Abstract
In this paper, we develop an optimised state-of-the-art 2D U-Net model by studying the effects of the individual deep learning model components in performing prostate segmentation. We found that for upsampling, the combination of interpolation and convolution is better than the use of transposed convolution. For combining feature maps in each convolution block, it is only beneficial if a skip connection with concatenation is used. With respect to pooling, average pooling is better than strided-convolution, max, RMS or L2 pooling. Introducing a batch normalisation layer before the activation layer gives further performance improvement. The optimisation is based on a private dataset as it has a fixed 2D resolution and voxel size for every image which mitigates the need of a resizing operation in the data preparation process. Non-enhancing data preprocessing was applied and five-fold cross-validation was used to evaluate the fully automatic segmentation approach. We show it outperforms the traditional methods that were previously applied on the private dataset, as well as outperforming other comparable state-of-the-art 2D models on the public dataset PROMISE12.
1. Introduction
Radiation therapy (radiotherapy) is a cancer treatment that uses ionizing radiation to kill cancer cells or control the growth of tumours. It is a very common procedure to treat all stages of prostate cancer. However, this procedure can also damage the normal cells around the cancer cells putting the surrounding organs at risk of post-treatment complications [1]. In the case of prostate cancer, the main objective is to deliver a maximum dose of radiation to the prostate and minimise the dose received by the bladder and rectum [2]. For this reason, accurate prostate segmentation is required.
Manual labelling of an organ can be a time consuming and very challenging process. It involves one or more experts scanning through the dataset and labelling the organ. As a result, labels produced by experts are usually subject to intra- and inter-expert variability as a result of the varying expertise levels [3], i.e., an expert may segment a specific image differently, if done more than once, or different experts may segment the same image differently [4].
Automatic segmentation can speed up the segmentation process as well as minimise the intra- and inter-expert variability problem [5,6]. In order to perform automatic prostate segmentation on MRI (Magnetic Resonance Imaging) images there are two main methods traditionally employed: atlas-based and deformable model-based methods [7]. In the atlas-based method, a set of images and their corresponding labels are combined together after non-rigid registration (NRR) to create a reference atlas and a corresponding labelled structure. The atlas image, in this case, contains the prostate and its surrounding tissue with the corresponding labelled structure representing the probability of a voxel being a part of the prostate. The NRR of the atlas to the new, unseen MRI scan is used to obtain the segmentation of the prostate of a new patient [7]. In the deformable model-based method, a good initialisation of the model is required. The model can be initialised by atlas-based segmentation [7,8], where a surface is extracted from a thresholded probabilistic segmentation and the model is deformed to closely match the organ boundary by the use of the grey-level information of the image. The grey-level model is developed offline with one-dimensional grey-level profiles taken along the normals of each vertex of the surface for the images. A distance metric is then used to match the profiles of the model and the profiles extracted from the case image [7]. As both methods either rely on the atlas-based method or good initialisation, they are prone to errors [9] and can be time consuming [7].
In recent years, machine learning-based algorithms have made positive contributions in prostate segmentation tasks [10,11,12,13]. Machine learning algorithms have the ability to automatically detect different patterns from the given data or information provided to the model [14]. Deep learning is a class of machine learning algorithms that model high-level abstraction by using several processing layers of transformations [15]. It uses an architecture of multi-level linear and non-linear operations (i.e., layers) to learn complex functions that can represent high-level abstractions [16]. It automatically learns hierarchical features of an input that carry different semantics on different levels [16]. Unlike the traditional machine learning algorithms, this feature-learning ability allows the system to learn complex functions that map the input to the output directly from the data without relying on handcrafted features [17].
Deep learning algorithms based on the convolutional neural network (CNN) such as the Fully Convolutional Network (FCN) [18], U-Net [19] and DenseNet [20] have achieved outstanding results in prostate segmentation tasks [21,22,23,24,25,26,27]. The CNN utilises a number of convolutional and pooling layers for extracting features automatically. The purpose of the convolutional layers is to apply different filters to produce different translation equivariance features [28]. Pooling layers apply non-linear filters to extract the most significant features and make the extracted features translation invariant [28].
Image segmentation based on deep learning algorithms use either patches of an input image or the entire image. Both approaches output a likelihood map that gives the probability of a given pixel being a part of the object to be segmented [26]. In some applications, a patched-based approach is preferable to reduce memory requirement and allow the user to provide a more balanced sample proportion for the training [29]. In other applications, it is more preferable to use the entire image as the input to give more contextual information [26]. Both approaches are able to produce state-of-the-art results on different organ segmentation applications [26].
Deep learning models incorporate a number of different layers and variables that we will denote in the sequel as components. All the components can be adjusted according to the application. It is important to understand which components are beneficial for the deep learning model to perform prostate segmentation. However, many studies often present their works in the final form [21,22,23,24,25] or incorporate post-processing methods into their pipeline to obtain more accurate segmentation results [26,30,31]. As a result, the effect of each component on the overall performance is hard to distinguish and/or unknown.
Many deep learning models have been used to perform medical segmentation tasks. In general, the most successful models are based on 3D neural networks, which include the fusion of 2D/3D models. There are many 3D networks such as, but not limited to, Hybrid Discriminative Network (HD-Net) [32], V-Net [33] and 3D Dense U-Net [34]. In this paper we are considering only 2D networks as the number of components to optimise over is significantly less than in the 3D case. Furthermore, a greater understanding of the contribution of each component is more easily obtained when the complexity of the model is kept as small as possible.
In this paper, we optimise a basic 2D U-Net model [19] for prostate segmentation based on a private dataset [35] of T2 weighted MR images with a fixed 2D resolution and voxel size across the whole dataset. This allow us to mitigate the possibility of artefacts caused by resizing operation in the data preparation process that may ultimately affect the optimisation process. Performance evaluation of different model architectures will be presented that provide an insight into the contribution of each deep learning model component in a prostate segmentation application.
Performance of the optimised model is evaluated on both a private dataset [35] and the public dataset, PROMISE12 [36], of T2 weighted MR images. With respect to the private dataset, we show the improvement achieved when compared to the traditional segmentation methods [35,37] that were previously applied. As well we compare the performance of the optimised model to other state-of-the-art 2D models on the PROMISE12 dataset. Furthermore, challenges due to inter-expert variability associated with the dataset are discussed to address a problem that is often overlooked in medical imaging segmentation. We provide suggestions on how the optimised model could be used to help with this problem.
In Section 2, we discuss background material for the deep learning model components that are considered in the optimised model. In Section 3, we describe the approach for the optimised network architecture and configuration. In Section 4, we train the optimised model and evaluate its performance in comparison to the traditional segmentation methods on the private dataset and to state-of-the-art 2D models on the public dataset, PROMISE12 [36]. In Section 5, we discuss the challenges associated with the dataset as well as make suggestions to address the problem and to further improve the segmentation results. Conclusions are drawn in Section 6.
2. Background
In this section, we describe the deep learning model components considered in the sequel for the development of a 2D model for performing prostate segmentation.
2.1. Structure
Here, we will refer to structure as the overall shape of the architecture. For the task of segmentation, the deep learning model structures that are often adopted are FCN and U-Net type structures. U-Net consists of two FCN-like structures that are cascaded in the form of an encoder-decoder (autoencoder) structure. The encoder is used for feature extraction and the decoder is used for feature mapping to the original input resolution. The main difference between the FCN and U-Net structures is that the FCN does not learn the mapping of the high-level features to the original input resolution in a step-by-step manner as it relies only on the feature extraction part of the network to make the final classification. Although both these structures are able to produce good results on different organ segmentation tasks [26], many works on prostate segmentation show success using U-Net as their base model [21,22,23,24,25]. Moreover the basic U-Net has been used successfully for the segmentation of different parts of the prostate [27].
2.2. The Convolution Layer
Convolution layers use a set of small parameterised filters, sometimes referred to as kernels, to perform convolution operations to produce different feature maps of their input [38]. Generally, in the state-of-the-art 2D models [18,19,39], the filter is based on a matrix. Larger filters, such as a matrix, can also be used in semantic segmentation to mitigate the contradiction between classification and localisation [40]. For example, in the classification problem, the model is required to be invariant to the transformation of the input image, while in the localisation problem, a model has to be transformation-sensitive to perform localisation [40].
The convolution layers can be used in the form of a transposed convolution or in combination with an interpolation layer. Transposed convolution (i.e., deconvolution) is obtained by reversing the forward and backward passes of a convolution [41]. Unlike the fixed interpolation methods (e.g., nearest-neighbour, bilinear interpolation), the transposed convolution filter weights can actually be learnt. Transposed convolution is known to produce checkerboard artifacts [42] that can be completely avoided by combining interpolation and convolution layers [43]. When a convolution layer is combined with an interpolation layer it can also improve upsampling. Note that transposed convolution can also improve the upsampling process.
A convolution layer with a stride greater than one (i.e., strided-convolution) can be used to replace a pooling layer (described in Section 2.3) to perform downsampling in CNN without loss in accuracy [44]. Here stride refers to the number of pixels the parameterised filter shifts at a time in a convolution operation.
2.3. The Pooling Layer
Pooling layers are used to reduce the dimension (i.e., downsample) of the input and introduce translational invariances in the network [38]. The most common type of pooling layer in CNN-based models is the max pooling layer which is used to extract significant features from the previous layer by taking the maximum value in each filter kernel. Max pooling can also be considered as a nonlinear filtering operation. Unlike linear filters that have poor performance in removing non-additive noise and tend to blur edges of an image [45], nonlinear filters usually remove noise as well as preserving the significance of the features of an image [45,46]. However, linear-filter-based pooling layers, such as average pooling or strided-convolution, are sometimes used in place of max pooling with success in certain applications [44]. Other pooling layers include the L2 and RMS, where the L2-norm and RMS value are the output of the layer respectively.
