Deep Subspace Clustering with Block Diagonal Constraint
Abstract
:1. Introduction
- A deep subspace clustering method based on an auto-encode is proposed, and block-diagonal constraints on representation matrix are used for better cluster performance.
- A separation strategy on the block diagonal constraint is proposed for more flexibility.
- Due to the existing of new block diagonal regularizer, an alternating optimization method to solve proposed model is developed.
- The proposed DSC-BDC is evaluated on four databases, and the results demonstrate the effectiveness of our model.
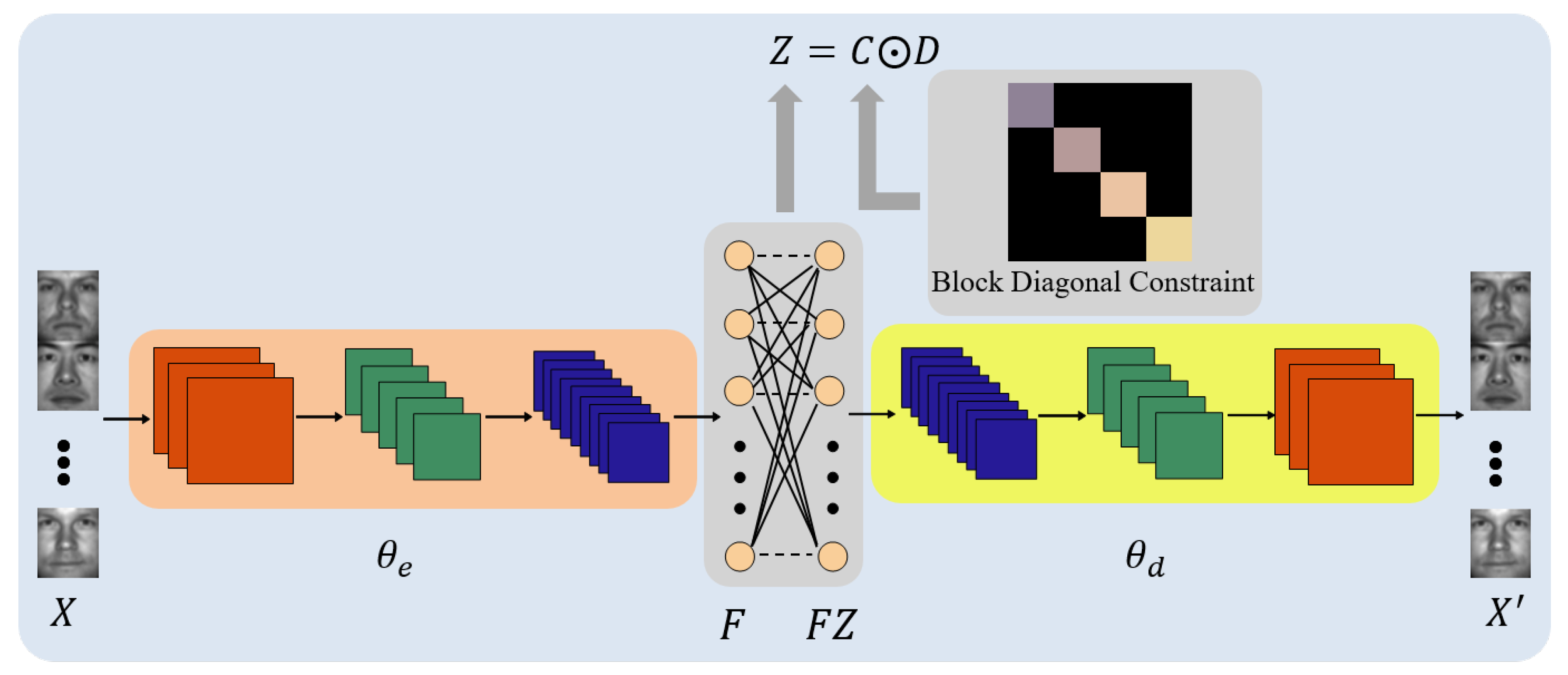
2. Deep Subspace Clustering Model with Block Diagonal Constraint
2.1. Model Formulation
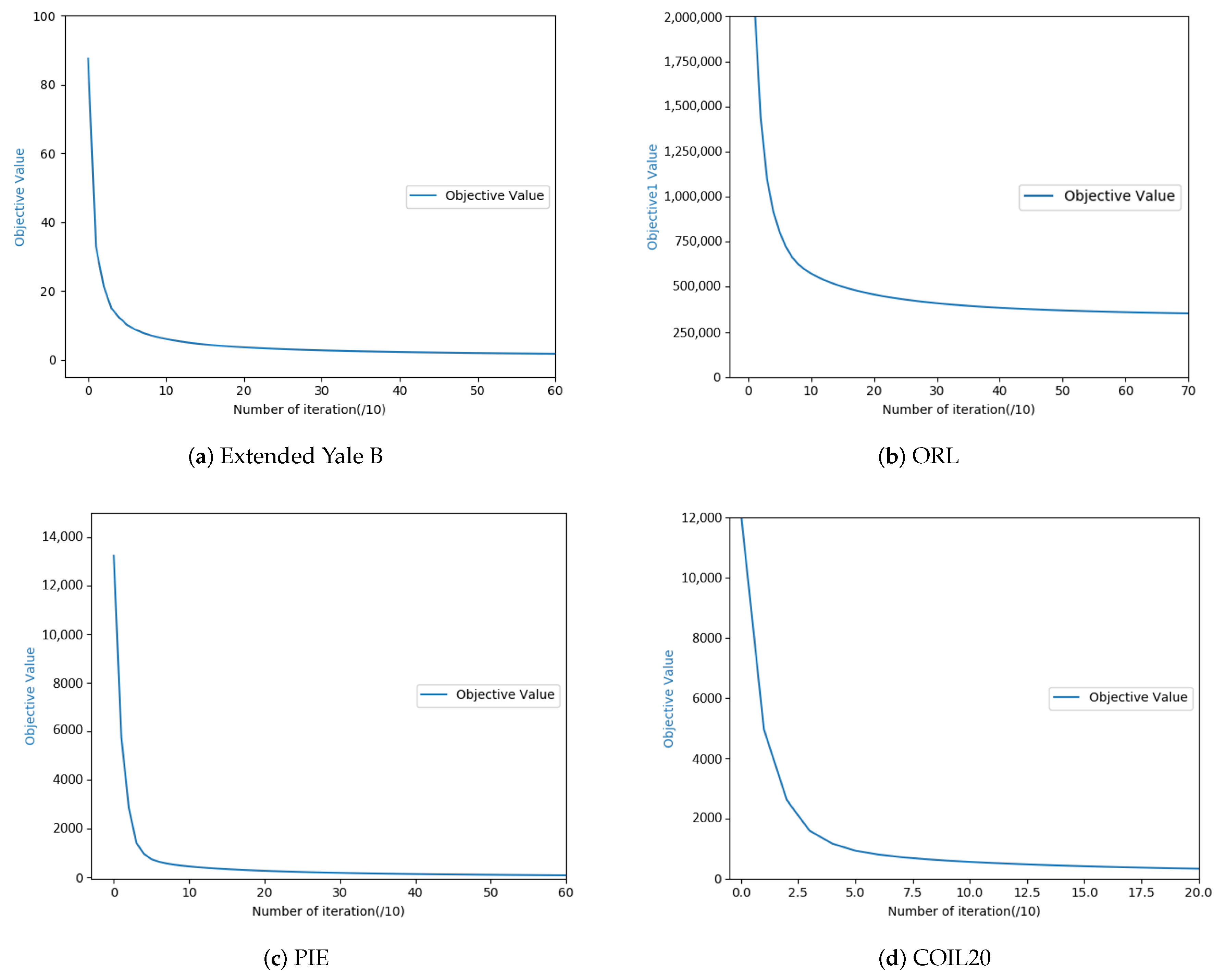
2.2. Model Optimization
| Algorithm 1 Optimization algorithm for DSC-BDC |
|
3. Experiments
3.1. Experimental Settings
3.1.1. Baseline Algorithms
3.1.2. Databases
- Extended Yale B database: Extended Yale B [50] consists of 2414 frontal face pictures of 38 subjects under 9 poses and 64 illumination conditions. For each subject, there are 64 images. Each cropped face image consists of pixels. In the experiment, we use the first 14 subjects with a total of 882 images for testing. All images are resized to pixels.
- ORL database: The ORL database [51] is composed of 400 photographs of size from 40 different individuals where each subject has 10 images taken under diverse variation of lighting conditions, poses and facial expressions. Following the literature, we downsample the images from their raw size to .
