Featured Application
Molecular barcoding NGS workflow for a high sensitive and accurate mutational profiling of circulating tumor DNA (ctDNA) in non-small cell lung cancer (NSCLC) patients.
Abstract
Targeted next-generation sequencing (NGS) based on molecular tagging technology allowed considerable improvement in the approaches of cell-free DNA (cfDNA) analysis. Previously, we demonstrated the feasibility of the OncomineTM Lung cell-free DNA Assay (OLcfA) NGS panel when applied on plasma samples of post-tyrosine kinase inhibitors (TKIs) non-small cell lung cancer (NSCLC) patients. Here, we explored in detail the coverage metrics and variant calling of the assay and highlighted strengths and challenges by analyzing 92 plasma samples collected from a routine cohort of 76 NSCLC patients. First, performance of OLcfA was assessed using Horizon HD780 reference standards and sensitivity and specificity of 92.5% and 100% reported, respectively. The OLcfA was consequently evaluated in our plasma cohort and NGS technically successful in all 92 sequenced libraries. We demonstrated that initial cfDNA amount correlated positively with library yields (p < 0.0001) and sequencing performance (p < 0.0001). In addition, 0.1% limit of detection could be achieved even when < 10 ng cfDNA was employed. In contrast, the cfDNA amount seems to not affect the EGFR mutational status (p = 0.16). This study demonstrated an optimal performance of the OLcfA on routine plasma samples from NSCLC patients and supports its application in the liquid biopsy practice for cfDNA investigation in precision medicine laboratories.
1. Introduction
The determination of the mutational status of cell-free DNA (cfDNA) is becoming a fundamental and attractive tool in the liquid biopsy field of cancer patients [1,2,3]. CfDNA is also released by normal cells during physiological processes (e.g., apoptosis and necrosis), thus discrimination between scant circulating tumor DNA (ctDNA) derived from tumor cells from normal cells can be challenging [4]. Additionally, since molecular findings by genomic characterization of ctDNA are able to drive treatment decisions, clinical laboratories have been adopting highly sensitive and accurate assays for a precise identification of different DNA alterations.
In this scenario, next-generation sequencing (NGS) approaches provide interesting application choices to deal with rare ctDNA molecules diluted within cfDNA and are rapidly gaining traction as a valid assay for cancer.
Since many tumors, including non-small cell lung cancer (NSCLC), may harbor subclonal mutations with molecular allele frequencies (MAFs) down to 0.1%, it is advisable to use NGS assays sensitive as well as accurate enough to detect such low-abundance alterations.
At present, some technology improvements have allowed researchers to develop NGS panels able to improve the performance levels of the NGS approach in liquid biopsy. Molecular barcoding is one of the possible solutions when dealing with challenging clinical samples since unique molecular tags, also known as unique molecular index (UMI), are added into every DNA molecule, prior to library amplification [5,6,7,8,9,10]. This has the great advantage to increase the sensitivity by keeping track of the DNA molecules intercepted, meanwhile reducing polymerase chain reaction (PCR) and/or sequencing artefacts [11,12,13,14], also when compared to amplicon-based NGS [15].
The OncomineTM Lung cell-free DNA Assay (OLcfA, ThermoFisher Scientific) is an NGS panel based on molecular tagging technical method, that we have already wet-lab tested with the main goal to detect low MAFs of EGFR Thr790Met mutation in plasma samples from tyrosine kinase inhibitors (TKIs)-treated NSCLC patients [16]. We also showed that the NGS results for the search of Thr790Met mutation were comparable with those obtained with a digital PCR (dPCR) approach, achieving a 100% of concordance in terms of sensitivity and specificity [16].
Here, we extended the assessment to a large cohort of 76 NSCLC patients and demonstrated, through a deep NGS data analysis of covered target regions, that the OLcfA performance makes this test suitable and reliable when used in the routine liquid biopsy practice in the NSCLC clinical setting.
2. Materials and Methods
2.1. Patients, Plasma Collection and Panel Sequencing
Ninety-two plasma samples from 76 advanced NSCLC post-TKI patients were collected and tested for EGFR Thr790Met mutation. The present study was approved by the Ethics Committee of Liguria Region (Italy) (P.R.273REG2016) and conducted according to the principle of the Declaration of Helsinky. For each patient included in the study a written informed consent was obtained.
