Big Data Analytics and Processing Platform in Czech Republic Healthcare
Abstract
:1. Introduction
1.1. Big Data Technology Perspective
1.2. Challenges and Opportunities
- improve the quality of personalised care and medical services;
- reduce cost of treatment;
- use predictive analytics for e.g., patients’ daily (loss of) income and disease progression;
- use real-time visualisation and analytics for immediate care and cases of readmission;
- to integrate small-data analytics and knowledge discovery that may also be integrated with big data [28];
- use healthcare data for identification of trends, strategic planning, governance, improved decision-making, and cost reduction [24].
- Is it possible to design and build a BDA platform for the Czech Republic healthcare service, in line with EU legislation, TPC-H [1] benchmarks, and other statutory requirements?
- If so, what BDA platform would provide optimal cost and performance features, while allowing installation of open-source software with various machine learning algorithms, development environments, and commercial visualisation and analytical tools?
- To what extend would such a BDA-based eSystem be future-proof for maintaining reliability, robustness, cost-effectiveness, and performance?
1.3. Industry Benchmarks
1.4. Big Data Analytics
1.5. SQL vs. NoSQL Approaches
2. Materials and Methods
- Phase I-compliant big data eSystem requirements and design decisions influencing architecture design;
- Decomposition of acceptance testing requirements;
- Hardware infrastructure optimisation to a set of the requirements (N = 119) for weighted scoring, including TPC-H decision support and minimal system performance; and
- Performance-driven eSystem optimisation from available test datasets.
2.1. IHIS Requirements
- Scalability: the eSystem must allow performance enhancement via additional and accessible computing technology, including commodity hardware products.
- Modularity, Openness, and Interoperability: the system components must be integrated via specified interfaces according to exact requirement specifications. It is also essential that a wide variety of vendors can readily utilise system components.
- Exchangeability: the eSystem solution must support installation of open-source operating systems and contain tools for non-profit and educational purposes. The eSystem must comply with standard Data Warehouse (DWH) systems. Some components must be interchangeable with Massive High Data Analytics (MHDA) system components.
- Extensibility: all tools and components of the eSystem must provide space for future upgrades, including functionality and capability advancements.
- Quality Assurance: a tool for validating data and metadata integrity is required to ensure that processed data remains accurate throughout the analysis procedure.
- Security: the eSystem must be operable on local servers, without reliance on cloud or outsourced backup systems. It is essential that the eSystem provides security for all data against external or internal threats. Therefore, authorisation, storage access and communication are of utmost concern. User access rights had to be set to the database, table, or column level to restrict data access to a limited number of advanced users. The eSystem must log all executions and read operations for future audits. The eSystem must support tools for version control and development, while meeting the requirements for metadata and data versioning, backup, and archiving.
- Simplicity: the eSystem must allow for parallel team collaboration on all processes, data flow, and database schemas. All tasks must be fully editable, allowing commit and revert changes in data and metadata. It is essential that the eSystem be simple and easy to use, as well as stable and resilient to subsystem outages.
- Performance: the eSystem must be designed for the specified minimum number of concurrent users. Batch processing of data sources and sophisticated data mining analyses are considered essential. Complete data integration processing of quarterly data increments must not exceed one hour.
- All tools, licenses, and environmental features used in the Proof of Concept (PoC) tests must match the eSystem offer submitted and documented in the public contract. To meet contractual obligations, the proposed solution cannot have: insincerely increased system performance, altered available license terms or otherwise improved or modified results vis-à-vis the delivery of the final solution. The environment configuration must satisfy the general requirements of the proposed eSystem (usage types, input data size, processing speed requirements).
- The proposed eSystem cannot be explicitly (manually) optimised for specific queries and individual task steps within a test. The test queries are not to be based on general metadata (cache, partitioning, supplemental indexes, derived tables, and views), except in exceptional cases where optimising the loading of large amounts of data is needed. The techniques based on general metadata may be used in future for enhancing performance but are not required as a precondition for system availability. To load large amounts of data, the environment configuration can be manually adjusted to a non-standard configuration for further test steps (Figure 1).
- The configuration must not be manually changed during the test to optimise individual tasks—the eSystem is required to be universal for tasks that may overlap in time.
