar] [CrossRef] [Green Version]
- Etiope, G.; Klusman, R.W. Geologic emissions of methane to the atmosphere. Chemosphere 2002, 49, 777–789. [Google Scholar] [CrossRef]
- Etiope, G.; Lollar, B.S. Abiotic methane on earth. Rev. Geophys. 2013, 51, 276–299. [Google Scholar] [CrossRef]
- Intergovernmental Panel on Climate Change. Carbon and other biogeochemical cycles. In Climate Change 2013: The Physical Science Basis; Cambridge Univ. Press: Cambridge, UK, 2014; pp. 465–570. [Google Scholar]
- Hall, E.K.; Bernhardt, E.S.; Bier, R.L.; Bradford, M.A.; Boot, C.M.; Cotner, J.B.; Del Giorgio, P.A.; Evans, S.E.; Graham, E.B.; Jones, S.E.; et al. Understanding how microbiomes influence the systems they inhabit. Nat. Microbiol. 2018, 3, 977–982. [Google Scholar] [CrossRef]
- Op den Camp, H.J.M.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.M.; Birkeland, N.-K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef]
- Butterfield, C.N.; Li, Z.; Andeer, P.F.; Spaulding, S.; Thomas, B.C.; Singh, A.; Hettich, R.L.; Suttle, K.B.; Probst, A.J.; Tringe, S.G.; et al. Proteogenomic analyses indicate bacterial methylotrophy and archaeal heterotrophy are prevalent below the grass root zone. PeerJ 2016, 4, e2687. [Google Scholar] [CrossRef] [Green Version]
- Hutchens, E.; Radajewski, S.; Dumont, M.G.; McDonald, I.R.; Murrell, J.C. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 2004, 6, 111–120. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, L.; Boden, R.; Hillebrand, A.; Kumaresan, D.; Moussard, H.; Baciu, M.; Lu, Y.; Murrell, J.C. Life without light: Microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 2009, 3, 1093–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumaresan, D.; Wischer, D.; Stephenson, J.; Hillebrand-Voiculescu, A.; Murrell, J.C. Microbiology of Movile Cave—A chemolithoautotrophic ecosystem. Geomicrobiol. J. 2014, 31, 186–193. [Google Scholar] [CrossRef]
- Kumaresan, D.; Stephenson, J.; Doxey, A.C.; Bandukwala, H.; Brooks, E.; Hillebrand-Voiculescu, A.; Whiteley, A.S.; Murrell, J.C. Aerobic proteobacterial methylotrophs in Movile Cave: Genomic and metagenomic analyses. Microbiome 2018, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiz-Jimenez, C. The microbiology of show caves, mines tunnels and tombs: Implications for management and conservation. In Microbial Life of Cave Systems; Engel, A.S., Ed.; De Gruiter: Berlin, Germany, 2015; pp. 231–261. [Google Scholar]
- Gonzalez-Pimentel, J.L.; Miller, A.Z.; Jurado, V.; Laiz, L.; Pereira, M.F.C.; Saiz-Jimenez, C. Yellow coloured mats from lava tubes of La Palma (Canary Islands, Spain) are dominated by metabolically active Actinobacteria. Sci. Rep. 2018, 8, 1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Cortes, A.; Cuezva, S.; Alvarez-Gallego, M.; Garcia-Anton, E.; Pla, C.; Benavente, D.; Jurado, V.; Saiz-Jimenez, C.; Sanchez-Moral, S. Subterranean atmospheres may act as daily methane sinks. Nat. Commun. 2015, 6, 7003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, I.R.; Murrell, J.C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol. Lett. 1997, 156, 205–210. [Google Scholar] [CrossRef]
- Tveit, A.T.; Hestnes, A.G.; Robinson, S.L.; Schintlmeister, A.; Dedysh, S.N.; Jehmlich, N.; Von Bergen, M.; Herbold, C.; Wagner, M.; Richter, A.; et al. Widespread soil bacterium that oxidizes atmospheric methane. Proc. Natl. Acad. Sci. USA 2019, 116, 8515–8524. [Google Scholar] [CrossRef] [Green Version]
- Schimmelmann, A.; Fernandez-Cortes, A.; Cuezva, S.; Streil, T.; Lennon, J.T. Radiolysis via radioactivity is not responsible for rapid methane oxidation in subterranean air. PLoS ONE 2018, 13, e0206506. [Google Scholar] [CrossRef]
- Webster, K.D.; Drobniak, A.; Etiope, G.; Mastalerz, M.; Sauer, P.E.; Schimmelmann, A. Subterranean karst environments as a global sink for atmospheric methane. Earth Planet. Sci. Lett. 2018, 485, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Lennon, J.T.; Nguyễn-Thùy, D.; Phạm, T.M.; Drobniak, A.; Tạ, P.H.; Phạm, N.Ð.; Streil, T.; Webster, K.D.; Schimmelmann, A. Microbial contributions to subterranean methane sinks. Geobiology 2016, 15, 254–258. [Google Scholar] [CrossRef]