2.4. Feature Maps
Convolution layers produce feature maps that consist of local features at each pixel location. The spatial resolution of the feature maps tend to reduce in size when more convolution and pooling layers are added. Reduction of spatial resolution in the feature maps can be compensated by a progressive increase in the number of feature maps, i.e., representations [47]. The number of feature maps reflects the capacity of the network to implement useful feature extractors (i.e., filters) for certain applications. However, the larger the number of feature maps, the more expensive the memory requirement and computation time becomes. Therefore, the number of feature maps should be adjusted according to the complexity of the task to be completed and the resources available.
2.5. The Activation Layer
Activation layers are typically applied after convolution layers in order to decide whether a particular neuron should be activated or not. Nonlinear activation functions, e.g., a rectified linear unit (ReLU), a leaky ReLU or a parametric ReLU [48], are used to enable the network to approximate most nonlinear functions.
The ReLU is a nonlinear function defined by
The ReLU restricts the activation of the neuron when the input is less than or equal to zero, which simplifies the network and reduces the computational time during the training process. This advantage however, comes with an issue, i.e., since the gradient of the ReLU function in the negative region of x is zero, once the neuron is inactive, the neuron will not be activated again throughout the training process (i.e., dying ReLU problem [49]).
A Leaky ReLU (LReLU) attempts to solve the dying ReLU problem by setting the gradient of the negative region as a small constant value, c, i.e.,
where, for example, c = 0.1.
Although the LReLU (and other modified ReLU activation functions) are shown to be superior to ReLU [48], many state-of-the-art models [18,19,39] still use ReLU as it produces satisfactory results and is simple to implement.
2.6. Dropout Layer
Dropout is a regularisation technique to prevent overfitting. The idea is to train different models simultaneously and use the average of the predictions to improve the generalisation of the model. The operation involves randomly removing neurons at a defined rate during training so the weights of the network are tuned based on different connectivity variations of the network [50].
2.7. Batch Normalisation Layer
The batch normalisation layer normalises the input by subtracting the mean and dividing by the standard deviation for each training batch. This operation reduces the need of Dropout as well as speeding up the training process [51]. However, applying normalisation to the input of each layer may change the representation of the original input, e.g., normalising the input of a sigmoid function may constrain the input to be within the linear region of the sigmoid function. Therefore, extra parameters that control the scale and shift of the normalised value are implemented in the batch normalisation layer to be learnt along with the other model parameters. These extra parameters enable the restoration of the original value if it produces better results than the normalised value [51].
Batch normalisation layers can be applied before or after an activation layer [51]. However, it is suggested [51] that applying the batch normalisation layer before the activation layer produces a more stable distribution. An earlier study has applied batch normalisation successfully to the output of the activation layer [52].
2.8. Skip Connections
Skip connections were originally implemented by ResNet [53] to address the degradation problem of training accuracy in deep networks. They help to prevent the deep model from having a high training error when compared to a shallow counterpart, as it simplifies the training when the additional complexity introduced by redundant layers in the network is not required [53]. An element-wise summation layer is used at the end of a skip connection hence keeping the dimension of the output layer fixed such that it adds neither additional parameters nor computational complexity [53].
In U-Net [19], the skip connections are used to pass the features from one layer to another layer which are then combined with a concatenation instead of summation. The idea is to maintain the preceding layer information and re-use it in the later layer to achieve better performance.
3. Materials and Methods
In this section, we investigate several deep learning model architectures to determine the components that can improve the performance of a 2D U-Net for prostate segmentation. The deep learning model components discussed in Section 2 are considered in this section.
3.1. Dataset
For the optimisation of the 2D U-Net model, we use a private dataset [35], which is collected following ethical approval and informed consents. The dataset is obtained without an endorectal coil, using a Siemens Skyra 3.0 Tesla magnet located at the Calvary Mater Newcastle Hospital, Australia. The dataset consisted of 41 prostate, T2 weighted, MRI scans with three expert manual delineations on each scan. Furthermore, each scan contains voxels with a voxel size of mm. For the performance evaluation in Section 3.2, the second expert label is used for both training and testing as this expert has the highest mean Dice’s similary coefficient (DSC) score against the majority voting [54]. This will also reduce the labelling bias on the smaller volumes of the organ to be segmented, as majority voting tends to be biased.
3.2. Performance Evaluation
In this study, Dice’s similary coefficient (DSC) is used to evaluate the performance of each model. The DSC is the intersection of the predicted regions and the ground truth over their average size [55], given by
Five-fold cross-validation is used for the evaluation of the model on the entire dataset. First, one scan is extracted to be used as the validation set and the remaining 40 scans are shuffled randomly and then divided into 5 folds. Every fold consists of 32 scans for the training set and 8 scans for the test set. The average (Avg) DSC score is the mean score of the five-fold cross-validation.
3.3. Model Architecture
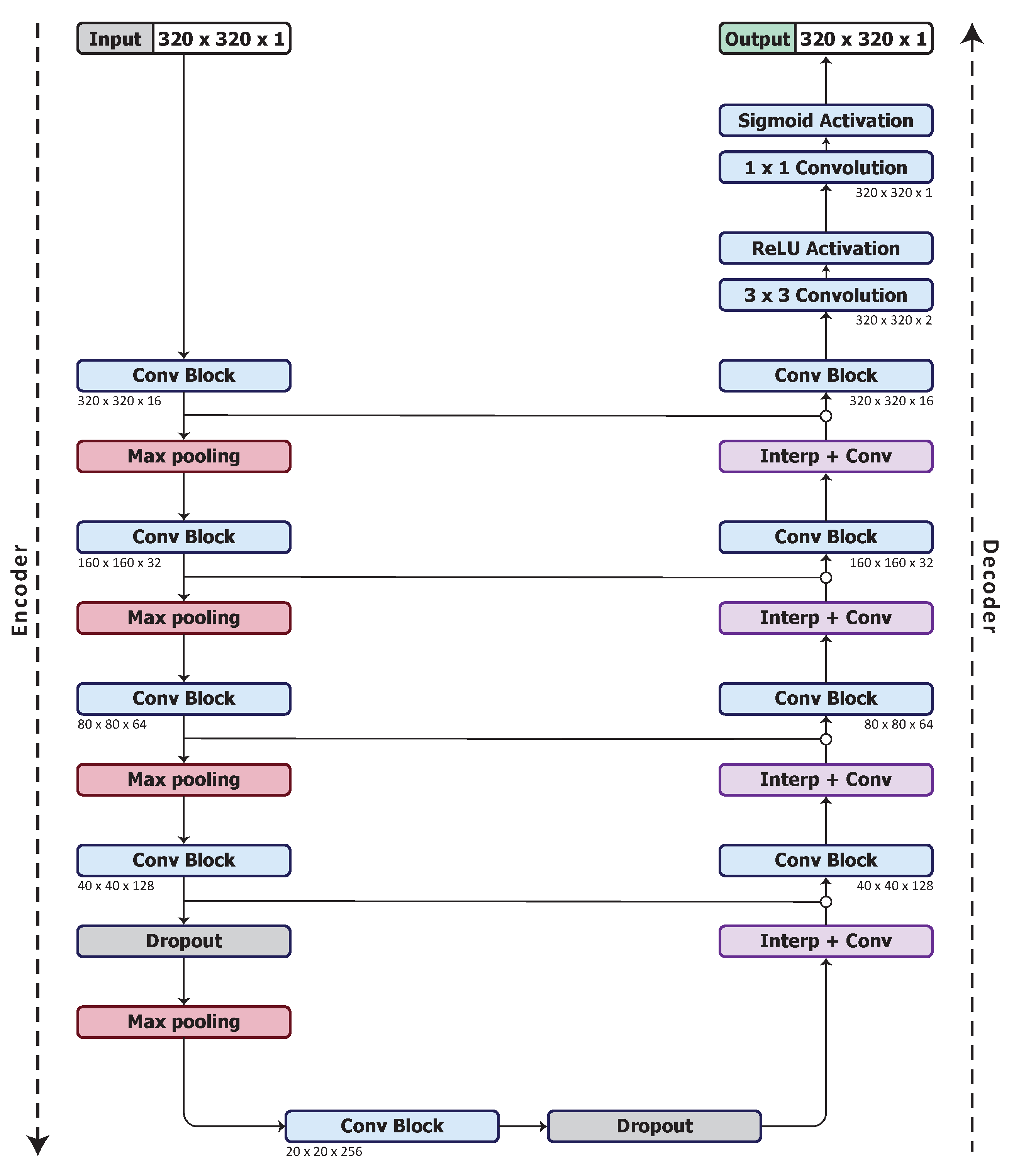
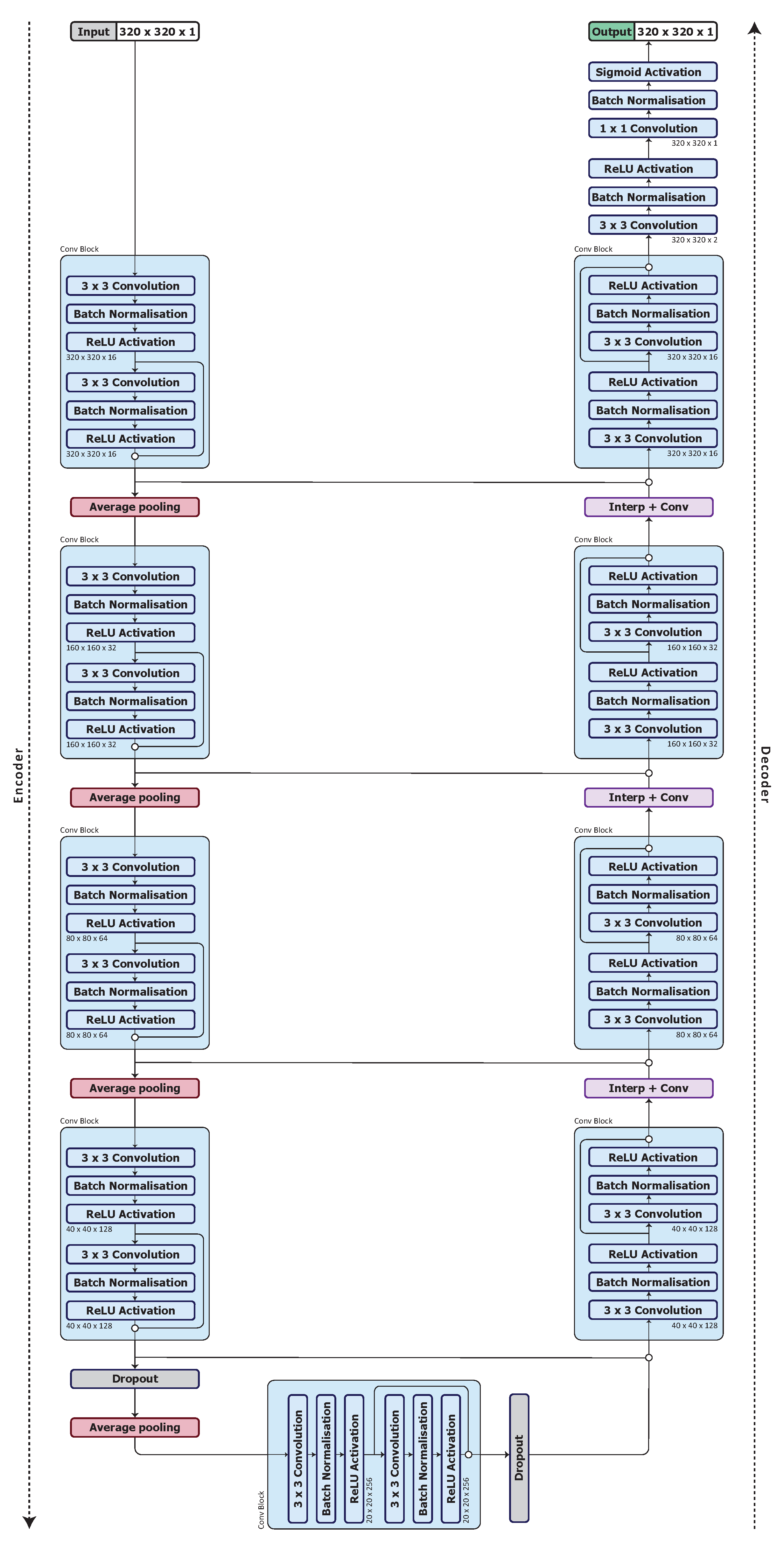
We use a U-Net architecture [56] for our base model. As discussed in Section 2.1, U-Net has equal downsampling and upsampling layer pairs in the network forming an autoencoder network structure that can be beneficial for organ segmentation. We begin with what we denote as UNet_S, a simplified U-Net model that uses a quarter of the number of feature maps used in the U-Net of [56]. The model architecture is shown in Figure 1.