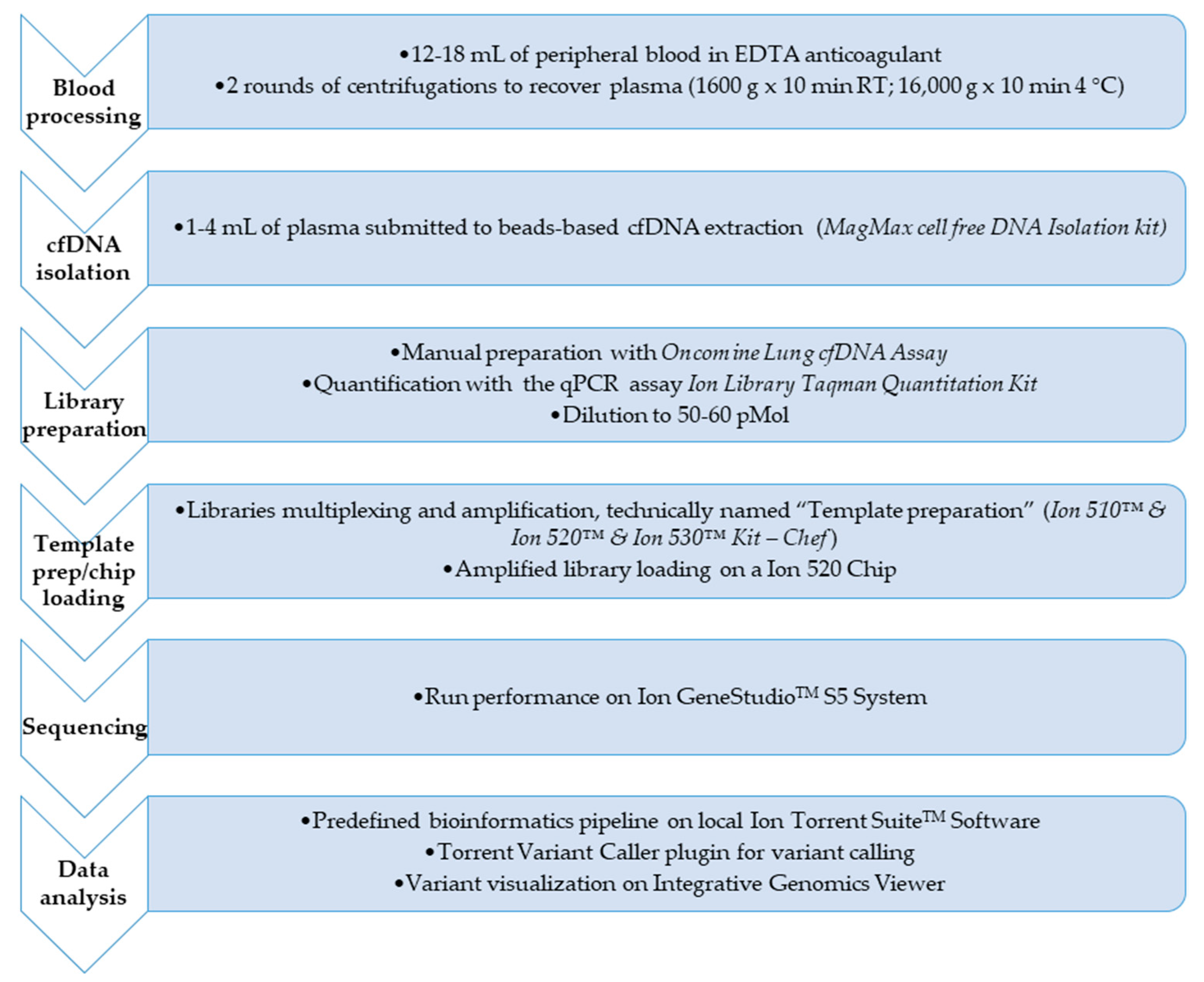
Procedures for molecular tagging-based NGS testing are described in details in reference 16 and here briefly retraced and displayed in Figure 1.
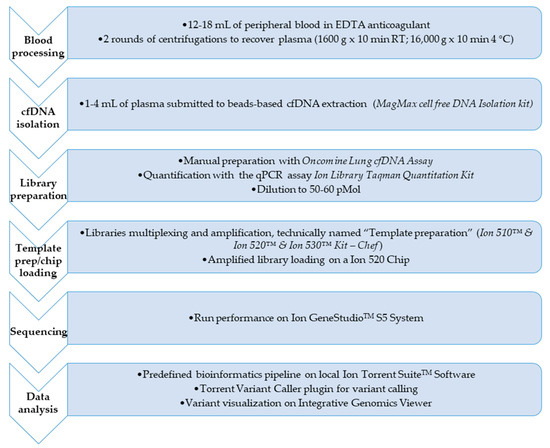
Figure 1.
Flowchart of the complete workflow for circulating tumor DNA (ctDNA) next-generation sequencing (NGS) analysis. The figure shows the main steps for the mutation profile of the ctDNA in the 76 non-small cell lung cancer (NSCLC) patients of the experimental design from blood sample processing to the application of the bioinformatics pipeline to call the variant/s eventually found.
Plasma samples were isolated from 12–18 mL of peripheral blood collected into EDTA-containing tubes and up to 4 mL processed for cfDNA extraction using the MagMAX™ Cell-Free DNA Isolation Kit (ThermoFisher Scientific, Waltham, MA, USA), followed by quantification with a Qubit 3TM Fluorometer (ThermoFisher Scientific).
Genomic profiling of samples by targeted NGS was performed by using the above cited OLcfA, covering DNA target regions containing hotspots variants relevant in pathogenetic and resistance molecular mechanisms in NSCLC (169 key hotspot mutations across 11 genes such as ALK, BRAF, EGFR, ERBB2, KRAS, MAP2K1, MET, NRAS, PIK3CA, ROS1 and TP53).
This NGS assay applies UMI to improve the sensitivity by decreasing the amount of sequencing artifacts. With the recommended input of 20 ng cfDNA, the use of UMI enables a limit of detection (LoD) as low as 0.1%. However, a range between 1–50 ng of cfDNA amount can be used with consequent higher LoD reached as indicated by the manufacturer (http://tools.thermofisher.com/content/sfs/brochures/ngs-analysis-mutations-cfdna-app-note.pdf).
Oncomine library quantities were determined by qPCR using the Ion Library TaqMan Quantitation Kit (ThermoFisher Scientific) and then diluted to a final concentration of 50–60 pmol/L. Next, sample barcoded libraries were pooled together for template preparation on the Ion ChefTM Instrument and loaded onto a Ion 520 chip (Ion 520TM Chip Kit, ThermoFisher Scientific).
The chip was sequenced on a Ion GeneStudio TM S5 System (ThermoFisher Scientific), and the unaligned BAM (Binary Alignment Map) files generated by the sequencer were mapped against the human reference genome (GRCh37/hg19) using the Torrent Mapping Alignment Program (TMAP), with default parameters and Ion Torrent Suite™ Software (TSS, version 5.10).
Analysis and annotation of variants were locally carried out with Torrent Variant Caller (TVC, version 5.10) plugin using preconfigured parameter settings for liquid biopsy application.