- Waring, C.L.; Hankin, S.I.; Griffith, D.W.T.; Kertesz, M.A.; Kobylski, V.; Wilson, N.L.; Coleman, N.V.; Kettlewell, G.; Zlot, R.; Bosse, M.; et al. Seasonal total methane depletion in limestone caves. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda, L.; Vadillo, I.; Etiope, G.; Benavente, J.; Liñán, C.; del Rosal, Y.; Tapia, S.T.; Moríñigo, M.Á.; Carrasco, F. Methane sources and sinks in karst systems: The Nerja cave and its vadose environment (Spain). Geochim. Cosmochim. Acta 2019, 259, 302–315. [Google Scholar] [CrossRef]
- Cuezva, S.; Martin-Pozas, T.; Fernandez-Cortes, A.; Cañaveras, J.C.; Janssens, I.; Sanchez-Moral, S. On the role of cave-soil in the carbon cycle. A first approach. In Proceedings of the EGU General Assembly 2020, Online, 4–8 May 2020. Abstract 21793. [Google Scholar] [CrossRef]
- Jurado, V.; Gonzalez-Pimentel, J.L.; Miller, A.Z.; Hermosin, B.; D’Angeli, I.M.; Tognini, P.; De Waele, J.; Saiz-Jimenez, C. Microbial communities in vermiculation deposits from an Alpine cave. Front Earth Sci. 2020, in press. [Google Scholar]
- Martin-Pozas, T.; Sánchez-Moral, S.; Cuezva, S.; Jurado, V.; Saiz-Jimenez, C.; López, R.P.; Carrey, R.; Otero, N.; Giesemann, A.; Well, R.; et al. Biologically mediated release of endogenous N2O and NO2 gases in a hydrothermal, hypoxic subterranean environment. Sci. Total Environ. 2020, 747, 141218. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cortes, A.; Perez-Lopez, R.; Cuezva, S.; Calaforra, J.M.; Cañaveras, J.C.; Sanchez-Moral, S. Geochemical fingerprinting of rising deep endogenous gases in an active hypogenic karst system. Geofluids 2018, 2018, 1–19. [Google Scholar] [CrossRef]
- He, Z.; Cai, C.; Wang, J.; Xu, X.; Zheng, P.; Jetten, M.S.M.; Hu, B. A novel denitrifying methanotroph of the NC10 phylum and its microcolony. Sci. Rep. 2016, 6, srep32241. [Google Scholar] [CrossRef]
- Isobe, K.; Bouskill, N.J.; Brodie, E.L.; Sudderth, E.A.; Martiny, J.B.H. Phylogenetic conservation of soil bacterial responses to simulated global changes. Philos. Trans. R. Soc. B 2020, 375, 20190242. [Google Scholar] [CrossRef]
- Cappelletti, M.; Ghezzi, D.; Zannoni, D.; Capaccioni, B.; Fedi, S. Diversity of methane-oxidizing bacteria in soils from “Hot Lands of Medolla” (Italy) featured by anomalous high-temperatures and biogenic CO2 emission. Microbes Environ. 2016, 31, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Kenney, G.E.; Rosenzweig, A.C. Methanobactins: Maintaining copper homeostasis in methanotrophs and beyond. J. Biol. Chem. 2018, 293, 4606–4615. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Chen, Y.; Jiang, P.; Zhang, C.; Smith, T.J.; Murrell, J.C.; Xing, X.-H. Methanotrophs: Multifunctional bacteria with promising applications in environmental bioengineering. Biochem. Eng. J. 2010, 49, 277–288. [Google Scholar] [CrossRef]
- Pandey, V.C.; Singh, J.S.; Singh, D.P.; Singh, R.P. Methanotrophs: Promising bacteria for environmental remediation. Int. J. Environ. Sci. Technol. 2014, 11, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Iguchi, H.; Yurimoto, H.; Sakai, Y. Stimulation of methanotrophic growth in cocultures by cobalamin excreted by Rhizobia. Appl. Environ. Microbiol. 2011, 77, 8509–8515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stock, M.; Hoefman, S.; Kerckhof, F.-M.; Boon, N.; De Vos, P.; De Baets, B.; Heylen, K.; Waegeman, W. Exploration and prediction of interactions between methanotrophs and heterotrophs. Res. Microbiol. 2013, 164, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Veraart, A.J.; Garbeva, P.; Van Beersum, F.; Ho, A.; Hordijk, C.A.; Meima-Franke, M.; Zweers, A.J.; Bodelier, P.L.E. Living apart together—Bacterial volatiles influence methanotrophic growth and activity. ISME J. 2018, 12, 1163–1166. