Interferometric Measurement of TGF-β Induced Epithelial-Mesenchymal Transition of Tumor Cells
Abstract
:1. Introduction
2. Materials and Methods
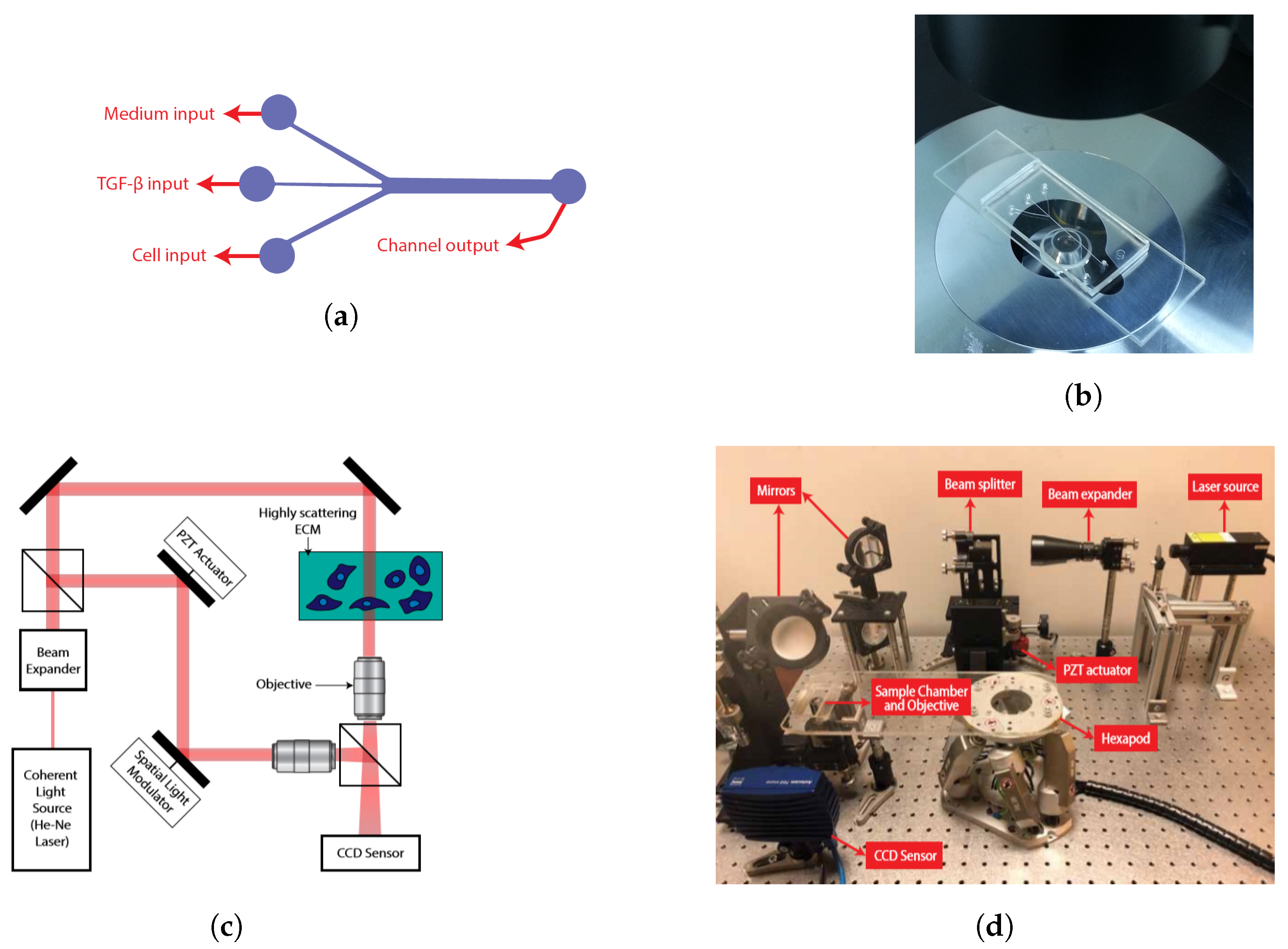
2.1. Cell Culturing and Manufacturing of Fluidic Chambers
2.2. Quantitative Phase Imaging Setup and Phase Reconstruction
2.3. Noise Reduction Methodology
3. Results and Discussion
4. Conclusions and Future Studies
Author Contributions
Funding
Conflicts of Interest
References
- Zernike, F. Phase contrast, a new method for the microscopic observation of transparent objects. Physica 1942, 9, 686–698. [Google Scholar] [CrossRef]
- Marquet, P.; Rappaz, B.; Magistretti, P.J.; Cuche, E.; Emery, Y.; Colomb, T.; Depeursinge, C. Digital holographic microscopy: A noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Opt. Lett. 2005, 30, 468. [Google Scholar] [CrossRef] [Green Version]
- Langehanenberg, P.; Kemper, B.; Dirksen, D.; Von Bally, G. Autofocusing in digital holographic phase contrast microscopy on pure phase objects for live cell imaging. Appl. Opt. 2008, 47, D176–D182. [Google Scholar] [CrossRef]
- Wang, Y.M.; Judkewitz, B.; Dimarzio, C.A.; Yang, C. Deep-tissue focal fluorescence imaging with digitally time-reversed ultrasound-encoded light. Nat. Commun. 2012, 3, 928. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Lai, P.; Ma, C.; Xu, X.; Grabar, A.A.; Wang, L.V. Optical focusing deep inside dynamic scattering media with near-infrared time-reversed ultrasonically encoded (TRUE) light. Nat. Commun. 2015, 6, 5904. [Google Scholar] [CrossRef] [PubMed]
- Asghar, W.; El Assal, R.; Shafiee, H.; Pitteri, S.; Paulmurugan, R.; Demirci, U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today 2015, 18, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Carl, D.; Kemper, B.; Wernicke, G.; Von Bally, G. Parameter-optimized digital holographic microscope for high-resolution living-cell analysis. Appl. Opt. 2004, 43, 6536–6544. [Google Scholar] [CrossRef] [PubMed]
- Bukowska, D.M.; Derzsi, L.; Tamborski, S.; Szkulmowski, M.; Garstecki, P.; Wojtkowski, M. Assessment of the flow velocity of blood cells in a microfluidic device using joint spectral and time domain optical coherence tomography. Opt. Express 2013, 21, 24025. [Google Scholar] [CrossRef] [PubMed]