- PIE database: This face image database [52,53] consists of 40,000 photographs of 68 individuals, illustrating 13 pose conditions for each person, 43 with illumination levels and four expressions. It is worth noting that the pictures in the PIE database are all color pictures. In the experiment, we select 1428 samples of 68 subjects, and the sample size is 32 × 32. For convenience, the color images are converted to gray-scale images.
- COIL20 database: COIL20 [54] is a color object image database with 1440 images of 20 objects. Each object was placed on a turntable against a black background, and 72 images were taken at pose intervals of 5 degrees. For convenience, the color images are converted to gray-scale images; however, this is not necessary. The images are down-sampled to 32 × 32.
3.1.3. Training Strategy and Parameter Settings
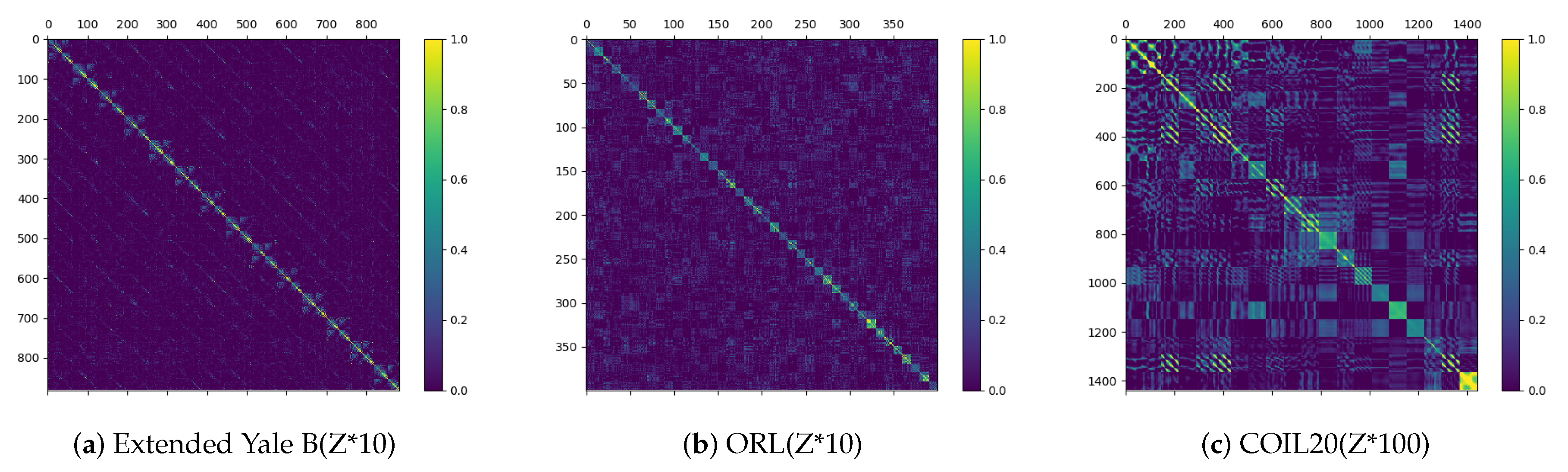
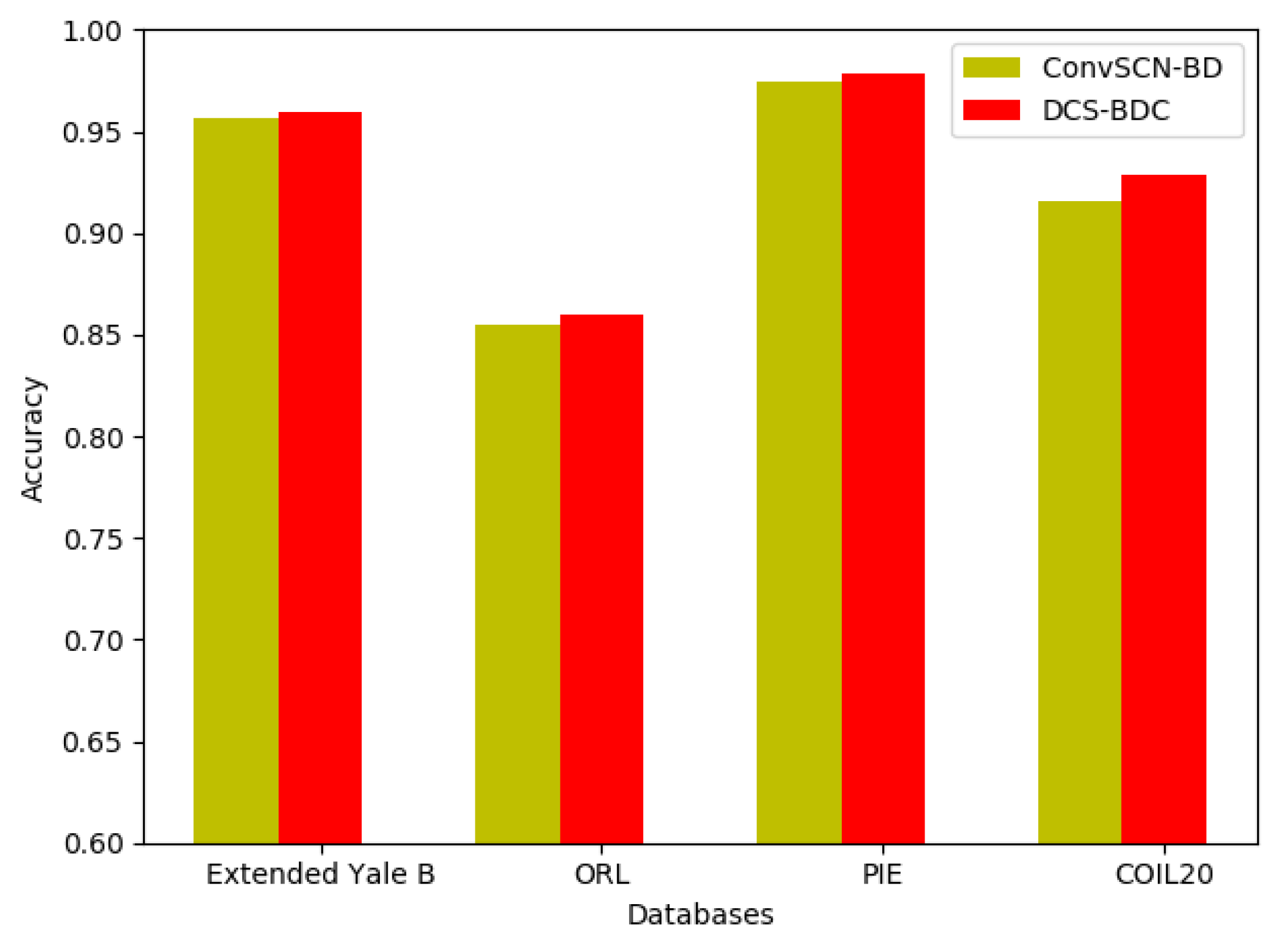
3.2. Experimental Results and Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Proof of Updating the Variable C
References
- Hong, W.; Wright, J.; Huang, K.; Ma, Y. Multiscale Hybrid Linear Models for Lossy Image Representation. IEEE Trans. Image Process. 2006, 15, 3655–3671. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Z.; Yan, S.; Sun, J.; Yu, Y.; Ma, Y. Robust Recovery of Subspace Structures by Low-Rank Representation. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 171–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eldar, Y.C.; Mishali, M. Robust Recovery of Signals From a Structured Union of Subspaces. IEEE Trans. Inf. Theory 2009, 55, 5302–5316. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Zhang, C.; Fu, H.; Liu, S.; Zhang, H. Diversity-induced Multi-view Subspace Clustering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 586–594. [Google Scholar]
- Zhang, C.; Fu, H.; Liu, S.; Liu, G.; Cao, X. Low-Rank Tensor Constrained Multiview Subspace Clustering. IEEE Int. Conf. Comput. Vis. 2015, 1582–1590. [Google Scholar]
- Pham, D.; Saha, B.; Phung, D.Q.; Venkatesh, S. Improved Subspace Clustering via Exploitation of Spatial Constraints. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 550–557. [Google Scholar]
- Rao, S.R.; Tron, R.; Vidal, R.; Ma, Y. Motion Segmentation in the Presence of Outlying, Incomplete, or Corrupted Trajectories. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 1832–1845. [Google Scholar] [CrossRef]
- Aldroubi, A.; Sekmen, A. Nearness to Local Subspace Algorithm for Subspace and Motion Segmentation. IEEE Signal Process. Lett. 2012, 19, 704–707. [Google Scholar] [CrossRef] [Green Version]