NGS data of library performance and variant calling quality were considered and presented for all plasma samples analyzed. The manufacturer recommends a Median Read Coverage (MedReadCov) >25,000 and Median Molecular Coverage (MedMolCov) >2500 to detect a variant with a MAF of 0.1%.
The output variant caller format (VCF) files from each sequenced sample were reviewed for further visual analysis on the Integrative Genomics Viewer (IGV, version 2.6, Broad Institute). VCF files were uploaded on IGV and the wild type/mutated molecular alleles aligned to the reference genome hg19 (GRCh37).
Reproducibility of the Oncomine Lung cfDNA panel was already evaluated in our previous paper [16]. Specifically, a high sensitivity tool such as the dPCR was tested on 26 cfDNA samples: 10 T790M-negative and 16 T790M-positive, with a 100% of concordance in both samples demonstrated.
2.2. Statistical Analysis
Statistical evaluation of the data in this study was performed using GraphPad Prism version 6 software. Threshold for statistical significance was considered to be p < 0.05.
3. Results
We initially evaluated the OLcfA performance using the Multiplex I cfDNA Reference Standard set–Horizon Discovery 780 (HD780). This commercial kit contains low allele frequencies variants (5%, 1% and 0.1%) at 8 hotspot genomic positions in genes EGFR, KRAS, NRAS and PIK3CA, covered by the Oncomine panel. Notably, 4 key alterations were investigated for EGFR gene (Leu858Arg, Thr790Met, Glu746_Ala750del, Val769_Asp770insAlaSerVal), 1 for KRAS (Gly12Asp), 2 for NRAS (Ala59Thr, Gln61Lys) and 1 for PIK3CA (Glu545Lys) and results shown in Table 1.

Table 1.
Frequency variants detected in Multiplex cfDNA Reference Standard for each of the 8 hotspot variants occurring in 4 genes (EGFR, KRAS, NRAS and PIK3CA). In bold, variants filtered out by the bioinformatic pipeline in EGFR gene and a failed call in PIK3CA gene.
Each reference standard was tested by using 30 ng of cfDNA, as described by the manufacturer (ThermoFisher Scientific, https://assets.thermofisher.com/TFS-Assets/LSG/brochures/verification-oncomine-lung-cfdna-ion-s5-white-paper.pdf). Additionally, we checked reproducibility in critical samples (i.e., those with cfDNA input <30 ng) and tested the HD780 reference standard at 0.1% MAF by using 20 ng of cfDNA in a duplicate experiment. Detailed data on MAFs are reported in Table 1. The overall variant-based sensitivity was 92.5%. Specifically, only in one case did the system fail to call a low frequency variant at 0.1% and it was relative to the hotspot Glu545Lys in PIK3CA gene. In addition, two hotspots, i.e., the deletion Glu746_Ala750del and the missense mutation Leu858Arg were filtered out by the bioinformatics pipeline in the 20 ng cfDNA replicates at 0.1% MAF, because not enough molecular families had been reached to enable a call.
Furthermore, reference cfDNA HD780 at 0% MAF, i.e., the standard characterized by the absence of low allele frequencies variants, was examined and no false positives were found at the genomic target regions analyzed, not even visualized on IGV. This indicates a 100% specificity of OLcfA and confirms its reliability to be used in a clinical setting.
3.1. Cell-Free DNA (CfDNA) Quantity and Library Yields in Clinical Specimens
Overall, 76 NSCLC patients and a total of 92 plasma samples were included in our NGS analysis. CfDNA was isolated from 1–4 mL of plasma and a range of 2.6–309 ng /mL of plasma was obtained (median 10.40 ng/mL).
It was possible to proceed with NGS library preparation in all 92 samples and the range of cfDNA used for the NGS reaction was 6.37–59.80 ng, being aware that 23/92 (25%) samples were below the suggested 20 ng cut-off needed to reach the 0.1% LoD.
In addition, when qualitative assessment of cfDNA was performed on a TapeStation 2200 (Agilent Technologies, Santa Clara, USA), it turned out that DNA fragments of about 150–180 bp and multiples were predominant compared to genomic DNA (high molecular weight), thus demonstrating the reliability of the magnetic extraction method in isolating the fragmented cfDNA fraction with a higher chance to be tumor-derived (data not shown).