2.2. TPC-H Performance Requirements and Tests
3. Results
3.1. Big Data Analytcs (BDA) Platform and Vertica-Based Architecture
- Provision for an SQL layer as well as support connection to Hadoop and fast data access to ORC, Parquet, Avro, and JSON as column-oriented data;
- High data compression ratio, including high-degree concurrency and massively parallel processing (MPP) system for processing tasks;
- Analytical database support for Kafka, Spark;
- Pricing model of enterprise solution optimised for IHIS requirements;
- Potential for huge demands of future analytical workloads;
- Cloud integration for future development; and
- Compression capabilities that can handle and deliver high-speed results for petabyte scale datasets.
3.2. Overview of Key Components
3.2.1. Data Integration (DI) Layer
3.2.2. Data Storage (DS)
3.2.3. Data Quality Management (DQM)
3.2.4. Metadata Management (MDM)
3.2.5. Ad-hoc Analysis Preparation (AAP)
3.2.6. Data Visualisation (DV)
3.3. TPC-H Tests Configuration
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
- Data warehouse contains ‘Hospital stay’ table with 10 linked tables (Figure A1) as: ‘Patient dimension’, ‘Patient’s diagnosis’, ‘Recommended hospitalization’, ‘Termination of treatment’, ‘Age dimension’, ‘Workplace’, ‘Performance group’, ‘Group exercising bridge’, ‘EHR dimension’, and ‘Date dimension’.
- Multiple compressed warehouse image archives are also included in a CSV semi-column delimited format.
- File sizes for import into the NoSQL database ranged from 100 GB up to 10 TB.
- Three standard quarterly datasets, which have been processed on quarter time base per each insurance company, as well as one corrective (simulates a situation where one insurance company supplied bad data).
- Standard input data size is less than 1 TB per packet of data.
- Each data package is in a ZIP format with metadata included in the package name as insurance company and package serial number.
- The Anatomical Therapeutic Chemical Classification System (ATC).
- Code lists and performance group, which are separated in the files to assign attributes of dynamically loaded dimensions.


References
- Transaction Processing Performance Council. TPC BenchmarkTM Standard Specification Revision 2.18.0.; Transaction Processing Performance Council (TPC): San Francisco, CA, USA, 2018. [Google Scholar]
- BigMedilytics. Big Data Project. Available online: https://www.bigmedilytics.eu/big-data-project/ (accessed on 29 December 2019).
- European Commission. Big Data: Digital Single Market Policy. Available online: https://ec.europa.eu/digital-single-market/en/policies/big-data (accessed on 28 December 2019).
- Srivastava, S. Top 10 Countries & Regions Leading the Big Data Adoption in 2019. Available online: https://www.analyticsinsight.net/top-10-countries-regions-leading-the-big-data-adoption-in-2019/ (accessed on 31 January 2020).
- The Big Data Value Association (BDVA). Data Protection Policy. Available online: http://www.bdva.eu/?q=data-protection-policy (accessed on 28 December 2019).
- Carnicero, R.; Rojas, D.; Elicegui, I.; Carnicero, J. Proposal of a learning health system to transform the national health system of Spain. Processes 2019, 7, 613. [Google Scholar] [CrossRef] [Green Version]
- Bhuiyan, M.A.R.; Ullah, M.R.; Das, A.K. iHealthcare: Predictive model analysis concerning big data applications for interactive healthcare systems. Appl. Sci. 2019, 9, 3365. [Google Scholar] [CrossRef] [Green Version]
- Saiful, I.M.; Mahmudul, H.M.; Xiaoyi, W.; Germack, D.H. A systematic review on healthcare analytics: Application and theoretical perspective of data mining. Healthcare 2018, 6, 54. [Google Scholar]
- Lyko, K.; Nitzschke, M.; Ngomo, A.-C.N. Big data acquisition. In New Horizons for a Data-Driven Economy; Cavanillas, J.M., Curry, E., Wahlster, W., Eds.; Springer: Cham, Switzerland, 2016; pp. 39–61. [Google Scholar] [CrossRef] [Green Version]