- Tari, L.; Baral, C.; Kim, S. Fuzzy C-means Clustering with Prior Biological Knowledge. J. Biomed. Inform. 2009, 42 1, 74–81. [Google Scholar] [CrossRef] [Green Version]
- Militello, C.; Vitabile, S.; Rundo, L.; Russo, G.; Midiri, M.; Gilardi, M. A Fully Automatic 2D Segmentation Method for Uterine Fibroid in MRgFUS Treatment Evaluation. Comput. Biol. Med. 2015, 62, 277–292. [Google Scholar] [CrossRef]
- Parsons, L.; Haque, E.; Liu, H. Subspace Clustering for High Dimensional Data: A Review. ACM Sigkdd Explor. Newsl. 2004, 6, 90–105. [Google Scholar] [CrossRef]
- Agrawal, R.; Gehrke, J.; Gunopulos, D.; Raghavan, P. Automatic Subspace Clustering of High Dimensional Data for Data Mining Applications. In Proceedings of the International Conference on Management of Data, Seattle, WA, USA, 2–4 June 1998; pp. 94–105. [Google Scholar]
- Elhamifar, E.; Vidal, R. Sparse Subspace Clustering: Algorithm, Theory, and Applications. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2765–2781. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Lerman, G. Spectral Curvature Clustering (SCC). Int. J. Comput. Vis. 2009, 81, 317–330. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Lin, Z.; Yu, Y. Robust Subspace Segmentation by Low-Rank Representation. In Proceedings of the International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010; pp. 663–670. [Google Scholar]
- Lu, C.; Min, H.; Zhao, Z.; Zhu, L.; Huang, D.; Yan, S. Robust and Efficient Subspace Segmentation via Least Squares Regression. In Proceedings of the European Conference on Computer Vision, Florence, Italy, 7–13 October 2012; pp. 347–360. [Google Scholar]
- Elhamifar, E.; Vidal, R. Sparse Subspace Clustering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 2790–2797. [Google Scholar]
- Vidal, R.; Favaro, P. Low Rank Subspace Clustering (LRSC). Pattern Recognit. Lett. 2014, 43, 47–61. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Lin, Z.; Xu, H.; Yan, S. Robust Subspace Segmentation with Block-diagonal Prior. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 3818–3825. [Google Scholar]
- Lu, C.; Feng, J.; Lin, Z.; Mei, T.; Yan, S. Subspace Clustering by Block Diagonal Representation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 487–501. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Yin, W.; Sun, Y.; Hu, Y. Multi-View Subspace Clustering With Block Diagonal Representation. IEEE Access 2019, 7, 84829–84838. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Leng, C. Provable Subspace Clustering: When LRR Meets SSC. In Proceedings of the Neural Information Processing Systems, Daegu, Korea, 3–7 November 2013; pp. 64–72. [Google Scholar]