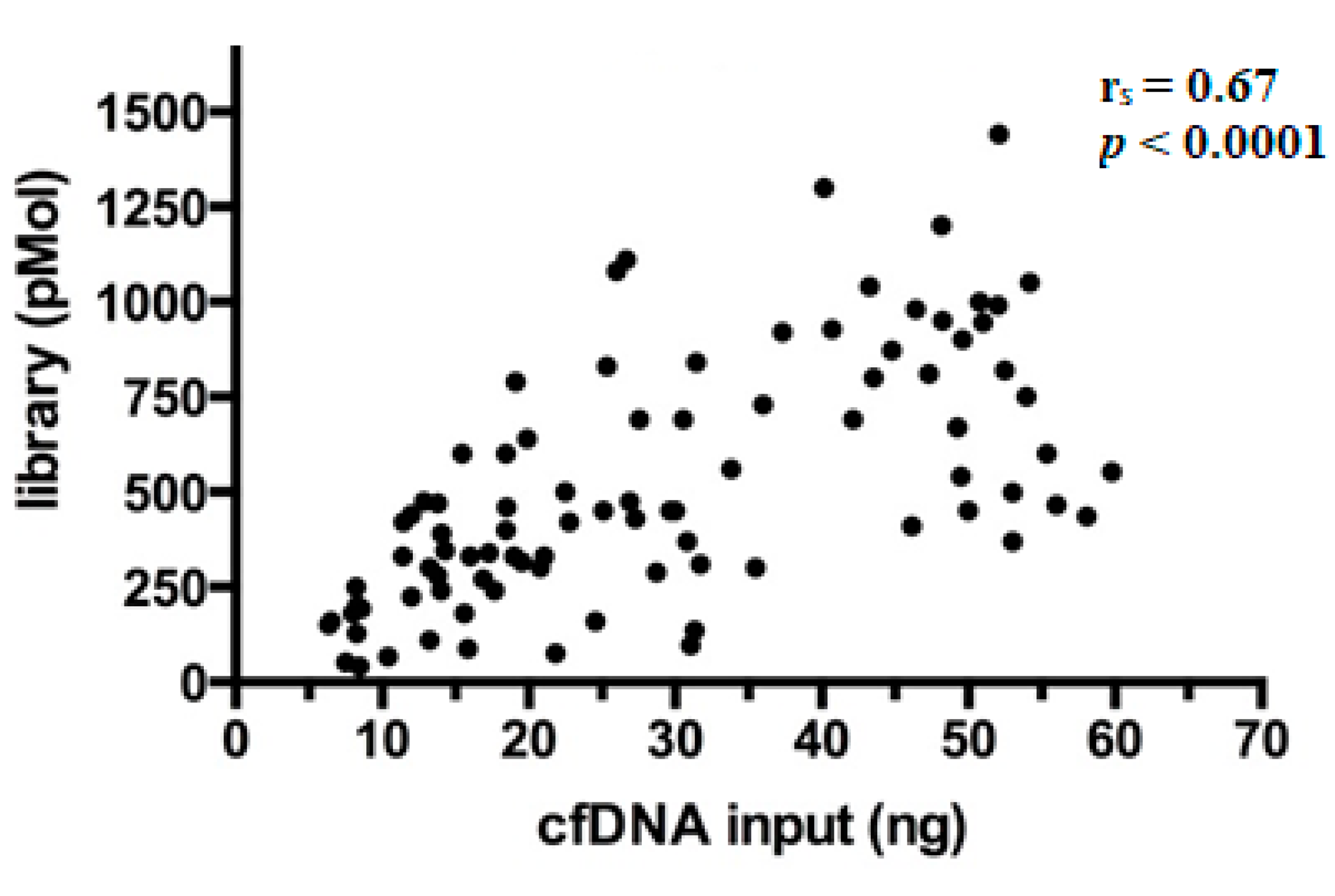
After preparation, libraries were quantified and showed a wide range of concentrations between 40.5–1440 pMol (median 450 pMol). As expected, it was observed that library yields increased proportionally with the amount of cfDNA ng employed per reaction (Spearman’s correlation coefficient (rs) = 0.67, p < 0.0001) (Figure 2), a finding in line with literature [17].
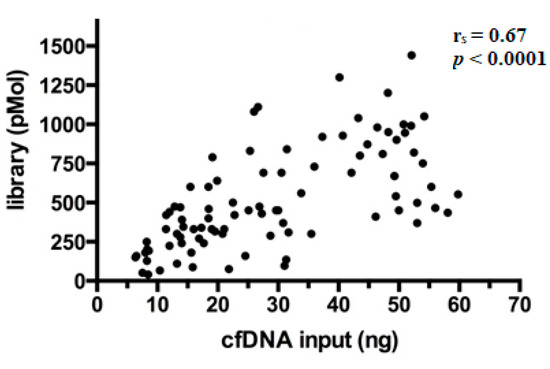
Figure 2.
Correlation between library yields and cfDNA input. Each dot in the scatterplot represents the single library prepared (n = 92) and the relative concentrations are expressed in pMol.
3.2. Overall Considerations of the OncomineTM Lung Cell-Free DNA Assay (OLcfA) Sequencing Performance Applied to a Real-Life Routine Plasma Cohort
3.2.1. Read Coverage through Panel Amplicons
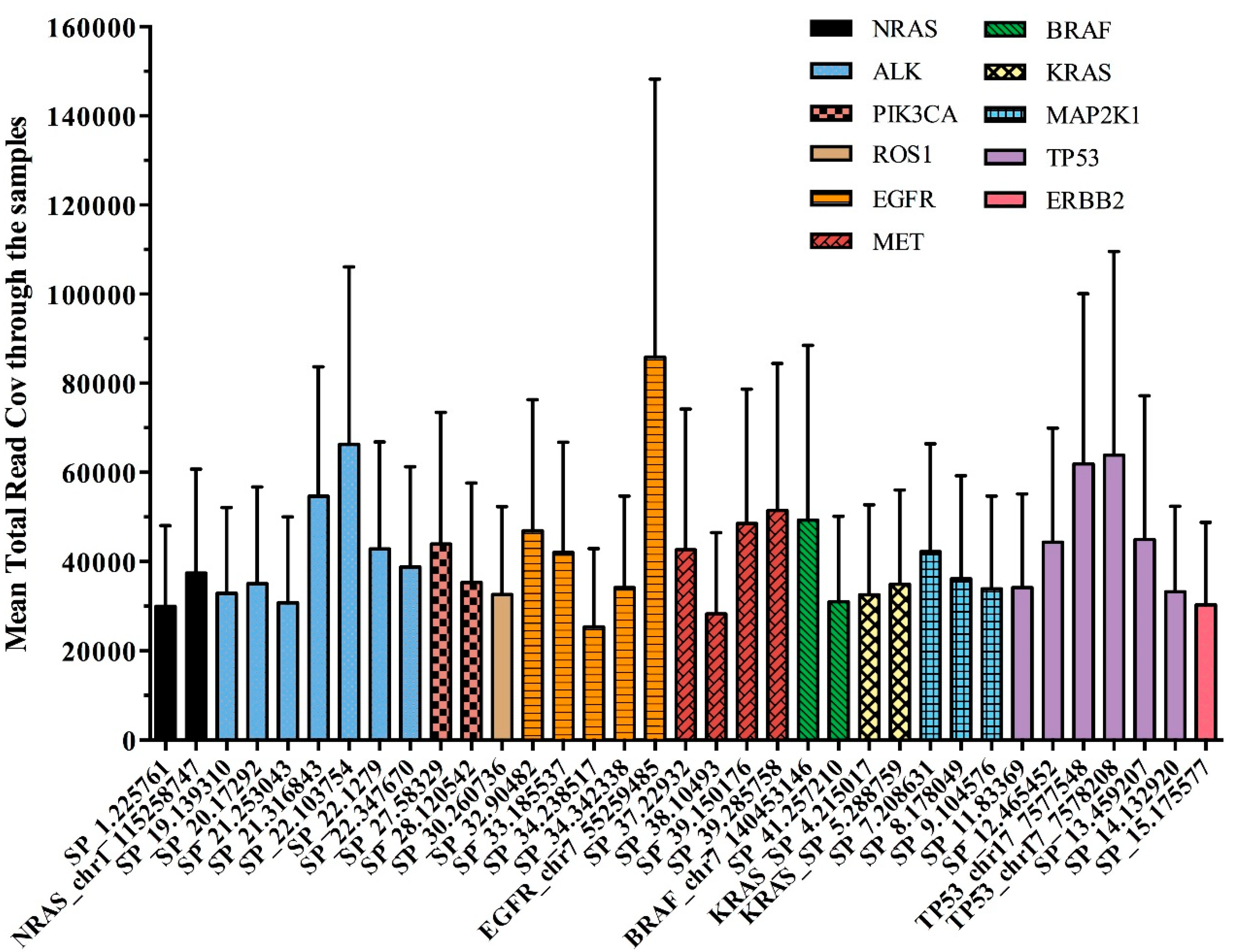
The overall panel performance was assessed by average amplicon coverage across all the 92 samples studied. All 35 amplicons were covered on average to a minimum of 25,352 reads and a maximum of 85,866 reads.
Figure 3 reported the performance of each single DNA region observed in our cohort of plasma samples, distributed across the 11 genes of the OLcfA. A high rate of variability in amplicon coverage was observed and it seems not to be related to cfDNA amounts used for libraries preparation (data not shown) but rather to the intrinsic technical characteristics of the panel. Indeed, the assay is based on a single tube multiplex PCR and primer pairs work under the same thermal conditions, and some differences in performance along the amplicons are expected.
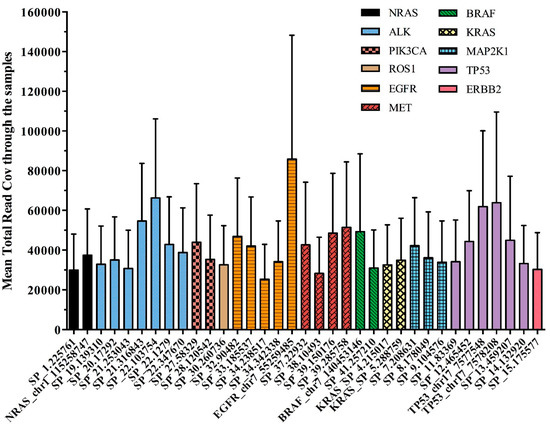
Figure 3.
Assessment of the 35 amplicons of the OLcfA among the 92 samples tested. Mean total read coverage was investigated across all clinical samples tested (n = 92) and reported on y axis.