- Sarnovsky, M.; Bednar, P.; Smatana, M. Big data processing and analytics platform architecture for process industry factories. Big Data Cogn. Comput. 2018, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Husamaldin, L.; Saeed, N. Big data analytics correlation taxonomy. Information 2019, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Ajah, I.A.; Nweke, H.F. Big data and business analytics: Trends, platforms, success factors and applications. Big Data Cogn. Comput. 2019, 3, 32. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Liu, W.; Tsai, S.-B.; Gao, J.; Bin, N.; Chen, Q. An empirical study on visualizing the intellectual structure and hotspots of big data research from a sustainable perspective. Sustainability 2018, 10, 667. [Google Scholar] [CrossRef] [Green Version]
- Ergüzen, A.; Ünver, M. Developing a file system structure to solve healthy big data storage and archiving problems using a distributed file system. Appl. Sci. 2018, 8, 913. [Google Scholar] [CrossRef] [Green Version]
- Lima, D.M.; Rodrigues-Jr, J.F.; Traina, J.A.M.; Pires, F.A.; Gutierrez, M.A. Transforming two decades of ePR data to OMOP CDM for clinical research. Stud. Health Technol. Inform. 2019, 264, 233–237. [Google Scholar]
- ur Rehman, M.H.; Jayaraman, P.P.; Malik, S.U.R.; Khan, A.U.R.; Gaber, M.M. RedEdge: A novel architecture for big data processing in mobile edge computing environments. Sens. Actuator Netw. 2017, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Sumbaly, R.; Kreps, J.; Shah, S. The big data ecosystem at LinkedIn. In Proceedings of the 2013 ACM SIGMOD International Conference on Management of Data, New York, NY, USA, 22–27 June 2013; pp. 1125–1134. [Google Scholar]
- Cohen, P.; Hahn, R.; Hall, J.; Levitt, S.; Metcalfe, R. Using Big Data to Estimate Consumer Surplus: The Case of Uber; 22627; National Bureau of Economic Research: Cambridge, MA, USA, 2016. [Google Scholar]
- Amatriain, X. Big & personal: Data and models behind Netflix recommendations. In Proceedings of the 2nd International Workshop on Big Data, Streams and Heterogeneous Source Mining: Algorithms, Systems, Programming Models and Applications, Chicago, IL, USA, 11 August 2013; pp. 1–6. [Google Scholar]
- Davoudi, S.; Dooling, J.A.; Glondys, B.; Jones, T.D.; Kadlec, L.; Overgaard, S.M.; Ruben, K.; Wendicke, A. Data quality management model (2015 update)—Retired. J. AHIMA 2015, 86, 62–65. [Google Scholar]
- Pavlo, A.; Paulson, E.; Rasin, A.; Abadi, D.J.; DeWitt, D.J.; Madden, S.; Stonebraker, M. A comparison of approaches to large-scale data analysis. In Proceedings of the 2009 ACM SIGMOD International Conference on Management of Data, Providence, RI, USA, 29 June–2 July 2009; pp. 165–178. [Google Scholar]
- Beam, A.L.; Kohane, I.S. Big data and machine learning in health care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef]
- Juddoo, S.; George, C.; Duquenoy, P.; Windridge, D. Data governance in the health industry: Investigating data quality dimensions within a big data context. Appl. Syst. Innov. 2018, 1, 43. [Google Scholar] [CrossRef] [Green Version]
- Kruse, C.S.; Goswamy, R.; Raval, Y.; Marawi, S. Challenges and opportunities of big data in health care: A systematic review. Med. Inform. 2016, 4, e38. [Google Scholar] [CrossRef]
- Bouzillé, G.; Morival, C.; Westerlynck, R.; Lemordant, P.; Chazard, E.; Lecorre, P.; Busnel, Y.; Cuggia, M. An automated detection system of drug-drug interactions from electronic patient records using big data analytics. In MEDINFO 2019: Health and Wellbeing e-Networks for All; Ohno-Machado, L., Séroussi, B., Eds.; IOS Press Ebooks: Amsteerdam, Netherland, 2019; Volume 264, pp. 45–49. [Google Scholar]
- Narayanan, A.; Greco, M. Patient experience of australian general practices. Big Data 2016, 4, 31–46. [Google Scholar] [CrossRef]
- Narayanan, A.; Greco, M.; Powell, H.; Coleman, L. The reliability of big “Patient Satisfaction” data. Big Data 2013, 1, 141–151. [Google Scholar] [CrossRef]
- Ohsawa, Y. Modeling the process of chance discovery. In Chance Discovery; Ohsawa, Y., McBurney, P., Eds.; Springer: Heidelberg, Germany, 2003; pp. 2–15. [Google Scholar] [CrossRef]
- Bačić, B. Predicting golf ball trajectories from swing plane: An artificial neural networks approach. Expert Syst. Appl. 2016, 65, 423–438. [Google Scholar] [CrossRef]