- Ji, P.; Salzmann, M.; Li, H. Shape Interaction Matrix Revisited and Robustified: Efficient Subspace Clustering with Corrupted and Incomplete Data. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 4687–4695. [Google Scholar]
- Yang, Z.; Xu, Q.; Zhang, W.; Cao, X.; Huang, Q. Split Multiplicative Multi-View Subspace Clustering. IEEE Trans. Image Process. 2019, 28, 5147–5160. [Google Scholar] [CrossRef]
- Chen, G.; Atev, S.; Lerman, G. Kernel Spectral Curvature Clustering (KSCC). In Proceedings of the IEEE International Conference on Computer Vision, Kyoto, Japan, 27 September–4 October 2009; pp. 765–772. [Google Scholar]
- Patel, V.M.; Van Nguyen, H.; Vidal, R. Latent Space Sparse Subspace Clustering. In Proceedings of the IEEE International Conference on Computer Vision, Sydney, Australia, 1–8 December 2013; pp. 225–232. [Google Scholar]
- Patel, V.M.; Vidal, R. Kernel Sparse Subspace Clustering. In Proceedings of the IEEE International Conference on Image Processing, Paris, France, 27–30 October 2014; pp. 2849–2853. [Google Scholar]
- Yin, M.; Guo, Y.; Gao, J.; He, Z.; Xie, S. Kernel Sparse Subspace Clustering on Symmetric Positive Definite Manifolds. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 5157–5164. [Google Scholar]
- Peng, X.; Xiao, S.; Feng, J.; Yau, W.; Yi, Z. Deep Subspace Clustering with Sparsity Prior. In Proceedings of the International Joint Conference on Artificial Intelligence, New York, NY, USA, 9–15 July 2016; pp. 1925–1931. [Google Scholar]
- Ji, P.; Zhang, T.; Li, H.; Salzmann, M.; Reid, I. Deep Subspace Clustering Networks. In Proceedings of the Conference and Workshop on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 24–33. [Google Scholar]
- Peng, X.; Feng, J.; Xiao, S.; Yau, W.; Zhou, J.T.; Yang, S. Structured AutoEncoders for Subspace Clustering. IEEE Trans. Image Process. 2018, 27, 5076–5086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; You, C.; Qi, X.; Zhang, H.; Guo, J.; Lin, Z. Self-Supervised Convolutional Subspace Clustering Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 5473–5482. [Google Scholar]
- Zhang, T.; Ji, P.; Harandi, M.; Huang, W.; Li, H. Neural Collaborative Subspace Clustering. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 7384–7393. [Google Scholar]
- Zhou, P.; Hou, Y.; Feng, J. Deep Adversarial Subspace Clustering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1596–1604. [Google Scholar]