The variability was also identified at an intragenic level, as reported in Figure 3. For example, the EGFR gene, represented by 5 amplicons covering exons 18–21 (orange histograms), contained both the best and the worst performing amplicons of the panel (i.e., 85,866 and 25,352 reads, respectively). Notably, the first corresponds to a target region covering hotspot positions in exon 21, instead the second one partly covers exon 20 of the EGFR gene.
3.2.2. Molecular and Read Coverage of Sequenced Libraries
In all sequenced samples it was possible to proceed with NGS data analysis to validate both library quality and variant call.
Library performance was in particular evaluated by two TSS parameters such as MedReadCov and MedMolCov, i.e., the median number of reads across target regions and original DNA molecules identified (tagged), respectively.
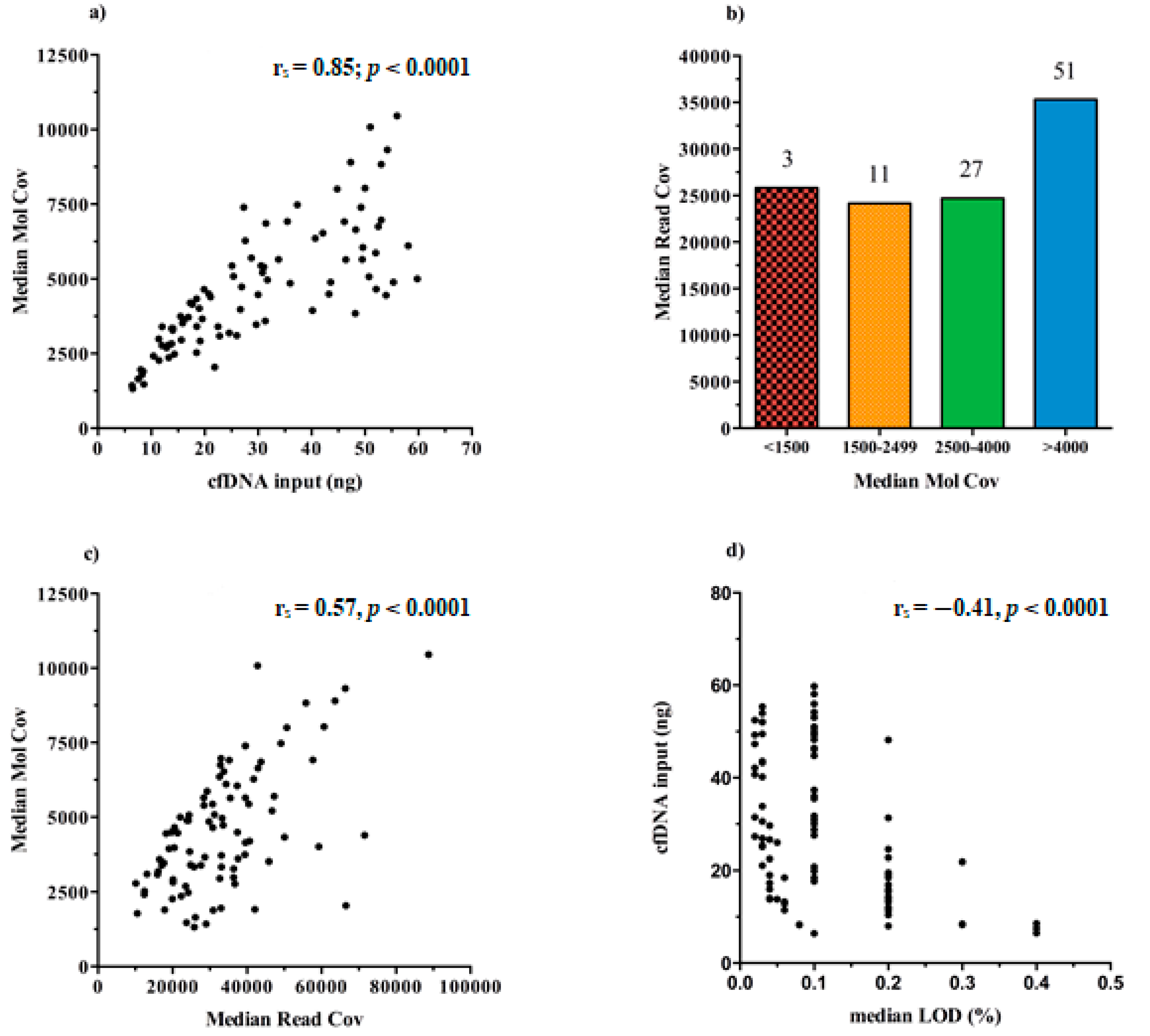
We found that the increasing cfDNA input (ng) used for test leads to higher rates of MedMolCov (rs = 0.85; p < 0.0001) and consequently to a more confident number of DNA molecules investigated for mutational search (Figure 4a).
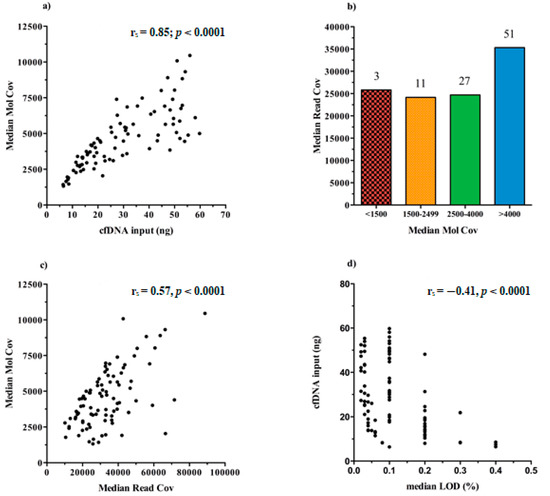
Figure 4.
Graphical representation of the Ion Torrent Suite™ Software (TSS) sequencing data about libraries performance in all samples (n = 92). (a) Scatter plot of the positive correlation between the cfDNA input and the median number of DNA molecules tagged (MedMolCov) (rs = 0.85; p < 0.0001). (b) Bar chart of the MedMolCov values divided into four ranges (<1500, 1500–2499, 2500–4000, >4000) and the relative MedReadCov values. (c) Scatter plot of the positive correlation between the MedReadCov and the Median Mol Cov (rs = 0.57, p < 0.0001). (d) Scatter plot of the negative correlation between the median LoD values of each library and the initial cfDNA input (rs = −0.41, p < 0.0001).