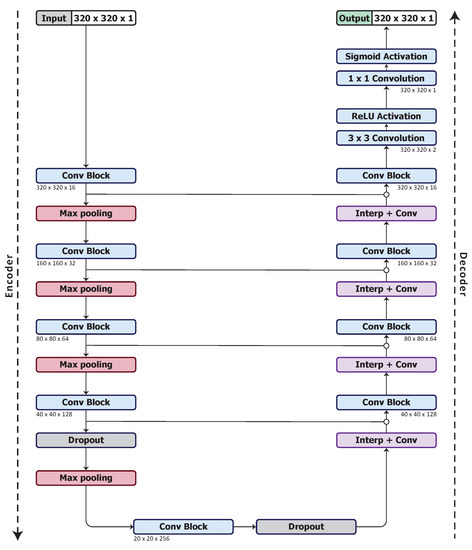
Figure 1.
UNet_S architecture, the base model.
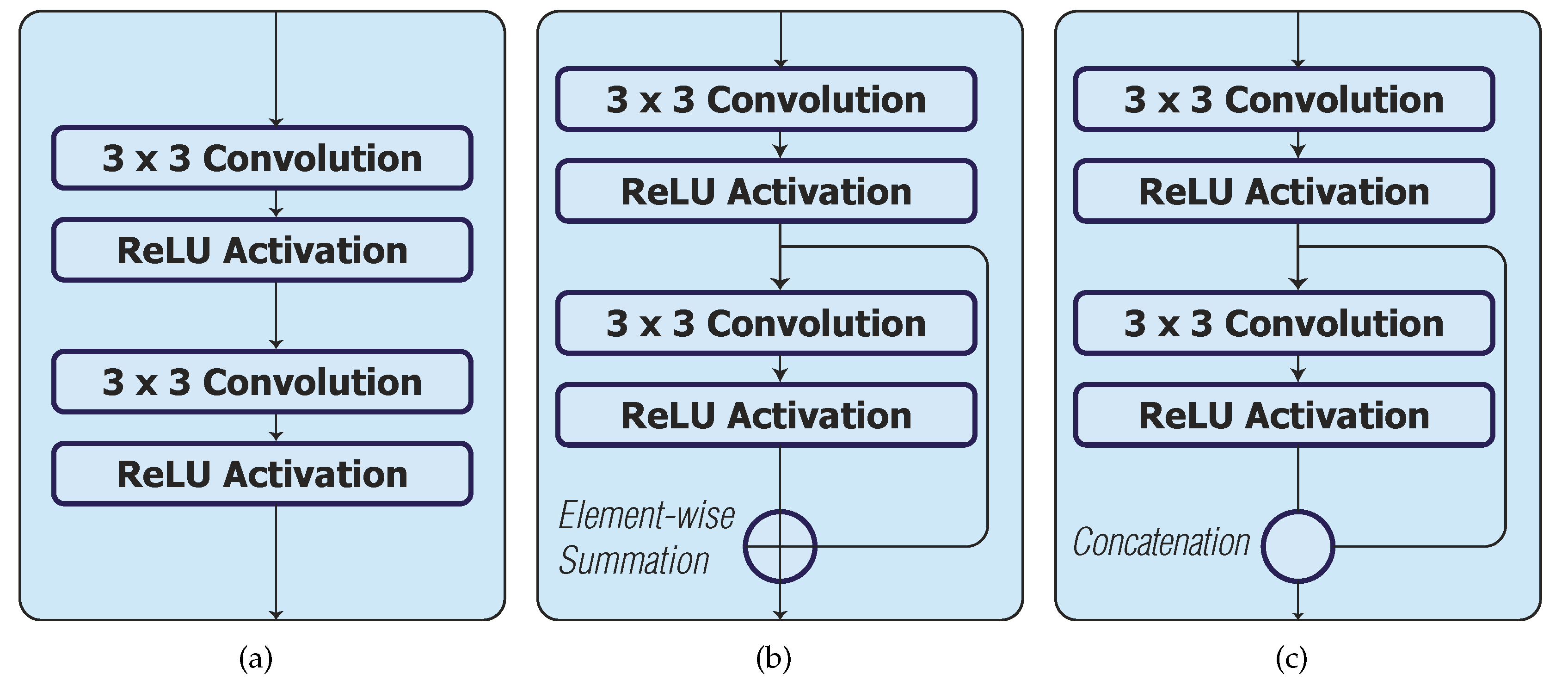
The UNet_S consists of convolution and ReLU activation layers in each convolution block (Conv Block), see Figure 2a. For downsampling, a max pooling layer with stride 2 is used. For upsampling, a combination of a nearest-neighbour interpolation and convolution with ReLU activation layers (Interp+Conv) are used. Skip connections concatenate the same resolution feature maps from the encoder to the decoder, but none are within the convolution block. There are 2 drop out layers with drop out rates of 0.5 after the fourth and fifth convolution block. Lastly, a convolution layer with sigmoid activation is used to produce a probability map.
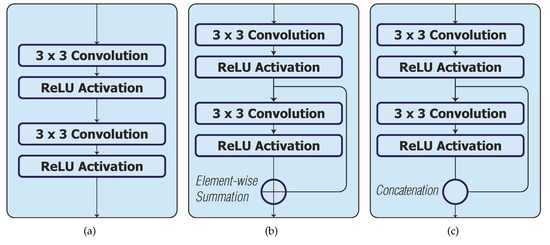
Figure 2.
Skip connection configurations in a convolution block. (a) Original convolution block in UNet_S, UNet_S1 and UNet_S2; (b) Skip connection with element-wise summation in UNet_S.1, UNet_S1.1 and UNet_S2.1; (c) Skip connection with concatenation in UNet_S.2, UNet_S1.2 and UNet_S2.2.
We use 6 phases of evaluation to determine the optimised model: downsampling and upsampling component modification, skip connection implementation, drop out rate adjustment, batch normalisation implementation, pooling layer selection, activation function selection and batch normalisation placement. Table 1 shows the main components of each model being compared in this section.

Table 1.
Model architecture details.
In phase 1 of the evaluation, we investigate the performance of the convolution layers in performing upsampling and downsampling. Model UNet_S1 is UNet_S where nearest-neighbour interpolation and convolution (Interp+Conv) is replaced with transposed convolution, while model UNet_S2 is UNet_S where max pooling is replaced with , 2-strided-convolution. As can be seen from the DSC results shown in Table 2 phase 1, the UNet_S downsampling and upsampling components produce the highest DSC score and hence are still preferable.

Table 2.
Model architecture optimisation results.
For phase 2, we considered the use of a skip connection in each convolution block to improve the performance of the models in phase 1 (i.e., UNet_S, UNet_S1 and UNet_S2). Two configurations of the skip connection are considered in each convolution block of the model. Models UNet_S.1, UNet_S1.1 and UNet_S2.1 have skip connections with element-wise summation (Figure 2b), whilst models UNet_S.2, UNet_S1.2 and UNet_S2.2 have skip connections with concatenation (Figure 2c).