- Yang, S.; Zhu, W.; Zhu, Y. Residual Encoder-Decoder Network for Deep Subspace Clustering. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Abu Dhabi, UAE, 25–28 October 2020. [Google Scholar]
- Peng, X.; Zhu, H.; Feng, J.; Shen, C.; Zhang, H.; Zhou, J.T. Deep Clustering With Sample-Assignment Invariance Prior. IEEE Trans. Neural Netw. Learn. Syst. 2020, 31, 4857–4868. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Jayendran, A.; Prathosh, A.P. Effect of the Latent Structure on Clustering with GANs. IEEE Signal Process. Lett. 2020, 27, 900–904. [Google Scholar] [CrossRef]
- Mukherjee, S.; Asnani, H.; Lin, E.; Kannan, S. ClusterGAN: Latent Space Clustering in Generative Adversarial Networks. arXiv 2019, arXiv:1809.03627. [Google Scholar] [CrossRef]
- Zhao, S.; Song, J.; Ermon, S. InfoVAE: Information Maximizing Variational Autoencoders. arXiv 2017, arXiv:1706.02262. [Google Scholar]
- Han, C.; Murao, K.; Noguchi, T.; Kawata, Y.; Uchiyama, F.; Rundo, L.; Nakayama, H.; Satoh, S. Learning More with Less: Conditional PGGAN-based Data Augmentation for Brain Metastases Detection Using Highly-Rough Annotation on MR Images. In Proceedings of the 28th ACM International Conference on Information and Knowledge Management, Beijing, China, 3–7 November 2019. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. arXiv 2014, arXiv:1312.6114. [Google Scholar]
- Wolterink, J.; Leiner, T.; Viergever, M.; Išgum, I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans. Med. Imaging 2017, 36, 2536–2545. [Google Scholar] [CrossRef]
- Masci, J.; Meier, U.; Ciresan, D.C.; Schmidhuber, J. Stacked Convolutional Auto-Encoders for Hierarchical Feature Extraction. In International Conference on Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Chung, F.R.; Graham, F.C. Spectral Graph Theory; Number 92; American Mathematical Society: Providence, RI, USA, 1997. [Google Scholar]
- Kheirandishfard, M.; Zohrizadeh, F.; Kamangar, F. Multi-Level Representation Learning for Deep Subspace Clustering. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, CO, USA, 1–5 March 2020. [Google Scholar]
- Lin, Z.; Liu, R.; Su, Z. Linearized Alternating Direction Method with Adaptive Penalty for Low-Rank Representation. Adv. Neural Inf. Process. Syst. 2011, 612–620. [Google Scholar]
- Ji, P.; Salzmann, M.; Li, H. Efficient Dense Subspace Clustering. In Proceedings of the Workshop on Applications of Computer Vision, Steamboat Springs, CO, USA, 24–26 March 2014; pp. 461–468. [Google Scholar]