Taking into account all the 92 sequenced libraries, it emerged that 51/92 achieved a MedMolCov >4000, with a median value of MedReadCov across the samples of 37,405 (range 18,252–88,731), whereas 27/92 samples obtained a MedMolCov between 2500–4000 with a median value of MedReadCov of 24,801 (range 10,116–45,861). Of the remaining 14 samples, 11 showed a MedMolCov between 1500–2500 and only 3 samples <1500, with a median MedReadCov of 24,170 and 25,826, respectively (Figure 4b).
Since the assay was designed to reach as optimal yield a MedReadCov >25,000 and a MedMolCov >2500 in order to obtain reliable sequencing results and reach a LoD of 0.1%, we showed that in our setting, a complete fulfillment of both parameters was found in 56/92 samples (60.9%). Separately, 81/92 (88%) libraries reached outperforming values of MedMolCov >2500 whereas 63/92 (68.5%) achieved values of MedReadCov >25,000.
An overall moderate positive correlation was revealed between the values of MedMolCov and MedReadCov (rs = 0.57, p < 0.0001) (Figure 4c), thus confirming an optimal NGS workflow performance involving both library generation and amplification steps.
In regard to LoD values of the assay, i.e., the lowest variant allelic frequency that could be reliably detected, in our 92 plasma samples we obtained a median LoD of 0.10%, (range 0.02–0.40%), and as expected, these data negatively correlated with cfDNA input amount (rs = −0.41, p < 0.0001) (Figure 4d). Notably, it was observed that 37/92 samples (40.22%) reached at least 0.1% LoD and among these, 16 (43.24%) initially presented <20 ng of cfDNA input for NGS library preparation.
3.3. cfDNA Input Does Not Determine the EGFR Mutational Status
The relationship between detection of EGFR mutations and cfDNA concentrations, a potential indicator of tumor shedding, was also examined.