- Zakrisson, J.; Schedin, S.; Andersson, M. Cell shape identification using digital holographic microscopy. Appl. Opt. 2015, 54, 7442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, V.K.; Nguyen, T.C.; Chung, B.M.; Nehmetallah, G.; Raub, C.B. Quantitative assessment of cancer cell morphology and motility using telecentric digital holographic microscopy and machine learning. Cytom. A 2018, 93, 334–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihailescu, M.; Popescu, R.C.; Matei, A.; Acasandrei, A.; Paun, I.A.; Dinescu, M. Investigation of osteoblast cells behavior in polymeric 3D micropatterned scaffolds using digital holographic microscopy. Appl. Opt. 2014, 53, 4850. [Google Scholar] [CrossRef] [PubMed]
- Dubey, V.; Singh, G.; Singh, V.; Ahmad, A.; Mehta, D.S. Multispectral quantitative phase imaging of human red blood cells using inexpensive narrowband multicolor LEDs. Appl. Opt. 2016, 55, 2521. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Di, J.; Wu, W.; Shang, P.; Zhao, J. Quantitative investigation on morphology and intracellular transport dynamics of migrating cells. Appl. Opt. 2019, 58, G162. [Google Scholar] [CrossRef] [PubMed]
- Hoang, V.T.; Stȩpniewski, G.; Czarnecka, K.H.; Kasztelanic, R.; Long, V.C.; Xuan, K.D.; Shao, L.; Śmietana, M.; Buczyński, R. Optical Properties of Buffers and Cell Culture Media for Optofluidic and Sensing Applications. Appl. Sci. 2019, 9, 1145. [Google Scholar] [CrossRef] [Green Version]
- Lam, V.K.; Nguyen, T.; Bui, V.; Chung, B.M.; Chang, L.C.; Nehmetallah, G.; Raub, C.B. Quantitative scoring of epithelial and mesenchymal qualities of cancer cells using machine learning and quantitative phase imaging. J. Biomed. Opt. 2020, 25, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, I.; Zhang, T. Phase-shifting digital holography. Opt. Lett. 1997, 22, 1268. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-β-induced epithelial to mesenchymal transition. Nat. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Baker, D.; ten Dijke, P. TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendt, M.K.; Allington, T.M.; Schiemann, W.P. Mechanisms of the epithelial-mesenchymal transition by TGF-β. Future Oncol. 2009, 5, 1145–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grau, V.; Mewes, A.U.; Alcañiz, M.; Kikinis, R.; Warfield, S.K. Improved watershed transform for medical image segmentation using prior information. IEEE Trans. Med. Imaging 2004, 23, 447–458. [Google Scholar] [CrossRef]