As can be seen from Table 2 phase 2, the skip connections with element-wise summation decreases the performance of UNet_S.1, UNet_S1.1 and UNet_S2.1 models as compared to the UNet_S model. On the other hand, the skip connections with concatenation improves the performance of UNet_S.2, UNet_S1.2 and UNet_S2.2 models as compared to UNet_S model.
From phases 1 and 2, we conclude that the original downsampling and upsampling configurations with a concatenation skip connection in each convolution block, UNet_S.2, performs better than the other configurations (see Table 2). Since the phase 2 results correspond with the phase 1 results, where the modified UNet_S is better than the corresponding UNet_S1 and UNet_S2, we perform the next modifications only on the best model.
In phase 3, we modify the dropout rate from a value of 0.5 to 0 to confirm the theory discussed in Section 2.6. As can be seen from Table 2 phases 2 and 3, the model with the original dropout rate of 0.5, UNet_S.2, still performs better when compared to the modified version, UNet_S.2.1, with a dropout rate of 0.
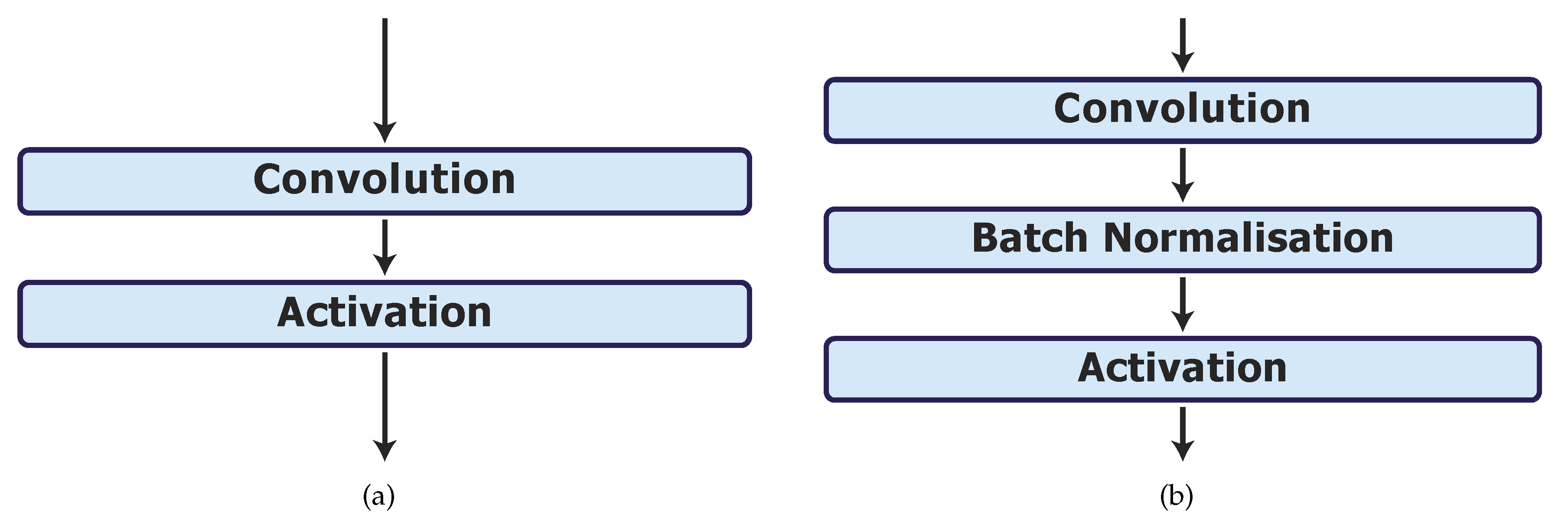
In phase 4, we investigate the effect of batch normalisation on the best model, UNet_S.2. In model UNet_S.2.0.1, the batch normalisation layers are placed in between the convolution and activation layers as shown in Figure 3b.
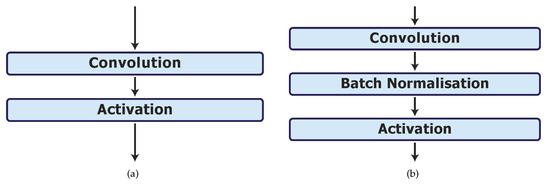
Figure 3.
Batch normalisation layer implementations. (a) Original convolution and activation layers in model UNet_S.2. (b) Batch normalisation layer implementation after each convolution layer in model UNet_S.2.0.1.
As can be seen from Table 2 phase 4, including batch normalisation layers in model UNet_S.2.0.1 improves on the previous best result, UNet_S.2. The use of the batch normalisation layer also increases the speed of training and reduces training problems related to local minima.
As discussed earlier, max pooling is a type of nonlinear filter that preserves the most significant features (lines, edges, etc.) of the previous layer. However, the prostate does not have a well-defined boundary, hence a different pooling layer may perform better segmentation. Therefore, in phase 5, we replace the max pooling layer in the UNet_S.2.0.1 model with average pooling (UNet_S.2.0.1.1), RMS pooling (UNet_S.2.0.1.2) and L2 pooling (UNet_S.2.0.1.3) layers.
The model with the average pooling layer, UNet_S.2.0.1.1, is shown to perform better than the UNet_S.2.0.1, UNet_S.2.0.1.2 and UNet_S.2.0.1.3 models as can be observed in Table 2 phase 5.
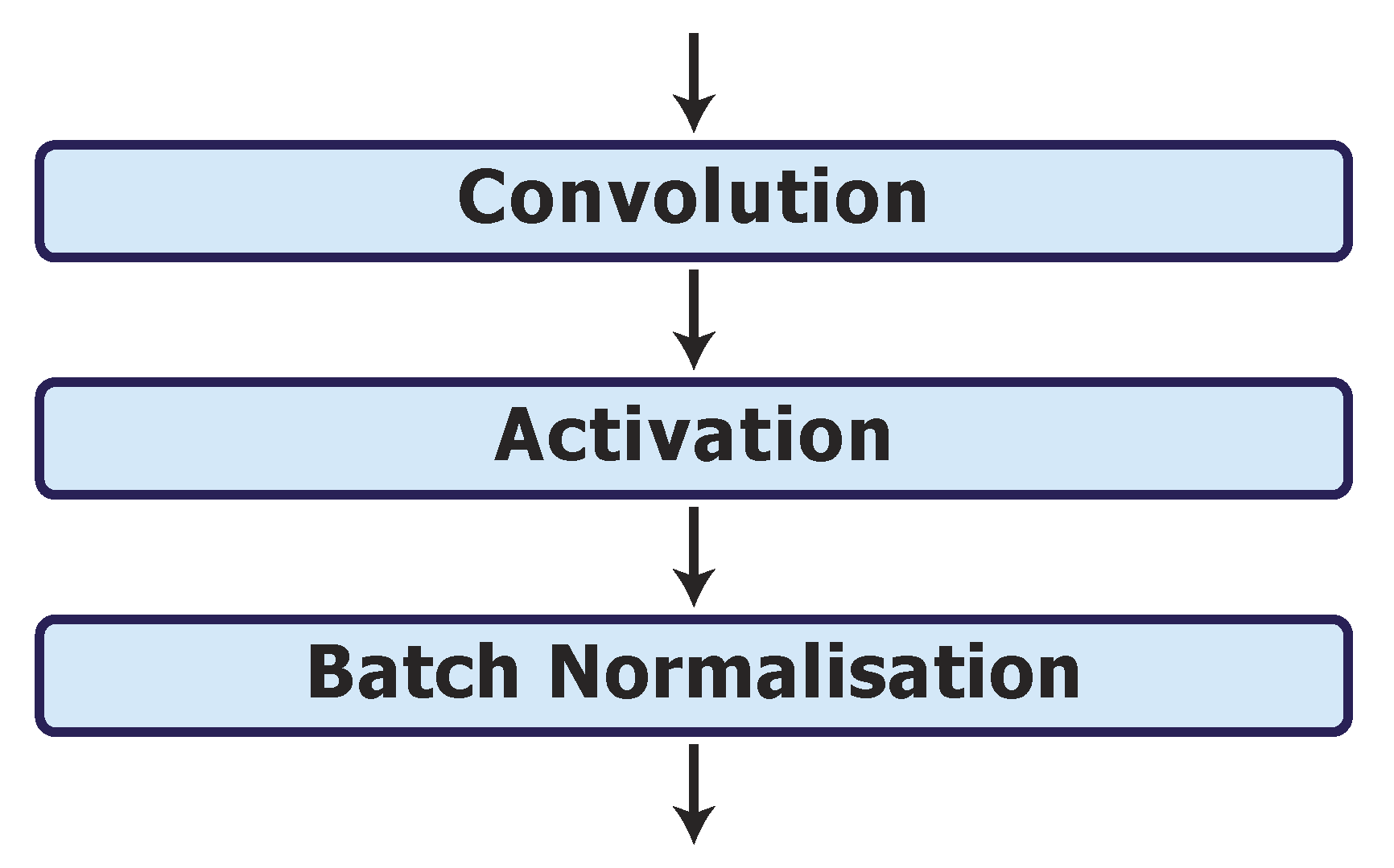
Finally, we investigate options for the activation and batch normalisation layers on the best model, UNet_S.2.0.1.1. As discussed in Section 2.5, the ReLU may cause the neurons in the model to die, which can be overcome by the LReLU. Hence, we replace the ReLU activation layers with LReLU activation layers in UNet_S.2.0.1.1.1. We also investigate the network performance when applying the batch normalisation layer after each activation layer (as shown in Figure 4) in model UNet_S.2.0.1.1.2. The results are shown in Table 2 phase 6. It can be observed that neither the modification of the ReLU to the LReLU nor the change in position of the batch normalisation layer with the ReLU layer improves on the performance of the UNet_S.2.0.1.1 model.