To this end, the variant calling analysis was performed by using the cfDNA variant caller plugin with parameters optimized for the “Oncology–Liquid Biopsy” application and in particular, variants were detected when it is found in at least 2 molecular families and when a minimum number of 3 reads with the same tag forms a functional family.
It was observed that 61/92 (66.3%) samples were mutated in hotspot genomic positions within EGFR gene (Table S1). In depth, 30/61 carried only one mutation, 30/61 two mutations, whereas only one patient (pt.43) had three mutations (Gly719Cys, Set768Ile and Thr790Met). In all the patients whose plasma resulted in being informative, i.e., positive for the sensitizing EGFR mutations, the variants matched between primary tissue and the corresponding cfDNA (data not shown; see [16]).
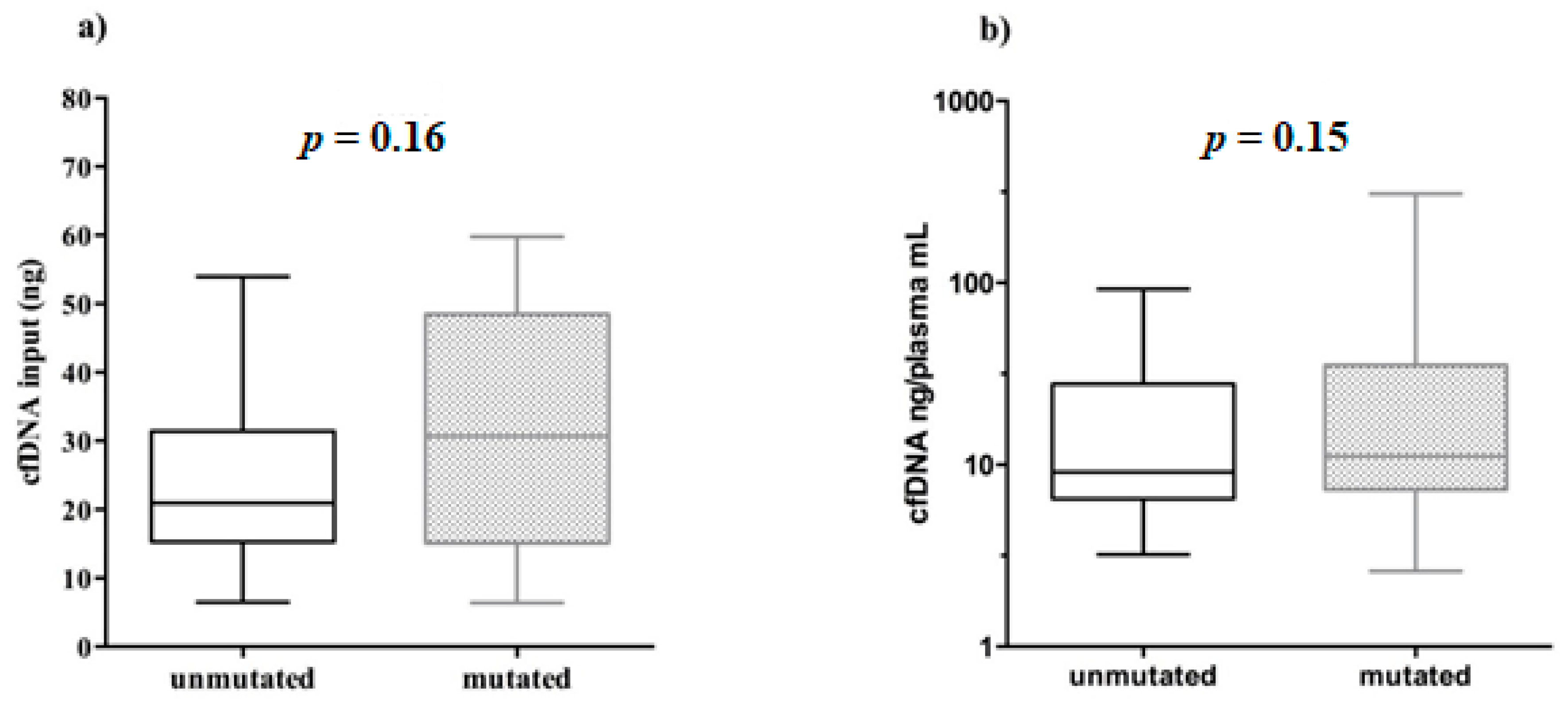
The cfDNA quantity, used for NGS libraries in mutated (6.37–59.80 ng) and unmutated (6.50–53.95 ng) samples was very similar. In addition, no significant correlation was found between the cfDNA input amount used for library preparation and the relative EGFR mutational status (Mann–Whitney test, p = 0.16, Figure 5a).
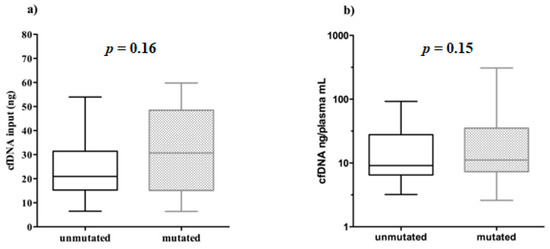
Figure 5.
Boxplots representing the EGFR mutational status behavior related to cfDNA input and abundance. Unmutated/mutated plasma samples were correlated with: (a) the cfDNA amount used for library preparation; (b) cfDNA abundance in plasma (cfDNA ng/plasma mL).
Furthermore, we showed that even the amount of cfDNA ng/mL plasma did not affect the positivity/negativity of EGFR mutational testing (Mann–Whitney test, p = 0.15, Figure 5b).
Together, these data demonstrate that although we need considerable yields of cfDNA to get more tagged DNA molecules and to intercept even single rare ctDNA molecules diluted within, the mutational detection rely possibly to different aspects mainly associated with biological variability intra- and inter-patients.