- Ji, P.; Reid, I.; Garg, R.; Li, H.; Salzmann, M. Low-Rank Kernel Subspace Clustering. arXiv 2017, arXiv:1707.04974. [Google Scholar]
- Zhang, J.; Li, C.; Du, T.; Zhang, H.; Guo, J. Convolutional Subspace Clustering Network with Block Diagonal Prior. IEEE Access 2020, 8, 5723–5732. [Google Scholar] [CrossRef]
- Georghiades, A.S.; Belhumeur, P.N.; Kriegman, D.J. From Few to Many: Illumination Cone Models for Face Recognition under Variable Lighting and Pose. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 643–660. [Google Scholar] [CrossRef] [Green Version]
- Samaria, F.S.; Harter, A. Parameterisation of a Stochastic Model for Human Face Identification. In Proceedings of the 1994 IEEE Workshop on Applications of Computer Vision, Sarasota, FL, USA, 5–7 December 1994; pp. 138–142. [Google Scholar]
- Fan, C.N.; Zhang, F. Homomorphic Filtering Based Illumination Normalization Method for Face Recognition. Pattern Recognit. Lett. 2011, 32, 1468–1479. [Google Scholar] [CrossRef]
- Sim, T.; Baker, S.; Bsat, M. The CMU Pose, Illumination, and Expression (PIE) Database. In Proceedings of the Fifth IEEE International Conference on Automatic Face Gesture Recognition, Washington, DC, USA, 20–21 May 2002; pp. 53–58. [Google Scholar]
- Nene, S.A.; Nayar, S.K.; Murase, H. Columbia Object Image Library (COIL-20); Department of Computer Science, Columbia University: New York, NY, USA, 1996. [Google Scholar]
| Database | Sample Number | Size | Class |
|---|---|---|---|
| Extended Yale B | 882 | 32 × 32 | 14 |
| ORL | 400 | 32 × 32 | 40 |
| PIE | 1428 | 32 × 32 | 68 |
| COIL20 | 1440 | 32 × 32 | 20 |
| Methods | ACC | NMI | ARI |
|---|---|---|---|
| LRR | 0.5696 ± 0.0104 | 0.5535 ± 0.0060 | 0.2164 ± 0.0149 |
| LSR | 0.5652 ± 0.0100 | 0.5548 ± 0.0052 | 0.2175 ± 0.0092 |
| SSC | 0.5412 ± 0.0103 | 0.5336 ± 0.0036 | 0.2456 ± 0.0162 |
| AE+SSC | 0.5778 ± 0.0400 | 0.5793 ± 0.0096 | 0.1751 ± 0.0373 |
| LR-kernel SC | 0.8016 ± 0.0101 | 0.8461 ± 0.0013 | 0.7352 ± 0.0108 |
| EDSC | 0.6138 ± 0.0196 | 0.5868 ± 0.0093 | 0.2722 ± 0.0220 |
| AE+EDSC | 0.6061 ± 0.0228 | 0.5910 ± 0.0163 | 0.2694 ± 0.0196 |
| BDR | 0.7253 ± 0.0111 | 0.7103 ± 0.0030 | 0.4927 ± 0.0046 |
| DSC-Net | 0.9548 ± 0.0003 | 0.9227 ± 0.0004 | 0.8987 ± 0.0008 |
| ConvSCN-BD | 0.9559 ± 0.0010 | 0.9260 ± 0.0008 | 0.9035 ± 0.0012 |
| Ours | 0.9592 ± 0.0008 | 0.9301 ± 0.0010 | 0.9060 ± 0.0018 |
| Methods | ACC | NMI | ARI |
|---|---|---|---|
| LRR | 0.6775 ± 0.0186 | 0.7881 ± 0.0160 | 0.4305 ± 0.0348 |
| LSR | 0.6800 ± 0.0260 | 0.7940 ± 0.0109 | 0.4288 ± 0.0473 |
| SSC | 0.7504 ± 0.0141 | 0.8578 ± 0.0071 | 0.5988 ± 0.0225 |