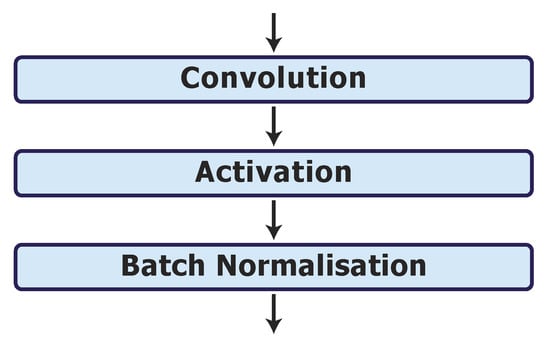
Figure 4.
Batch normalisation layer after each activation layer in model UNet_S.2.0.1.1.2.
Therefore, our final model is the UNet_S.2.0.1.1, which is a simplified UNet with a concatenation skip connection in each convolution block, with a average pooling layer used for downsampling and a batch normalisation layer placed between each convolution and ReLU activation layer.
3.4. Optimised Network Architecture
The configuration of the optimised network, model UNet_S.2.0.1.1, is shown in Figure 5. The network starts with a input layer followed by a batch normalisation layer. Each convolution block consists of convolution layers, batch normalisation layers and ReLU activation layers. A skip connection with a concatenation is used to pass the feature maps between the outputs of the activation layers in a convolution block so as to combine the feature maps. The concatenated feature maps are passed deeper into the encoder as well as to the decoder to improve the spatial information in the higher level feature maps. A average pooling layer is used to downsample the feature maps from a resolution of to . A dropout layer, with a dropout rate of 0.5, is applied after the fourth and fifth convolution blocks. The Interp+Conv layers consist of a nearest neighbour interpolation followed by convolution, batch normalisation and ReLU activation layers. The final classification layer consists of convolution, batch normalisation and sigmoid activation layers.
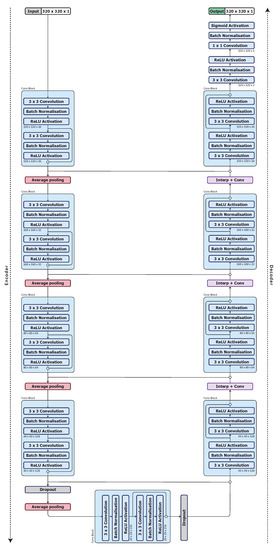
Figure 5.
Optimised model architecture (UNet_S.2.0.1.1).
4. Results
In this section, we explain the training process and the results of the optimised network on both the private and the PROMISE12 [36] datasets.
4.1. Implementation on the Private Dataset
The same dataset (scan dimension of voxels with voxel size of mm) used for the model evaluation (Section 3) is used for both training and testing in this section. However, instead of using labels from one expert as the label of the prostate, we use the majority voting of the labels from 3 experts, as used in the other studies [35,37] we are comparing with, to extract the consensus label of the prostate for training.
4.1.1. Training on the Private Dataset
The training process consists of 2 non-enhancing preprocessing steps, data portioning/balancing and normalisation. To minimise bias on the weight tuning in the training process, an equal portion of the data has to be used for training. In this case, we use an equal number of slices with and without prostate labels. We extract all the 2D scan slices in a volume that have a prostate label, then randomly select an equal number of the 2D scan slices that do not have a prostate label. For the normalisation, we subtract the mean of the whole training set from each voxel of the training, validation and test sets, then divide it by one standard deviation of the training set.
An Adam optimiser [57] is used with a learning rate and a binary cross entropy loss function [58], given by
where is the network prediction on sample i in a range between 0 and 1 and is the ground truth of sample i in binary (0 or 1). The model is developed with Keras [59]. Both training and testing are performed on 4 Gb GeForce GTX 960 GPU with Intel(R) Core(TM) i5-3550 CPU @3.30 GHz and 16 Gb RAM. The training time per epoch is approximately 275 s, while testing time is 2.85 s per case and 0.0474 s per slide.
4.1.2. Results on the Private Dataset
The performance difference between the optimised method and three traditional prostate segmentation methods [35,37] is presented in this section. The three prostate segmentation methods that were used in previous studies are the multi-atlas [35], multi-object weighted and standard (unweighted) deformable model approaches [37].
Five-fold cross-validation mean DSC, median DSC, average symmetric surface distance (ASD) and Hausdorff distance are used for the evaluation of the model performance to ensure that the segmentation errors are reasonable for the application, i.e., treatment planning.
The ASD is the average Euclidean distance from all the points on the predicted region boundary, , to the ground truth region boundary, , and from all the points on the to the [55], given by
where the Euclidean distance from a voxel x to a set of voxels A is given by
with the Euclidean distance between 2 voxels (e.g., voxel x and y) denoted by .
The Hausdorff distance measures the maximum distance from a point in set A to the nearest point in set B [55], given by
Examining the score for the whole dataset, the optimised model gives a mean DSC of 87.38%, median DSC of 88.19%, median ASD of 0.72 mm and median Hausdorff of 4 mm. As shown in the Table 3, the optimised model outperforms the traditional methods by at least 7% and 6% in mean and median DSC respectively, 1.32 mm in median ASD and 5.6 mm in median Hausdorff distance.

Table 3.
Performance comparison between the optimised model and traditional methods.
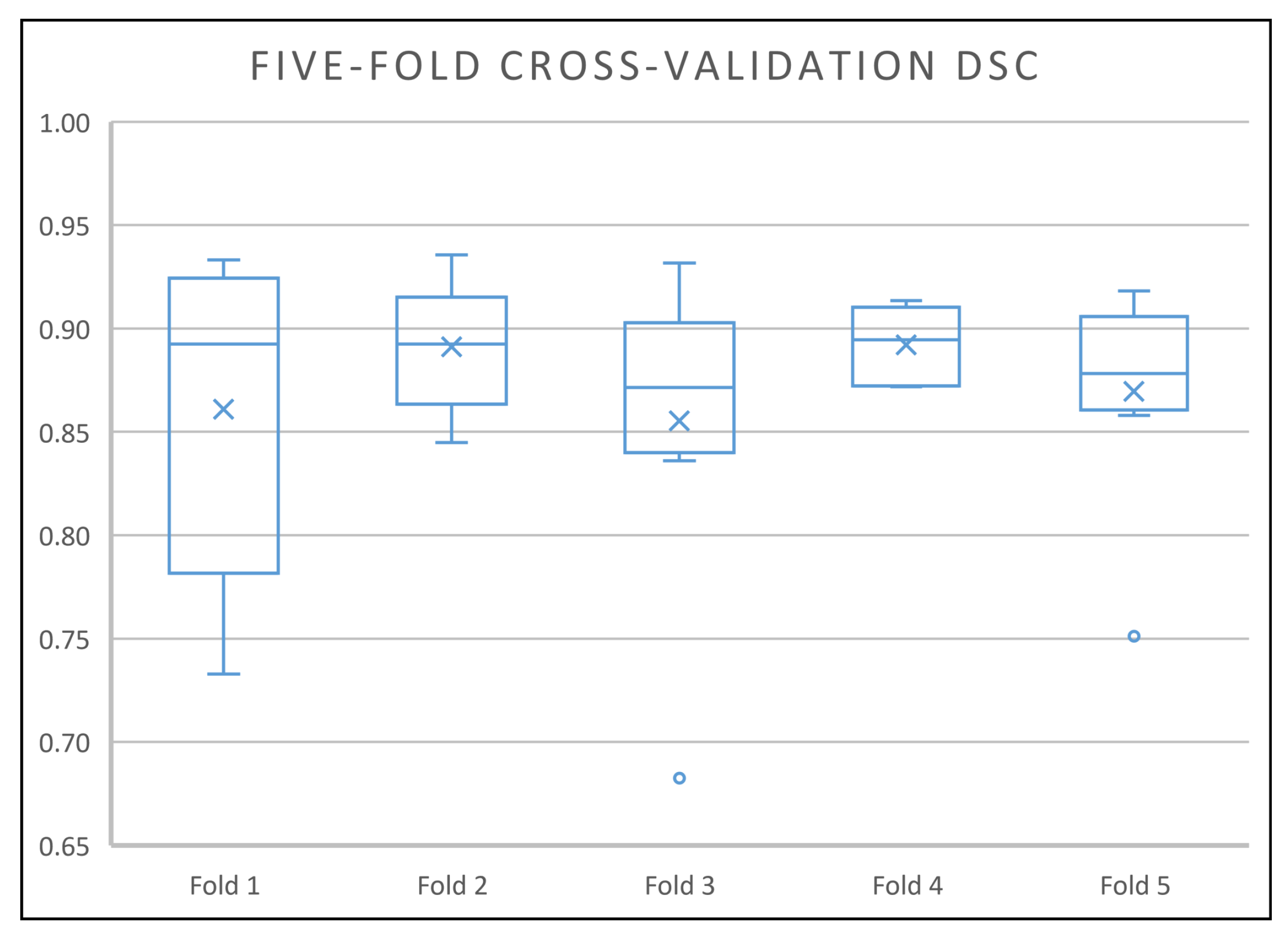
As can be seen in Figure 6, the optimised model performed well on all the cross-validation folds by having DSC scores in the range of 0.73 to 0.94, where the third quartile of the five folds are at least 0.9 and only 4 out of 40 predictions (2 in fold 1, 1 in fold 3, 1 in fold 5) get a DSC score below 0.81. All of the mean DSC scores are at least 0.86 and the median DSC scores are at least 0.87. Without the outliers, the minimum DSC scores of 4 cross-validation folds are all 0.84 and above (fold 2, 3, 4 and 5), which is above the mean and the median DSC scores achieved by the traditional methods.
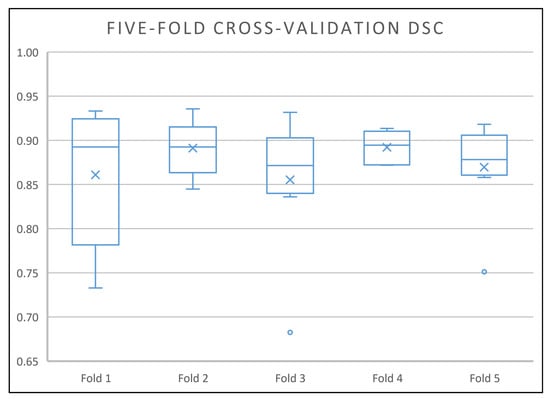
Figure 6.