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- De Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- World Health Organization. Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Theuretzbacher, U. Antibiotic innovation for future public health needs. Clin. Microbiol. Infect. 2017, 23, 713–717. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; Van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Mast, Y.; Stegmann, E. Actinomycetes: The antibiotics producers. Antibiotics 2018, 8, 105. [Google Scholar] [CrossRef] [Green Version]
- Waksman, S.A.; Woodruff, H.B. Bacteriostatic and bactericidal substances produced by a soil Actinomyces. Exp. Biol. Med. 1940, 45, 609–614. [Google Scholar] [CrossRef]
- Schatz, A.; Bugle, E.; Waksman, S.A. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Exp. Biol. Med. 1944, 55, 66–69. [Google Scholar] [CrossRef]
- Nakashima, T.; Anzai, K.; Suzuki, R.; Kuwahara, N.; Takeshita, S.; Kanamoto, A.; Ando, K. Productivity of bioactive compounds in Streptomyces species isolated from Nagasaki marine environments. Actinomycetologica 2009, 23, 16–20. [Google Scholar] [CrossRef]
- Koomsiri, W.; Inahashi, Y.; Kimura, T.; Shiomi, K.; Takahashi, Y.; Ōmura, S.; Thamchaipenet, A.; Nakashima, T. Bisoxazolomycin A: A new natural product from ‘Streptomyces subflavus subsp. irumaensis’ AM-3603. J. Antibiot. 2017, 70, 1142–1145. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nakashima, T. Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics 2018, 7, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, W.; Burke, D.C. Marine products. XXXIX. The nucleosides of sponges. III. Spongothymidine and spongouridine. J. Org. Chem. 1955, 20, 1501–1507. [Google Scholar] [CrossRef]
- Lindequist, U. Marine-derived pharmaceuticals—Challenges and opportunities. Biomol. Ther. 2016, 24, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, K.; Gupta, R.K. Diversity and isolation of rare actinomycetes: An overview. Crit. Rev. Microbiol. 2013, 39, 256–294. [Google Scholar] [CrossRef]
- Fang, B.-Z.; Salam, N.; Han, M.-X.; Jiao, J.-Y.; Cheng, J.; Wei, D.-Q.; Xiao, M.; Li, W.-J. Insights on the effects of heat pretreatment, pH, and calcium salts on isolation of rare Actinobacteria from karstic caves. Front. Microbiol. 2017, 8, 1535. [Google Scholar] [CrossRef] [Green Version]
- Cheeptham, N. Cave Microbiomes: A Novel Resource for Drug Discovery; Springer: New York, NY, USA, 2013. [Google Scholar]
- Cheeptham, N.; Saiz-Jimenez, C. New sources of antibiotics: Caves. In Antibiotics. Current Innovations and Future Trends; Sánchez, S., Demain, A.L., Eds.; Caister Academic Press: Portland, Oregon, 2015; pp. 213–227. [Google Scholar]
- Groth, I.; Vettermann, R.; Schuetze, B.; Schumann, P.; Saiz-Jimenez, C. Actinomycetes in Karstic caves of northern Spain (Altamira and Tito Bustillo). J. Microbiol. Methods 1999, 36, 115–122. [Google Scholar] [CrossRef]
- Jurado, V.; Laiz, L.; Rodríguez-Nava, V.; Boiron, P.; Hermosin, B.; Sanchez-Moral, S.; Saiz-Jimenez, C. Pathogenic and opportunistic microorganisms in caves. Int. J. Speleol. 2010, 39, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Cheeptham, N.; Sadoway, T.; Rule, D.; Watson, K.; Moote, P.; Soliman, L.C.; Azad, N.; Donkor, K.K.; Horne, D. Cure from the cave: Volcanic cave actinomycetes and their potential in drug discovery. Int. J. Speleol. 2013, 42, 35–47. [Google Scholar] [CrossRef] [Green Version]
- Belyagoubi, L.; Belyagoubi-Benhammou, N.; Jurado, V.; Dupont, J.; Lacoste, S.; Djebbah, F.; Ounadjela, F.Z.; Benaissa, S.; Habi, S.; Abdelouahid, D.E.; et al. Antimicrobial activities of culturable microorganisms (actinomycetes and fungi) isolated from Chaabe Cave, Algeria. Int. J. Speleol. 2018, 47, 189–199. [Google Scholar] [CrossRef] [Green Version]