- Bačić, B. Towards the next generation of exergames: Flexible and personalised assessment-based identification of tennis swings. In Proceedings of the IEEE World Congress on Computational Intelligence (IEEE WCCI) & International Joint Conference on Neural Networks (IJCNN), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–8. [Google Scholar]
- Chan, K.Y.; Bačić, B. Pseudo-3D binary silhouette for augmented golf coaching. In Proceedings of the XXXVI International Symposium on Biomechanics in Sports—ISBS 2018, Auckland, New Zealand, 10–14 September 2018; pp. 790–793. [Google Scholar]
- Bačić, B.; Meng, Q.; Chan, K.Y. Privacy preservation for eSports: A case study towards augmented video golf coaching system. In Proceedings of the 10th International Conference on Developments in e-Systems Engineering (DeSE), Paris, France, 14–16 June 2017; pp. 169–174. [Google Scholar]
- Bačić, B.; Iwamoto, S.; Parry, D. Open source software and interdisciplinary data management: Post-surgery rehabilitation case study. In Proceedings of the Health Informatics New Zealand (HINZ), Auckland, New Zealand, 10–12 November 2014; p. 48. [Google Scholar]
- Bačić, B.; Hume, P.A. Computational intelligence for qualitative coaching diagnostics: Automated assessment of tennis swings to improve performance and safety. Big Data 2018, 6, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Piot, P. What Ebola Teaches us about Controlling Coronavirus. Available online: https://time.com/5778998/ebola-coronavirus-lessons/ (accessed on 15 February 2020).
- Yong, E. The New Coronavirus is a Truly Modern Epidemic: New Diseases Are Mirrors that Reflect How a Society Works—And Where It Fails. Available online: https://www.theatlantic.com/science/archive/2020/02/coronavirus-very-2020-epidemic/605941/ (accessed on 16 February 2020).
- Klein, J.; Gorton, I.; Ernst, N.; Donohoe, P.; Pham, K.; Matser, C. Performance evaluation of NoSQL databases: A case study. In Proceedings of the 1st Workshop on Performance Analysis of Big Data Systems, Austin, TX, USA, 1 February 2015; pp. 5–10. [Google Scholar]
- Moniruzzaman, A.; Hossain, S.A. NoSQL database: New era of databases for big data analytics-classification, characteristics and comparison. Int. J. Database Theor. Appl. 2013, 6. [Google Scholar]
- Senthilkumar, S.A.; Bharatendara, K.R.; Amruta, A.M.; Angappa, G.; Chandrakumarmangalam, S. Big data in healthcare management: A review of literature. Am. J. Theor. Appl. Bus. 2018, 4, 57–69. [Google Scholar]
- Manyika, J.; Chui, M.; Brown, B.; Bughin, J.; Dobbs, R.; Roxburgh, C.; Byers, A.H. Big Data: The Next Frontier for Innovation, Competition, and Productivity; McKinsey Global Institute: New York, NY, USA, 2011; p. 156. [Google Scholar]
- Shafqat, S.; Kishwer, S.; Rasool, R.U.; Qadir, J.; Amjad, T.; Ahmad, H.F. Big data analytics enhanced healthcare systems: A review. J. Supercomput. 2018, 1–46. [Google Scholar] [CrossRef]
- Lo, E.; Cheng, N.; Lin, W.W.; Hon, W.-K.; Choi, B. MyBenchmark: Generating databases for query workloads. VLDB J. Int. J. Large Data Bases 2014, 23, 895–913. [Google Scholar] [CrossRef]
- Olson, D.R.; Konty, K.J.; Paladini, M.; Viboud, C.; Simonsen, L. Reassessing Google flu trends data for detection of seasonal and pandemic influenza: A comparative epidemiological study at three geographic scales. PLoS Comput. Biol. 2013, 9, e1003256. [Google Scholar] [CrossRef]
- Raghupathi, W.; Raghupathi, V. Big data analytics in healthcare: Promise and potential. Health Inf. Sci. Syst. 2014, 2, 3. [Google Scholar] [CrossRef]
- Ginsberg, J.; Mohebbi, M.H.; Patel, R.S.; Brammer, L.; Smolinski, M.S.; Brilliant, L. Detecting influenza epidemics using search engine query data. Nature 2009, 457, 1012. [Google Scholar] [CrossRef]
- Jacobson, S.H. Responding to the Coronavirus Threat Using Lessons Learned from Ebola: Assessing Risks and Transmission of Coronavirus. Available online: https://www.washingtontimes.com/news/2020/jan/30/responding-to-the-coronavirus-threat-using-lessons/ (accessed on 15 February 2020).