- Goodman, J.W. Introduction to Fourier Optics; Roberts and Company Publishers: Greenwood Village, CO, USA, 2005; p. 491. [Google Scholar]


| Slide without Medium | Slide with Medium | Beads without Medium | Beads with Medium | |
|---|---|---|---|---|
| Median filter | 35.67 | 34.78 | 35.79 | 34.66 |
| Wiener filter | 37.78 | 37.23 | 37.90 | 37.12 |
| Haar | 37.12 | 35.64 | 37.34 | 35.76 |
| Daubechies | 37.89 | 37.89 | 37.92 | 37.02 |
| Symlets | 39.12 | 38.56 | 39.19 | 38.43 |
| Biorthogonal | 37.96 | 36.54 | 37.88 | 36.34 |
| Noiselets | 38.87 | 37.91 | 38.89 | 37.72 |
| Fresnelet | 39.45 | 38.03 | 39.25 | 38.23 |
| Beamlet | 38.79 | 37.99 | 38.55 | 37.79 |
| Coiflet | 38.85 | 37.15 | 39.00 | 37.27 |
| Discrete Meyer | 37.75 | 37.04 | 38.14 | 37.14 |
| HCT-116 | ONCO-DG-1 | HUVEC | MDA-MB-231 | |
|---|---|---|---|---|
| Median filter | 34.67 | 34.62 | 34.86 | 33.80 |
| Wiener filter | 36.78 | 36.66 | 36.70 | 35.69 |
| Haar | 36.12 | 35.98 | 35.99 | 34.87 |
| Daubechies | 36.89 | 36.75 | 36.86 | 35.81 |
| Symlets | 38.12 | 37.92 | 38.23 | 37.06 |
| Biorthogonal | 36.96 | 36.78 | 37.06 | 36.03 |
| Noiselets | 37.87 | 37.76 | 38.05 | 36.92 |
| Fresnelet | 38.45 | 38.30 | 38.28 | 37.10 |
| Beamlet | 37.79 | 37.67 | 37.81 | 36.63 |
| Coiflet | 37.85 | 37.72 | 37.80 | 36.75 |
| Discrete Meyer | 36.75 | 36.57 | 36.68 | 35.63 |
| BM3D filter | 39.02 | 38.97 | 38.96 | 37.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varol, R.; Esmer, G.B.; Uvet, H. Interferometric Measurement of TGF-β Induced Epithelial-Mesenchymal Transition of Tumor Cells. Appl. Sci. 2020, 10, 9107. https://doi.org/10.3390/app10249107
Varol R, Esmer GB, Uvet H. Interferometric Measurement of TGF-β Induced Epithelial-Mesenchymal Transition of Tumor Cells. Applied Sciences. 2020; 10(24):9107. https://doi.org/10.3390/app10249107
Chicago/Turabian StyleVarol, Rahmetullah, Gokhan Bora Esmer, and Huseyin Uvet. 2020. "Interferometric Measurement of TGF-β Induced Epithelial-Mesenchymal Transition of Tumor Cells" Applied Sciences 10, no. 24: 9107. https://doi.org/10.3390/app10249107
APA StyleVarol, R., Esmer, G. B., & Uvet, H. (2020). Interferometric Measurement of TGF-β Induced Epithelial-Mesenchymal Transition of Tumor Cells. Applied Sciences, 10(24), 9107. https://doi.org/10.3390/app10249107