| AE+SSC | 0.7383 ± 0.0215 | 0.8428 ± 0.0088 | 0.5915 ± 0.0252 |
| LR-kernel SC | 0.7185 ± 0.0194 | 0.8540 ± 0.0068 | 0.6088 ± 0.0200 |
| EDSC | 0.6938 ± 0.0255 | 0.8089 ± 0.0126 | 0.5043 ± 0.0305 |
| AE+EDSC | 0.6974 ± 0.0171 | 0.8139 ± 0.0109 | 0.5026 ± 0.0365 |
| BDR | 0.7204 ± 0.0192 | 0.8350 ± 0.0117 | 0.4662 ± 0.0471 |
| DSC-Net | 0.8463 ± 0.0080 | 0.9172 ± 0.0021 | 0.7878 ± 0.0062 |
| ConvSCN-BD | 0.8360 ± 0.0090 | 0.9149 ± 0.0059 | 0.7791 ± 0.0131 |
| Ours | 0.8505 ± 0.0079 | 0.9182 ± 0.0027 | 0.7932 ± 0.0037 |
| Methods | ACC | NMI | ARI |
|---|---|---|---|
| LRR | 0.7392 ± 0.0206 | 0.8808 ± 0.0079 | 0.6041 ± 0.0239 |
| LSR | 0.7484 ± 0.0212 | 0.8812 ± 0.0100 | 0.5799 ± 0.0404 |
| SSC | 0.7652 ± 0.0184 | 0.9060 ± 0.0070 | 0.6510 ± 0.0285 |
| AE+SSC | 0.7820 ± 0.0234 | 0.9191 ± 0.0066 | 0.6850 ± 0.0204 |
| LR-kernel SC | 0.7520 ± 0.0174 | 0.9341 ± 0.0043 | 0.7238 ± 0.0199 |
| EDSC | 0.8148 ± 0.0143 | 0.9073 ± 0.0060 | 0.6231 ± 0.0641 |
| AE+EDSC | 0.8251 ± 0.0184 | 0.9164 ± 0.0062 | 0.6433 ± 0.0545 |
| BDR | 0.8094 ± 0.0200 | 0.9130 ± 0.0086 | 0.5980 ± 0.0712 |
| DSC-Net | 0.9686 ± 0.0072 | 0.9911 ± 0.0052 | 0.9620 ± 0.0120 |
| ConvSCN-BD | 0.9732 ± 0.0203 | 0.9890 ± 0.0041 | 0.9723 ± 0.0211 |
| Ours | 0.9760 ± 0.0035 | 0.9960 ± 0.0046 | 0.9807 ± 0.0031 |
| Methods | ACC | NMI | ARI |
|---|---|---|---|
| LRR | 0.6887 ± 0.0190 | 0.7714 ± 0.0097 | 0.5873 ± 0.0150 |
| LSR | 0.6871 ± 0.0142 | 0.7683 ± 0.0097 | 0.5927 ± 0.0204 |
| SSC | 0.7898 ± 0.0292 | 0.8917 ± 0.0117 | 0.0.6546 ± 0.0820 |
| AE+SSC | 0.8863 ± 0.0113 | 0.9205 ± 0.0074 | 0.7979 ± 0.0278 |
| LR-kernel SC | 0.6268 ± 0.0083 | 0.8056 ± 0.0035 | 0.5568 ± 0.0154 |
| EDSC | 0.8514 ± 0.0000 | - | - |
| AE+EDSC | 0.8521 ± 0.0000 | - | - |
| BDR | 0.7549 ± 0.0000 | 0.8862 ± 0.0000 | 0.6249 ± 0.0000 |
| DSC-Net | 0.9004 ± 0.0288 | 0.9562 ± 0.0095 | 0.8765 ± 0.0384 |
| ConvSCN-BD | 0.9160 ± 0.0037 | 0.9613 ± 0.0002 | 0.8989 ± 0.0032 |
| Ours | 0.9174 ± 0.0010 | 0.9632 ± 0.0021 | 0.8944 ± 0.0037 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Sun, Y.; Hu, Y. Deep Subspace Clustering with Block Diagonal Constraint. Appl. Sci. 2020, 10, 8942. https://doi.org/10.3390/app10248942
Liu J, Sun Y, Hu Y. Deep Subspace Clustering with Block Diagonal Constraint. Applied Sciences. 2020; 10(24):8942. https://doi.org/10.3390/app10248942
Chicago/Turabian StyleLiu, Jing, Yanfeng Sun, and Yongli Hu. 2020. "Deep Subspace Clustering with Block Diagonal Constraint" Applied Sciences 10, no. 24: 8942. https://doi.org/10.3390/app10248942