- Honigsbaum, M. How Our Modern World Creates Outbreaks Like Coronavirus. Available online: https://time.com/5779578/modern-world-epidemics-coronavirus/ (accessed on 16 February 2020).
- Kankanhalli, A.; Hahn, J.; Tan, S.; Gao, G. Big data and analytics in healthcare: Introduction to the special section. Inf. Syst. Front. 2016, 18, 233–235. [Google Scholar] [CrossRef] [Green Version]
- Codd, E.F. A relational model of data for large shared data banks. Commun. ACM 1970, 13, 377–387. [Google Scholar] [CrossRef]
- Jatana, N.; Puri, S.; Ahuja, M.; Kathuria, I.; Gosain, D. A survey and comparison of relational and non-relational database. IJERT 2012, 1, 1–5. [Google Scholar]
- Mehmet, Z.E.; Lane, M. Evaluation of NoSQL databases for EHR systems. In Proceedings of the 25th Australasian Conference on Information Systems, Auckland, New Zealand, 8–10 December 2014. [Google Scholar]
- Stonebraker, M.; Abadi, D.J.; Batkin, A.; Chen, X.; Cherniack, M.; Ferreira, M.; Lau, E.; Lin, A.; Madden, S.; O’Neil, E. C-store: A column-oriented DBMS. In Proceedings of the 31st International Conference on Very Large Data Bases, Trondheim, Norway, 30 Novemeber–2 Septermber 2005; pp. 553–564. [Google Scholar]
- Chen, H.; Chiang, R.H.; Storey, V.C. Business intelligence and analytics: From big data to big impact. J. MIS Q. 2012, 36, 1165–1188. [Google Scholar] [CrossRef]
- Badawi, O.; Brennan, T.; Celi, L.A.; Feng, M.; Ghassemi, M.; Ippolito, A.; Johnson, A.; Mark, R.G.; Mayaud, L.; Moody, G. Making big data useful for health care: A summary of the inaugural MIT critical data conference. JMIR Med. Inform. 2014, 2, e22. [Google Scholar] [CrossRef]
- Murdoch, T.B.; Detsky, A.S.-J. The inevitable application of big data to health care. JAMA 2013, 309, 1351–1352. [Google Scholar] [CrossRef]
- Niyizamwiyitira, C.; Lundberg, L. Performance evaluation of SQL and NoSQL database management systems in a cluster. IJDMS 2017, 9, 1–24. [Google Scholar] [CrossRef]
- Hong, L.; Luo, M.; Wang, R.; Lu, P.; Lu, W.; Lu, L. Big data in health care: Applications and challenges. DIM 2018, 2, 175–197. [Google Scholar] [CrossRef] [Green Version]
- Parker, Z.; Poe, S.; Vrbsky, S.V. Comparing NoSQL MongoDB to an SQL DB. In Communications of the ACM; ACM: New York, NY, USA, 2013; p. 5. [Google Scholar]