Box-and-whisker plot of the optimised model five-fold cross-validation Dice’s similary coefficient (DSC) scores. All the mean and median DSC scores are at least 0.86 and 0.87 respectively (the best mean and median DSC scores achieved by the traditional method are 0.80 and 0.82 respectively in this dataset).
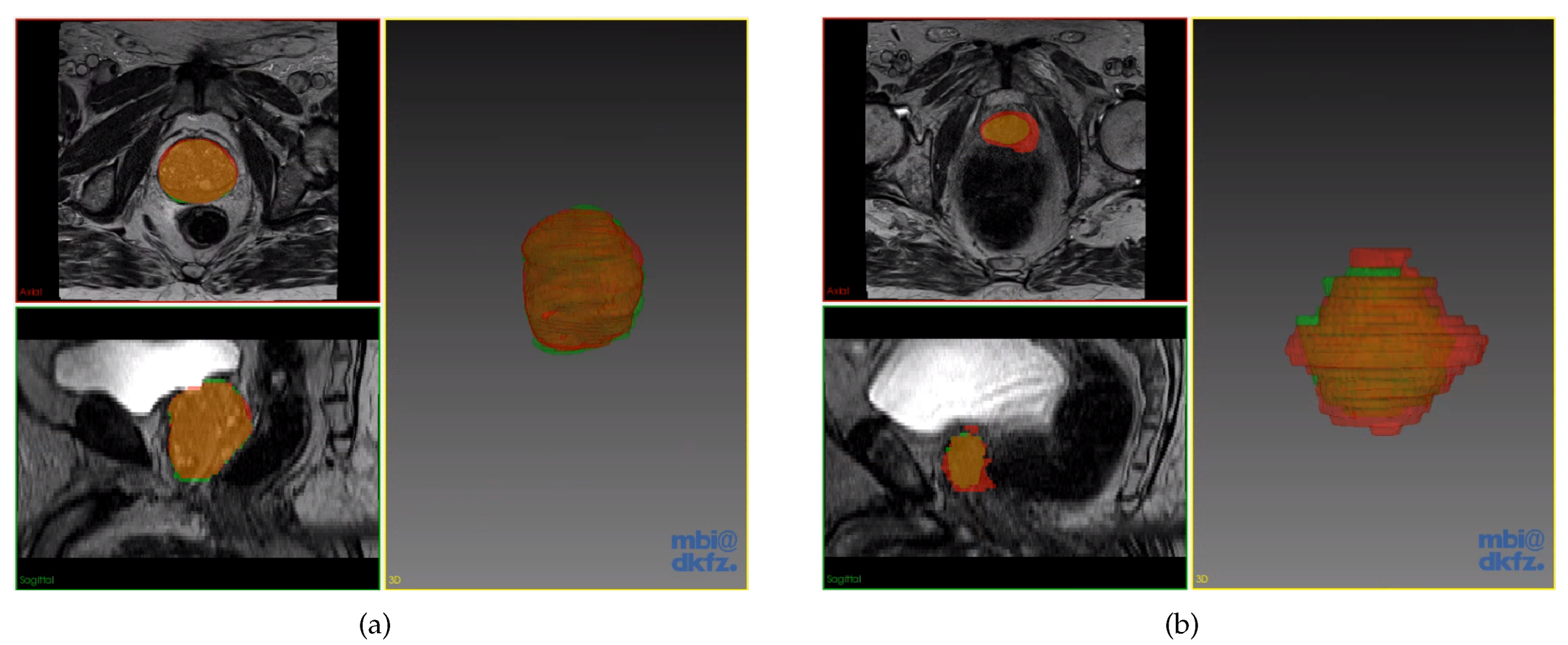
Excluding the obvious outliers in Figure 6, we present the best and worst predictions of the segmentation in Figure 7a,b respectively. These prediction volumes have DSC scores of 0.94 and 0.73. It is easily observed that the prediction volume with a DSC score of 0.94 is very similar to the ground truth segmented volume. We note that even for the worst case, the majority of the prediction volume still overlapped well with the ground truth segmented volume having a DSC score of 0.73.
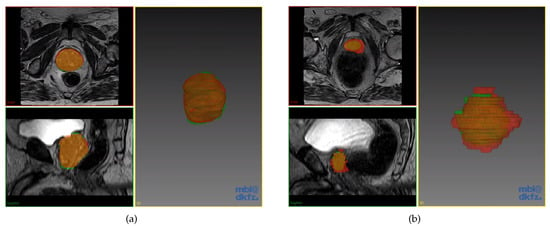
Figure 7.
Prostate segmentation results by the optimised model. Model prediction in red, while the ground truth in green. (a) Prediction with DSC score of 0.94; (b) Prediction with DSC score of 0.73.
4.2. Implementation on the PROMISE12 Dataset
The PROMISE12 dataset consists of 80 T2-weighted MR images of the prostate. It is collected with different acquisition protocols (e.g., different slice thickness, with/without endorectal coil) from multiple centres and vendors. The training set consists of 50 T2-weighted MRI scans with a single prostate label (i.e., reference segmentation). The test set consists of 30 T2-weighted MRI scans without any label.
4.2.1. Training on the PROMISE12 dataset
In addition to the 2 non-enhancing preprocessing steps, data portioning/balancing and normalisation, performed in Section 4.1.1, 2D resizing to a fixed size of has to be performed in this dataset as the MRI scans come in 2 different sizes (e.g., and ).
The same optimiser, learning rate and binary cross entropy loss function, as described in Section 4.1.1, are used for the training. The same development environment, as in Section 4.1.1, is used for both training and testing. The training time per epoch is approximately 200 s, while the testing time is 2.85 s per case and 0.0474 s per slide.
4.2.2. Results on the PROMISE12 Test Set
In this section, the performance of the optimised network was evaluated on the PROMISE12 test set and compared to the state-of-the-art models on the PROMISE12 leaderboard by submitting the model predictions to the MICCAI PROMISE12 grand-challenge website where the scores are generated by the organiser. Here, the test set mean DSC and PROMISE12 overall score [36] are used for the evaluation of the models. The performance results of different methods as well as the model type, preprocessing and postprocessing details are presented in the Table 4. Further details of the results can be obtained from the MICCAI PROMISE12 grand-challenge website [36].

Table 4.
Performance comparison between the optimised model and state-of-the-art models on the PROMISE12 test set.
In Table 4, the optimised model is compared with the top 12 state-of-the-art 2D models on the PROMISE12 leaderboard, all the 3D and combination of the 2D/3D models have been excluded from the table as they are not directly comparable to the model structure in this paper. From the models shown in the table, we will discuss in the sequel why some of these should be excluded from a direct comparison with our model due to a number of factors such as enhancing pre/post-processing and a stacked model structure.
As shown, both of the Z-Nets perform better as compared to our optimised model in terms of both mean DSC and overall score. However, both Z-Nets use either enhancing pre-processing or post-processing to improve the model performance. Therefore, the Z-Nets results are incomparable with our optimised model as we do not employ any enhancing pre- or post-processing. On the other hand, W-Net does not use any enhancing pre- or post-processing and it has a mean DSC of 0.8996 and overall score of 86.5028. However, W-Net is a stacked U-Net, i.e., it utilises a double U-Net to perform the segmentation, hence it is not comparable with our optimised model that consists of a single U-Net. Similar with our approach, sho89512 uses a single U-Net structure model and does not employ any enhancing pre- or post-processing method. However, although it performs better in the overall score (86.3676 vs. 85.4954), it performs slightly worse than the optimised model in the mean DSC (0.8898 vs. 0.89). As can be seen from Table 4, our optimised model performs significantly better compared to the rest of the 2D U-Net-based models with (e.g., MBIOS, UdeM 2D and AdaResU-Net) or without (e.g., U-Net with dense block and U-Net with skip connection) the use of enhancing pre- and post-processing. Note that, it also performs better than the adversial network.
Most importantly, the optimised model is seen to be better than U-Net (MBIOS), U-Net with residual connection (Udem 2D) and U-Net with skip connection (by wanlichen (WNet)) as a result of the optimisation. Another key point to note is that the optimised model performs better than the U-Net with dense block (by ppppppppjw), and only slightly worse than U-Net with dense dilated block (by sho89512 and fumin).