- Yasir, M. Analysis of bacterial communities and characterization of antimicrobial strains from cave microbiota. Braz. J. Microbiol. 2018, 49, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Rangseekaew, P.; Pathom-Aree, W. Cave Actinobacteria as producers of bioactive metabolites. Front. Microbiol. 2019, 10, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avguštin, J.A.; Petrič, P.; Pašić, L. Screening the cultivable cave microbial mats for the production of antimicrobial compounds and antibiotic resistance. Int. J. Speleol. 2019, 48, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Voytsekhovskaya, I.V.; Axenov-Gribanov, D.V.; Murzina, S.A.; Pekkoeva, S.N.; Protasov, E.S.; Gamaiunov, S.V.; Timofeyev, M. Estimation of antimicrobial activities and fatty acid composition of actinobacteria isolated from water surface of underground lakes from Badzheyskaya and Okhotnichya caves in Siberia. PeerJ 2018, 6, e5832. [Google Scholar] [CrossRef]
- Herold, K.; Gollmick, F.A.; Groth, I.; Roth, M.; Menzel, K.-D.; Möllmann, U.; Gräfe, U.; Hertweck, C. Cervimycin A-D: A polyketide glycoside complex from a cave bacterium can defeat vancomycin resistance. Chem. Eur. J. 2005, 11, 5523–5530. [Google Scholar] [CrossRef]
- Jiang, L.; Pu, H.; Xiang, J.; Su, M.; Yan, X.; Yang, D.; Zhu, X.; Shen, B.; Duan, Y.; Huang, Y. Huanglongmycin A-C, cytotoxic polyketides biosynthesized by a putative type ii polyketide synthase from Streptomyces sp. CB09001. Front. Chem. 2018, 6, 254. [Google Scholar] [CrossRef]
- Axenov-Gibanov, D.; Voytsekhovskaya, I.V.; Tokovenko, B.T.; Protasov, E.S.; Gamaiunov, S.V.; Rabets, Y.V.; Luzhetskyy, A.N.; Timofeyev, M.A. Actinobacteria isolated from an underground lake moonmilk speleothem from the biggest conglomeratic karstic cave in Siberia as sources of novel biologically active compounds. PLoS ONE 2016, 11, e0149216. [Google Scholar] [CrossRef]
- Bollmann, A.; Lewis, K.; Epstein, S.S. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 2007, 73, 6386–6390. [Google Scholar] [CrossRef] [Green Version]
- Hug, J.J.; Bader, C.D.; Remškar, M.; Cirnski, K.; Müller, R. Concepts and methods to access novel antibiotics from Actinomycetes. Antibiotics 2018, 7, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, K.; Epstein, S.S.; D’Onofrio, A.; Ling, L.L. Uncultured microorganisms as a source of secondary metabolites. J. Antibiot. 2010, 63, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Manteca, A.; Yagüe, P. Streptomyces differentiation in liquid cultures as a trigger of secondary metabolism. Antibiotics 2018, 7, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Bukelskis, D.; Dabkeviciene, D.; Lukoseviciute, L.; Bucelis, A.; Kriaučiūnas, I.; Lebedeva, J.; Kuisiene, N. Screening and transcriptional analysis of polyketide synthases and non-ribosomal peptide synthetases in bacterial strains from Krubera-Voronja Cave. Front. Microbiol. 2019, 10, 2149. [Google Scholar] [CrossRef]
- Giubergia, S.; Phippen, C.; Nielsen, K.F.; Gram, L. Growth on chitin impacts the transcriptome and metabolite profiles of antibiotic-producing Vibrio coralliilyticus S2052 and Photobacterium galatheae S2753. mSystems 2017, 2, e00141-16. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Y.; Rebets, Y.; Estévez, M.R.; Zapp, J.; Myronovskyi, M.; Luzhetskyy, A. Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters. Microb. Cell Fact. 2020, 19, 5. [Google Scholar] [CrossRef] [Green Version]
- Paun, V.I.; Icaza, G.; Lavin, P.; Marin, C.; Tudorache, A.; Perşoiu, A.; Dorador, C.; Purcarea, C. Total and potentially active bacterial communities entrapped in a late glacial through holocene ice core from Scarisoara Ice Cave, Romania. Front. Microbiol. 2019, 10, 1193. [Google Scholar] [CrossRef]
- Interreg project. 0483_PROBIOMA_5_E: Prospecting Underground Environments for Microbial Bioactive Compounds with Potential Use in Medicine, Agriculture and Environment. Available online: https://probioma.org (accessed on 15 November 2020).