- Labhansh, A.; Parth, N.; Sandeep, T.; Vasundhra, G. Business intelligence tools for big data. JBAR 2016, 3, 505–509. [Google Scholar]
- Bear, C.; Lamb, A.; Tran, N. The Vertica database: SQL RDBMS for managing big data. In Proceedings of the 2012 Workshop on Management of Big Data Systems; ACM: New York, NY, USA, 2012; pp. 37–38. [Google Scholar]
- McLernon, D.J.; Bond, C.M.; Hannaford, P.C.; Watson, M.C.; Lee, A.J.; Hazell, L.; Avery, A.; Collaboration, Y.C. Adverse drug reaction reporting in the UK. Drug Saf. 2010, 33, 775–788. [Google Scholar] [CrossRef]
- Lamb, A.; Fuller, M.; Varadarajan, R.; Tran, N.; Vandiver, B.; Doshi, L.; Bear, C. The Vertica analytic database: C-store 7 years later. Proc. VLDB Endow. 2012, 5, 1790–1801. [Google Scholar] [CrossRef]
- Chang, B.R.; Tsai, H.-F.; Lee, Y.-D. Integrated high-performance platform for fast query response in big data with Hive, Impala, and SparkSQL: A performance evaluation. Appl. Sci. 2018, 8, 1514. [Google Scholar] [CrossRef] [Green Version]
- Amato, F.; Marrone, S.; Moscato, V.; Piantadosi, G.; Picariello, A.; Sansone, C. HOLMeS: EHealth in the big data and deep learning era. Information 2019, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Nielsen, P.S. A hybrid ICT-solution for smart meter data analytics. Energy 2016, 115, 1710–1722. [Google Scholar] [CrossRef] [Green Version]
- Stonebraker, M. SQL databases v. NoSQL databases. Commun. ACM 2010, 53, 10–11. [Google Scholar] [CrossRef]
- Tauro, C.J.; Aravindh, S.; Shreeharsha, A. Comparative study of the new generation, agile, scalable, high performance NoSQL databases. Int. J. Comput. Appl. 2012, 48, 1–4. [Google Scholar]
- Hellerstein, J.M.; Stonebraker, M.; Hamilton, J. Architecture of a database system. Foundat. Trends Databases 2007, 1, 141–259. [Google Scholar] [CrossRef]
- Basu, A.; Toy, T. Real-Time Healthcare Analytics on Apache Hadoop* Using Spark* and Shark*; Intel® Distribution for Apache Hadoop* Software: New York, NY, USA, 2014. [Google Scholar]
- Harerimana, G.; Jang, B.; Kim, J.W.; Park, H.K. Health big data analytics: A technology survey. IEEE Access 2018, 6, 65661–65678. [Google Scholar] [CrossRef]