5. Discussion
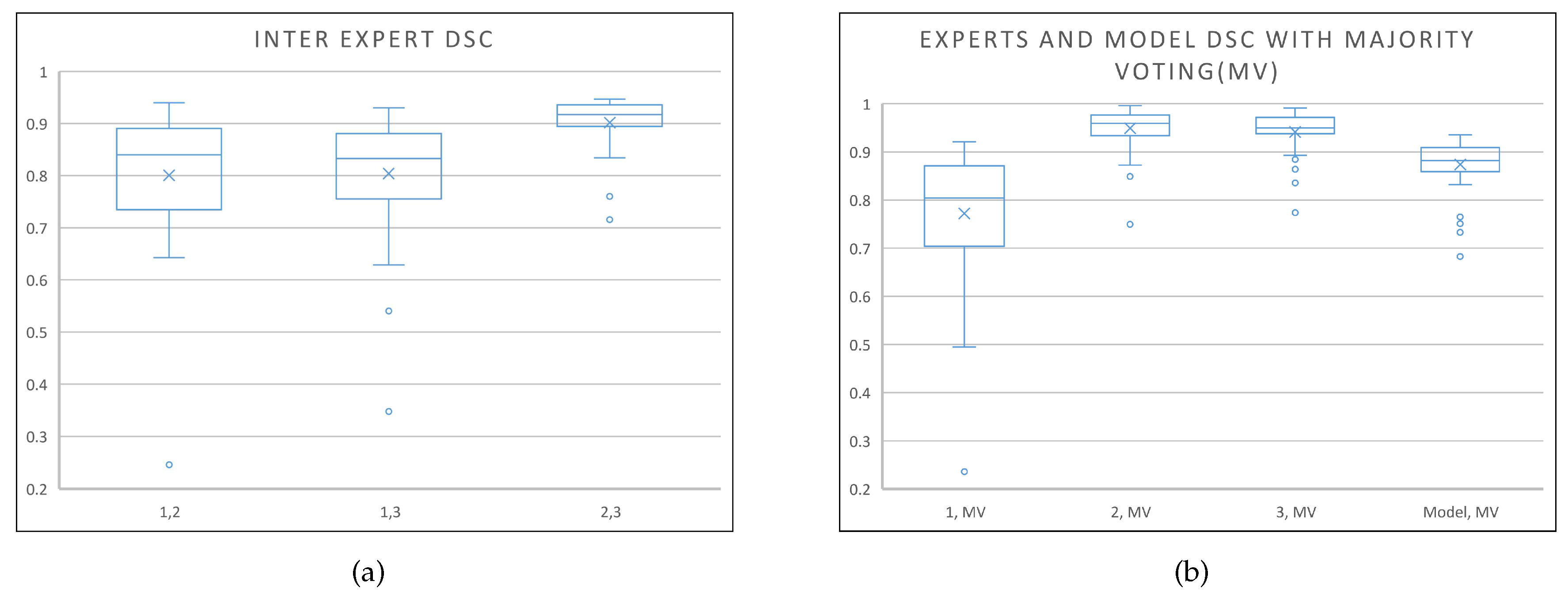
In deep learning applications, the model architecture tends to be the main focus as it defines the capability and capacity of a network to extract features and learn. Often a very complicated model is developed (e.g., stacked, ensemble, hybrid, etc.) without understanding the full capability of a single U-Net structure and the components within. For a deep learning model with supervised learning, data quantity and label quality should also be of a primary focus. For example, it is known that medical imaging data labelling is always subject to intra- and inter-expert variability, that cannot be avoided, hence larger data sets are required in order to obtain the best result. To highlight the inter-expert variability problem, we show the inter-expert DSC score (between experts 1 and 2, 1 and 3, and 2 and 3) for the segmentation of the prostate in the box-and-whisker plot, Figure 8a, of the private dataset where it can be observed that the range of variability is very large. Also, it can be seen from the box-and-whisker plot in Figure 8b that the optimised model is shown to perform better than any given expert and almost as well as the other experts with respect to the majority voting label.
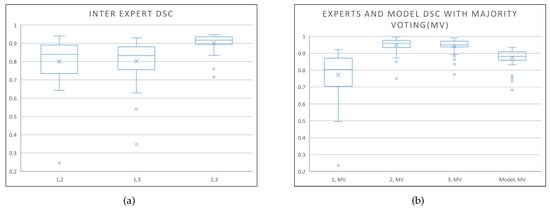
Figure 8.
Box-and-whisker plots. (a) Inter-expert DSC scores to highlight the inter-expert variability problem; (b) DSC scores of the 3 experts and the model against the majority voting label to show that the optimised model performance is within the performance range of the experts.
In future work, we suggest that an automatic segmentation model could be used to provide basic guidance for the expert to produce a larger dataset so that a more robust model can be developed. Furthermore, a combination of Multiparametric MRI (such as T1w, T2w, ADC and PDw) can be considered as input to the neural network model to provide more initial features (i.e., information) for the network to perform the segmentation as it has been shown to be significantly beneficial in prostate segmentation [27,62] and prostate cancer detection [63]. Furthermore, integration of Attention Gates (AGs) [64] and Squeeze-and-Excitation (SE) blocks [65,66] are shown to increase the performance of the U-Net model in performing segmentation [64,66] and could be considered as another component for optimisation in the U-Net structure as performed in this paper. In addition, optimisation of the components for 3D neural network models would be a logical and next step to further the work in this paper as it processes 3D input and extracts 3D features, which can be beneficial for performing volumetric medical image segmentation [34].
6. Conclusions
In this paper, we develop an optimised 2D U-Net model to perform prostate segmentation, without the need of any enhancing pre- or post-processing. We also establish the importance of individual components within the U-Net model to perform prostate segmentation. Compared to transposed convolution, we found that interpolation and convolution results in a better performance for upsampling. Within each convolution block, the combination of feature maps with a skip connection is only beneficial with a concatenation operation. For pooling, the use of average pooling brings significant improvement as compared to the strided-convolution, max, RMS or L2 pooling. Including a batch normalisation layer before the activation layer also brings further improvement in the model performance. We show that the optimised model in this paper outperforms traditional segmentation methods on the private dataset by approximately 6% and 7% in median and mean DSC score respectively. Furthermore, it outperforms (in terms of DSC) other comparable state-of-the-art 2D models on the PROMISE12 public dataset. In addition, we discuss the intra-and inter-expert label variability and the effect on the model performance, as well as provide suggestions to reduce the associated errors.
Author Contributions
Data, review, P.G.; Methodology, software, analysis, validation, writing, I.P.A.; Main supervision, review, editing, J.S.W.; Review, S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
I. Astono has been awarded a 2016 University of Newcastle Scholarship provided by UNIPRS and UNRSC 50:50.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mishra, N.; Petrovic, S.; Sundar, S. A Knowledge-Light Nonlinear Case-Based Reasoning Approach to Radiotherapy Planning. In Proceedings of the 2009 21st IEEE International Conference on Tools with Artificial Intelligence, Newark, NJ, USA, 2–4 November 2009; pp. 776–783. [Google Scholar] [CrossRef]
- Dowling, J.A.; Fripp, J.; Chandra, S.; Pluim, J.P.W.; Lambert, J.; Parker, J.; Denham, J.; Greer, P.B.; Salvado, O. Fast Automatic Multi-atlas Segmentation of the Prostate from 3D MR Images. In Prostate Cancer Imaging. Image Analysis and Image-Guided Interventions; Madabhushi, A., Dowling, J., Huisman, H., Barratt, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 10–21. [Google Scholar]
- Mahapatra, D. Semi-supervised learning and graph cuts for consensus based medical image segmentation. Pattern Recognit. 2017, 63, 700–709. [Google Scholar] [CrossRef]
- White, D.; Houston, A.S.; Sampson, W.F.D.; Wilkins, G.P. Intra- and Interoperator Variations in Region-of-Interest Drawing and Their Effect on the Measurement of Glomerular Filtration Rates. Clin. Nucl. Med. 1999, 24, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Dowling, J.; Shen, K.; Pluim, J.; Greer, P.; Salvado, O.; Fripp, J. Automatic Segmentation of the Prostate in 3D Magnetic Resonance Images Using Case Specific Deformable Models. In Proceedings of the 2011 International Conference on Digital Image Computing: Techniques and Applications, Noosa, QLD, Australia, 6–8 December 2011; pp. 7–12. [Google Scholar] [CrossRef]
- Shahedi, M.; Ma, L.; Halicek, M.; Guo, R.; Zhang, G.; Schuster, D.M.; Nieh, P.; Master, V.; Fei, B. A semiautomatic algorithm for three-dimensional segmentation of the prostate on CT images using shape and local texture characteristics. In Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling; Fei, B., Webster, R.J., III, Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2018; Volume 10576, pp. 280–287. [Google Scholar] [CrossRef]
- Chandra, S.S.; Dowling, J.A.; Shen, K.; Raniga, P.; Pluim, J.P.W.; Greer, P.B.; Salvado, O.; Fripp, J. Patient Specific Prostate Segmentation in 3-D Magnetic Resonance Images. IEEE Trans. Med. Imaging 2012, 31, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Troccaz, J.; Daanen, V. Automated segmentation of the prostate in 3D MR images using a probabilistic atlas and a spatially constrained deformable model. Med. Phys. 2010, 37, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.K.H.; Leung, L.H.T.; Kwong, D.L.W. Evaluation and optimization of the parameters used in multiple-atlas-based segmentation of prostate cancers in radiation therapy. Br. J. Radiol. 2016, 89, 20140732. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shao, Y.; Lian, J.; Wang, A.Z.; Chen, R.C.; Shen, D. Accurate Segmentation of CT Male Pelvic Organs via Regression-Based Deformable Models and Multi-Task Random Forests. IEEE Trans. Med. Imaging 2016, 35, 1532–1543. [Google Scholar] [CrossRef]
- Cheng, R.; Turkbey, B.; Gandler, W.; Agarwal, H.K.; Shah, V.P.; Bokinsky, A.; McCreedy, E.; Wang, S.; Sankineni, S.; Bernardo, M.; et al. Atlas based AAM and SVM model for fully automatic MRI prostate segmentation. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 2881–2885. [Google Scholar] [CrossRef]
- Yang, M.; Li, X.; Turkbey, B.; Choyke, P.L.; Yan, P. Prostate Segmentation in MR Images Using Discriminant Boundary Features. IEEE Trans. Biomed. Eng. 2013, 60, 479–488. [Google Scholar] [CrossRef]
- Gao, Q.; Asthana, A.; Tong, T.; Hu, Y.; Rueckert, D.; Edwards, P. Hybrid Decision Forests for Prostate Segmentation in Multi-channel MR Images. In Proceedings of the 2014 22nd International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014; pp. 3298–3303. [Google Scholar] [CrossRef]
- Kunjir, A.; Shaikh, B. A Survey on Machine Learning Algorithms for Building Smart Systems. Int. J. Innov. Res. Comput. Commun. Eng. 2017, 5, 1052–1058. [Google Scholar] [CrossRef]
- Fatima, M.; Pasha, M. Survey of Machine Learning Algorithms for Disease Diagnostic. J. Intell. Learn. Syst. Appl. 2017, 9, 1–16. [Google Scholar] [CrossRef]
- Yuan, Z.; Xu, C.; Sang, J.; Yan, S.; Hossain, M.S. Learning Feature Hierarchies: A Layer-Wise Tag-Embedded Approach. IEEE Trans. Multimed. 2015, 17, 816–827. [Google Scholar] [CrossRef]