- Sánchez España, J. Acid mine drainage in the Iberian Pyrite Belt: An overview with special emphasis on generation mechanisms, aqueous composition and associated mineral phases. Macla 2008, 10, 34–43. [Google Scholar]
- Porca, E.; Jurado, V.; Martin-Sanchez, P.M.; Hermosin, B.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Aerobiology: An ecological indicator for early detection and control of fungal outbreaks in caves. Ecol. Indic. 2011, 11, 1594–1598. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jünemann, S.; Kleinbölting, N.; Jaenicke, S.; Henke, C.; Hassa, J.; Nelkner, J.; Stolze, Y.; Albaum, S.P.; Schlüter, A.; Goesmann, A.; et al. Bioinformatics for NGS-based metagenomics and the application to biogas research. J. Biotechnol. 2017, 261, 10–23. [Google Scholar] [CrossRef]
- Alain, K.; Callac, N.; Ciobanu, M.C.; Reynaud, Y.; Duthoit, F.; Jebbar, M. DNA extractions from deep subsea floor sediments: Novel cryogenic mill-based procedure and comparison to existing protocols. J. Microbiol. Methods 2011, 87, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [Green Version]

| Origin | Tested Strains | Positive Strains | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|---|
| Terrestrial caves | 863 | 126 | 77 | 48 | 116 | 18 | 31 | 24 |
| Marine caves | 144 | 2 | 0 | 0 | 2 | 0 | 1 | 0 |
| Mines | 79 | 23 | 13 | 7 | 14 | 3 | 8 | 3 |
| Total (bacteria) | 1086 | 151 | 90 | 55 | 132 | 21 | 40 | 27 |
| Total (%) | 100% | 13.9% | 8.29% | 5.06% | 12.15% | 1.93% | 3.68% | 2.49% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin-Pozas, T.; Gonzalez-Pimentel, J.L.; Jurado, V.; Cuezva, S.; Dominguez-Moñino, I.; Fernandez-Cortes, A.; Cañaveras, J.C.; Sanchez-Moral, S.; Saiz-Jimenez, C. Microbial Activity in Subterranean Ecosystems: Recent Advances. Appl. Sci. 2020, 10, 8130. https://doi.org/10.3390/app10228130
Martin-Pozas T, Gonzalez-Pimentel JL, Jurado V, Cuezva S, Dominguez-Moñino I, Fernandez-Cortes A, Cañaveras JC, Sanchez-Moral S, Saiz-Jimenez C. Microbial Activity in Subterranean Ecosystems: Recent Advances. Applied Sciences. 2020; 10(22):8130. https://doi.org/10.3390/app10228130
Chicago/Turabian StyleMartin-Pozas, Tamara, Jose Luis Gonzalez-Pimentel, Valme Jurado, Soledad Cuezva, Irene Dominguez-Moñino, Angel Fernandez-Cortes, Juan Carlos Cañaveras, Sergio Sanchez-Moral, and Cesareo Saiz-Jimenez. 2020. "Microbial Activity in Subterranean Ecosystems: Recent Advances" Applied Sciences 10, no. 22: 8130. https://doi.org/10.3390/app10228130
APA StyleMartin-Pozas, T., Gonzalez-Pimentel, J. L., Jurado, V., Cuezva, S., Dominguez-Moñino, I., Fernandez-Cortes, A., Cañaveras, J. C., Sanchez-Moral, S., & Saiz-Jimenez, C. (2020). Microbial Activity in Subterranean Ecosystems: Recent Advances. Applied Sciences, 10(22), 8130. https://doi.org/10.3390/app10228130