- Aboudi, N.E.; Benhlima, L. Big data management for healthcare systems: Architecture, requirements, and implementation. Adv. Bioinform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, V.; Thirunavukarasu, R. Implications of big data analytics in developing healthcare frameworks–A review. J. King Saud. Univ. Sci. 2019, 31, 415–425. [Google Scholar] [CrossRef]
- Shoro, A.G.; Soomro, T.R. Big data analysis: Apache Spark perspective. Glob. J. Comput. Sci. Technol. 2015, 15. [Google Scholar]
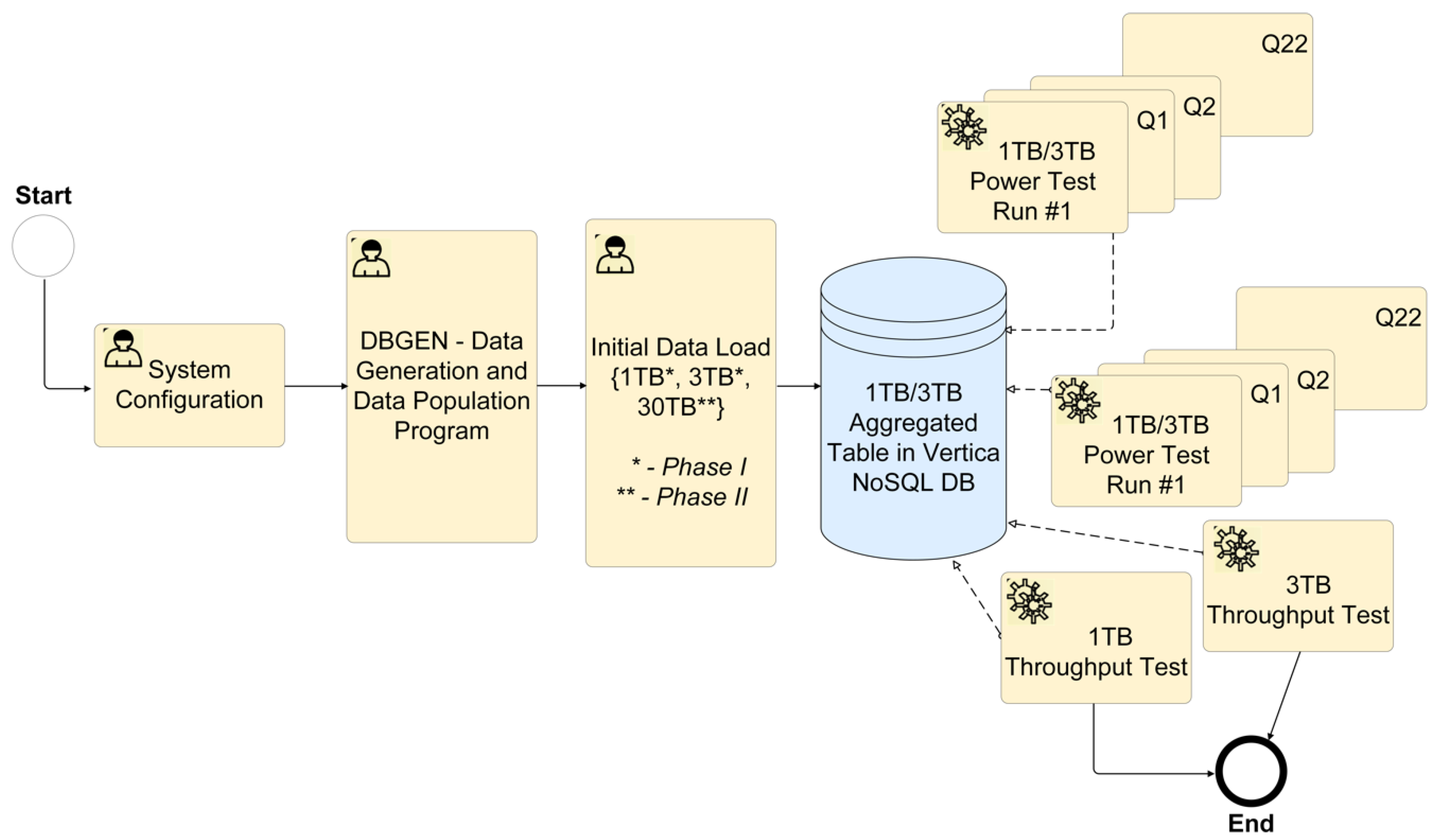









| Parameter | Limit [Hours] | Achieved Results [Hours] |
|---|---|---|
| Initial import TPC-H 1 TB | 24 | 2.94 * |
| Initial import TPC-H 3 TB | 96 | 5.99 * |
| Power test TPC-H 1 TB—1st run | 1.5 | 1.4 ** |
| Power test TPC-H 1 TB—2nd run | 1.5 | 1.36 ** |
| Power test TPC-H 3 TB—1st run | 5 | 4.2 ** |
| Power test TPC-H 3 TB—2nd run | 5 | 4.17 ** |
| Data Size | 1 TB Data | 3 TB Data | ||||
|---|---|---|---|---|---|---|
| Table Name | No. of Rows | Duration in [s] | Duration in Hours | No. of Rows | Duration in [s] | Duration in Hours |
| Customer | 150,000,000 | 1185.00 | 0.33 | 450,000,000 | 4,100.00 | 1.14 |
| Nation | 25 | 0.10 | 0.00 | 25 | 0.20 | 0.00 |
| Orders | 1,500,000,000 | 2533.00 | 0.70 | 4,500,000,000 | 5423.00 | 1.51 |
| Part | 200,000,000 | 272.00 | 0.08 | 600,000,000 | 865.00 | 0.24 |
| Part supp. | 800,000,000 | 1342.00 | 0.37 | 2,400,000,000 | 4240.00 | 1.18 |
| Region | 5 | 0.07 | 0.00 | 5 | 0.07 | 0.00 |
| Supplier | 10,000,000 | 105.00 | 0.03 | 30,000,000 | 266.00 | 0.07 |
| Line item | 5,999,989,709 | 10,594.00 | 2.94 | 18,000,048,306 | 21,548.00 | 5.99 |
| Total load duration | 2.94 * [h] (16,031.17 [s]) | 5.99 ** [h] (36,442.27 [s]) | ||||
| Data Size | 1 TB Data | 3 TB Data | ||||||
|---|---|---|---|---|---|---|---|---|
| Query No. | 3 Nodes | 3 Nodes | 5 Nodes | 5 Nodes | 3 Nodes | 3 Nodes | 5 Nodes | 5 Nodes |
| 1st Run | 2nd Run | 1st Run | 2nd Run | 1st Run | 2nd Run | 1st Run | 2nd Run | |
| Q1 | 267 | 51 | 232 | 161 | 427 | 383 | 441 | 457 |
| Q2 | 23 | 22 | 19 | 15 | 52 | 42 | 36 | 40 |
| Q3 | 64 | 65 | 55 | 40 | 128 | 109 | 121 | 125 |
| Q4 | 470 | 480 | 287 | 320 | 918 | 897 | 914 | 900 |