- Bengio, Y. Learning Deep Architectures for AI; Essence of knowledge; Now Publishers: Delft The Netherlands, 2009; pp. 4–6. [Google Scholar]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Zhu, Q.; Du, B.; Turkbey, B.; Choyke, P.L.; Yan, P. Deeply-supervised CNN for prostate segmentation. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, Alaska, AK, USA, 14–19 May 2017; pp. 178–184. [Google Scholar] [CrossRef]
- Xiangxiang, Q.; Yu, Z.; Bingbing, Z. Automated Segmentation Based on Residual U-Net Model for MR Prostate Images. In Proceedings of the 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Beijing, China, 13–15 October 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Zhu, Y.; Wei, R.; Gao, G.; Ding, L.; Zhang, X.; Wang, X.; Zhang, J. Fully automatic segmentation on prostate MR images based on cascaded fully convolution network. J. Magn. Reson. Imaging 2018, 49, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, T.; Hamey, L.G.C.; Ho-Shon, K. Convolutional Neural Networks for Prostate Magnetic Resonance Image Segmentation. IEEE Access 2019, 7, 36748–36760. [Google Scholar] [CrossRef]
- Yuan, Y.; Qin, W.; Guo, X.; Buyyounouski, M.; Hancock, S.; Han, B.; Xing, L. Prostate Segmentation with Encoder-Decoder Densely Connected Convolutional Network (Ed-Densenet). In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 434–437. [Google Scholar] [CrossRef]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef]
- Zabihollahy, F.; Schieda, N.; Krishna Jeyaraj, S.; Ukwatta, E. Automated segmentation of prostate zonal anatomy on T2-weighted (T2W) and apparent diffusion coefficient (ADC) map MR images using U-Nets. Med. Phys. 2019, 46, 3078–3090. [Google Scholar] [CrossRef]
- Pattanayak, S. Pro Deep Learning with TensorFlow: A Mathematical Approach to Advanced Artificial Intelligence in Python; Apress: Berkeley, CA, USA, 2017; pp. 188–190. [Google Scholar] [CrossRef]
- Hou, L.; Samaras, D.; M Kurc, T.; Gao, Y.; E Davis, J.; Saltz, J. Patch-Based Convolutional Neural Network for Whole Slide Tissue Image Classification. In Proceedings of the 2016 IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Vegas, NV, USA, 27–30 June 2016; Volume 2016, pp. 2424–2433. [Google Scholar] [CrossRef]
- Yan, K.; Li, C.; Wang, X.; Li, A.; Yuan, Y.; Feng, D.; Khadra, M.; Kim, J. Automatic prostate segmentation on MR images with deep network and graph model. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 635–638. [Google Scholar] [CrossRef]
- He, B.; Xiao, D.; Hu, Q.; Jia, F. Automatic Magnetic Resonance Image Prostate Segmentation Based on Adaptive Feature Learning Probability Boosting Tree Initialization and CNN-ASM Refinement. IEEE Access 2018, 6, 2005–2015. [Google Scholar] [CrossRef]
- Haozhe, J.; Song, Y.; Huang, H.; Cai, W.; Xia, Y. HD-Net: Hybrid Discriminative Network for Prostate Segmentation in MR Images. In Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention –MICCAI 2019, Shenzhen, China, 13–17 October 2019; pp. 110–118. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Kolařík, M.; Burget, R.; Uher, V.; Riha, K.; Dutta, M. Optimized High Resolution 3D Dense-U-Net Network for Brain and Spine Segmentation. Appl. Sci. 2019, 9, 404. [Google Scholar] [CrossRef]
- Dowling, J.A.; Sun, J.; Pichler, P.; Rivest-Hénault, D.; Ghose, S.; Richardson, H.; Wratten, C.; Martin, J.; Arm, J.; Best, L.; et al. Automatic Substitute Computed Tomography Generation and Contouring for Magnetic Resonance Imaging (MRI)-Alone External Beam Radiation Therapy From Standard MRI Sequences. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 1144–1153. [Google Scholar] [CrossRef]
- Litjens, G.; Toth, R.; van de Ven, W.; Hoeks, C.; Kerkstra, S.; van Ginneken, B.; Vincent, G.; Guillard, G.; Birbeck, N.; Zhang, J.; et al. Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge. Med. Image Anal. 2014, 18, 359–373. [Google Scholar] [CrossRef]
- Chandra, S.S.; Dowling, J.A.; Greer, P.B.; Martin, J.; Wratten, C.; Pichler, P.; Fripp, J.; Crozier, S. Fast automated segmentation of multiple objects via spatially weighted shape learning. Phys. Med. Biol. 2016, 61, 8070–8084. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Für Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation from CT Volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Zhang, X.; Yu, G.; Luo, G.; Sun, J. Large Kernel Matters – Improve Semantic Segmentation by Global Convolutional Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1743–1751. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar] [CrossRef]
- Sugawara, Y.; Shiota, S.; Kiya, H. Checkerboard artifacts free convolutional neural networks. APSIPA Trans. Signal Inf. Process. 2019, 8, e9. [Google Scholar] [CrossRef]
- Odena, A.; Dumoulin, V.; Olah, C. Deconvolution and Checkerboard Artifacts. Distill 2016. [Google Scholar] [CrossRef]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M.A. Striving for Simplicity: The All Convolutional Net. arXiv 2014, arXiv:1412.6806. [Google Scholar]
- Bhat, S.S.; Hanumantharaju, M.C.; Gopalakrishna, M.T. An Exploration on Various Nonlinear Filters to Preserve the Edges of a Digital Image in Spatial Domain. In Proceedings of the 2015 International Conference on Advanced Research in Computer Science Engineering & Technology (ICARCSET 2015), ICARCSET ’15, Fukuoka, Japan, 27 July–1 August 2015; ACM: New York, NY, USA, 2015; pp. 51:1–51:7. [Google Scholar] [CrossRef]
- Burger, W.; Burge, M.J. Principles of Digital Image Processing: Fundamental Techniques, 1st ed.; Springer Publishing Company, Incorporated: Berlin/Heidelberg, Germany, 2009; pp. 116–130. [Google Scholar]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object recognition with gradient-based learning. In Shape, Contour and Grouping in Computer Vision; Lecture Notes in Computer Science; Springer: Berlin, Germany, 1999; Volume 1681, pp. 319–345. [Google Scholar] [CrossRef]
- Lau, M.M.; Hann Lim, K. Review of Adaptive Activation Function in Deep Neural Network. In Proceedings of the 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Sarawak, Malaysia, 3–6 December 2018; pp. 686–690. [Google Scholar] [CrossRef]
- Douglas, S.C.; Yu, J. Why RELU Units Sometimes Die: Analysis of Single-Unit Error Backpropagation in Neural Networks. In Proceedings of the 2018 52nd Asilomar Conference on Signals, Systems, and Computers, Pacific Grove, CA, USA, 28–31 October 2018; pp. 864–868. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015; Bach, F., Blei, D., Eds.; PMLR: Lille, France, 2015; Volume 37, pp. 448–456. [Google Scholar]
- Gülçehre, Ç.; Bengio, Y. Knowledge Matters: Importance of Prior Information for Optimization. J. Mach. Learn. Res. 2016, 17, 1–32. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Iglesias, J.E.; Sabuncu, M.R. Multi-atlas segmentation of biomedical images: A survey. Med. Image Anal. 2015, 24, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Yeghiazaryan, V.; Voiculescu, I. An Overview of Current Evaluation Methods Used in Medical Image Segmentation; Technical Report RR-15-08; Department of Computer Science: Oxford, UK, 2015. [Google Scholar]
- Zhi, X. Unet. 2017. Available online: https://github.com/zhixuhao/unet (accessed on 3 July 2019).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. CoRR 2014. abs/1412.6980. [Google Scholar]
- Jain, A.; Fandango, A.; Kapoor, A. TensorFlow Machine Learning Projects: Build 13 Real-World Projects With Advanced Numerical Computations Using the Python Ecosystem; Packt Publishing: Birmingham, UK, 2018; pp. 59–60. [Google Scholar]
- Chollet, F. Keras. 2015. Available online: https://keras.io (accessed on 27 June 2019).
- Chen, W.; Zhang, Y.; He, J.; Qiao, Y.; Chen, Y.; Shi, H.; Tang, X. W-net: Bridged U-net for 2D Medical Image Segmentation. arXiv 2018, arXiv:1807.04459. [Google Scholar]
- Baldeon-Calisto, M.; Lai-Yuen, S.K. AdaResU-Net: Multiobjective adaptive convolutional neural network for medical image segmentation. Neurocomputing 2019. [Google Scholar] [CrossRef]
- Rundo, L.; Militello, C.; Russo, G.; Garufi, A.; Vitabile, S.; Gilardi, M.C.; Mauri, G. Automated Prostate Gland Segmentation Based on an Unsupervised Fuzzy C-Means Clustering Technique Using Multispectral T1w and T2w MR Imaging. Information 2017, 8, 49. [Google Scholar] [CrossRef]
- Lapa, P.; Castelli, M.; Gonçalves, I.; Sala, E.; Rundo, L. A Hybrid End-to-End Approach Integrating Conditional Random Fields into CNNs for Prostate Cancer Detection on MRI. Appl. Sci. 2020, 10, 338. [Google Scholar] [CrossRef]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 1. [Google Scholar] [CrossRef]
- Rundo, L.; Han, C.; Nagano, Y.; Zhang, J.; Hataya, R.; Militello, C.; Tangherloni, A.; Nobile, M.; Ferretti, C.; Besozzi, D.; et al. USE-Net: Incorporating squeeze-and-excitation blocks into U-net for prostate zonal segmentation of multi-institutional MRI datasets. Neurocomputing 2019, 365, 31–43. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).