| Q5 | 177 | 114 | 71 | 70 | 484 | 462 | 465 | 454 |
| Q6 | 0.6 | 0.7 | 0.8 | 0.5 | 1.2 | 1 | 1.4 | 1 |
| Q7 | 119 | 129 | 65 | 65 | 144 | 129 | 140 | 140 |
| Q8 | 37 | 34 | 40 | 22 | 375 | 361 | 270 | 263 |
| Q9 | 2576 | 2551 | 1555 | 1397 | 16,015 | 15,173 | 3791 | 3824 |
| Q10 | 52 | 130 | 44 | 72 | 65 | 58 | 64 | 64 |
| Q11 | 6 | 7 | 5 | 3.8 | 13 | 10 | 10 | 11 |
| Q12 | 13 | 13 | 8 | 11 | 24 | 20 | 23 | 23 |
| Q13 | 221 | 237 | 180 | 136 | 296 | 251 | 325 | 301 |
| Q14 | 49 | 55 | 41 | 36 | 111 | 102 | 105 | 105 |
| Q15 | 7 | 7 | 4 | 4 | 9 | 7 | 9 | 10 |
| Q16 | 42 | 42 | 29 | 30 | 87 | 78 | 86 | 88 |
| Q17 | 11 | 12 | 8 | 6 | 23 | 19 | 23 | 22 |
| Q18 | 376 | 380 | 517 | 554 | 741 | 721 | 743 | 746 |
| Q19 | 58 | 58 | 41 | 41 | 112 | 104 | 111 | 110 |
| Q20 | 129 | 131 | 87 | 73 | 156 | 137 | 150 | 147 |
| Q21 | 2850 | 1763 | 1703 | 1787 | 7278 | 7021 | 7196 | 7085 |
| Q22 | 67 | 60 | 62 | 35 | 107 | 88 | 110 | 106 |
| ∑ of the query execution times in seconds | 7614.6 | 6341.7 | 5053.8 | 4879.3 | 27,566.2 | 26,173 | 15,134.4 | 15,022 |
| ∑ of the query execution times in hours | 2.12 | 1.76 | 1.4 | 1.36 | 7.66 | 7.3 | 4.2 | 4.17 |
| 3 Nodes | 3 Nodes | 5 Nodes | 5 Nodes | |
|---|---|---|---|---|
| 1st Run | 2nd Run | 1st Run | 2nd Run | |
| Results in [s] for 1 TB [s] | 7614.6 | 6341.7 | 5053.8 | 4879.3 |
| Results in [s] for 3 TB [s] | 33,099.26 | 27,566.2 | 15,134.4 | 15,022.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Štufi, M.; Bačić, B.; Stoimenov, L. Big Data Analytics and Processing Platform in Czech Republic Healthcare. Appl. Sci. 2020, 10, 1705. https://doi.org/10.3390/app10051705
Štufi M, Bačić B, Stoimenov L. Big Data Analytics and Processing Platform in Czech Republic Healthcare. Applied Sciences. 2020; 10(5):1705. https://doi.org/10.3390/app10051705
Chicago/Turabian StyleŠtufi, Martin, Boris Bačić, and Leonid Stoimenov. 2020. "Big Data Analytics and Processing Platform in Czech Republic Healthcare" Applied Sciences 10, no. 5: 1705. https://doi.org/10.3390/app10051705
APA StyleŠtufi, M., Bačić, B., & Stoimenov, L. (2020). Big Data Analytics and Processing Platform in Czech Republic Healthcare. Applied Sciences, 10(5), 1705. https://doi.org/10.3390/app10051705