3.4. Thr790Met Detection and Torrent Variant Caller (TVC) Metrics Evaluation
The EGFR Thr790Met variant was detected in 27/61 (44.3%) driver EGFR mutated plasma samples and co-occurred mostly with exon 19 deletions in 17/27 (63%) and Leu858Arg in 8/27 (29.6%) cases (Table S1).
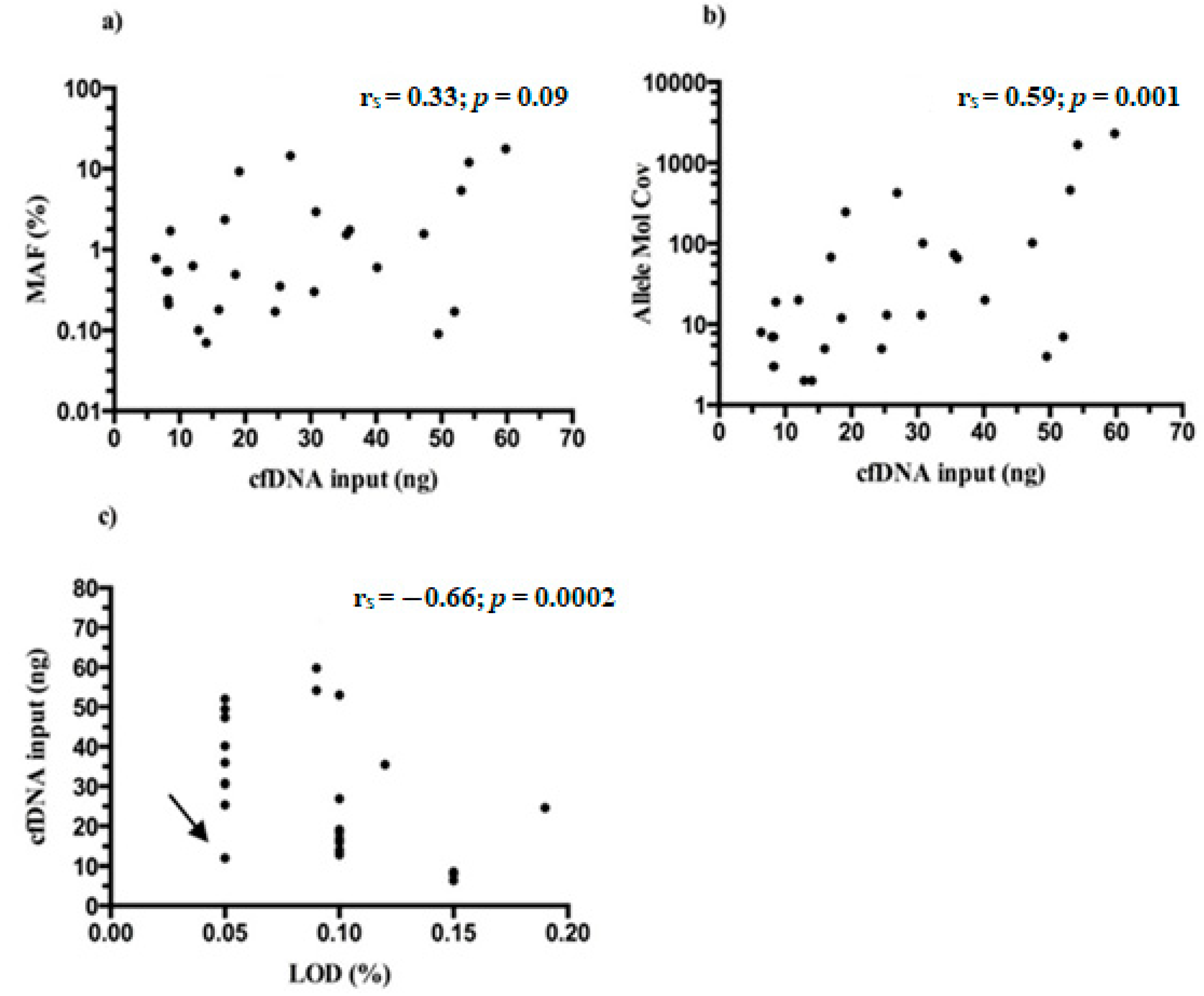
MAFs of Thr790Met detected in mutated samples ranged between 0.06–17.67% (median 0.60%) and almost always were lower than those detected for driver mutation/s in the same patients (Table S1). No statistical significance was found between the starting NGS input of cfDNA and MAFs found for Thr790Met (rs = 0.33; p = 0.09) (Figure 6a), whereas a moderate correlation was reported when cfDNA amounts were related with allele molecular coverage of tagged DNA molecules containing the variant (rs = 0.59; p = 0.001) (Figure 6b).
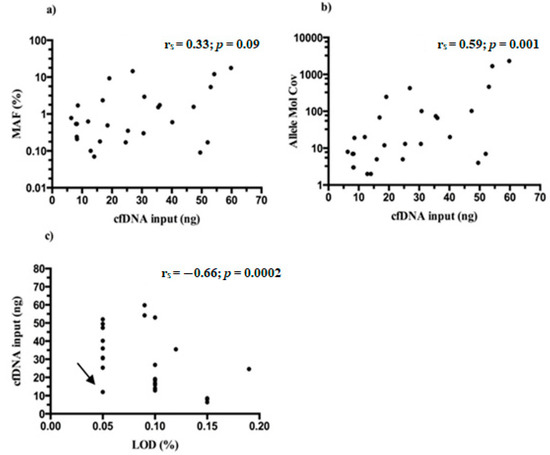
Figure 6.
CfDNA input and TVC parameters from Thr790Met positive samples. (a) No significant correlation between cfDNA and MAFs was reported (rs = 0.33; p = 0.09). Moderate correlations were found between the cfDNA input and: (b) the number of mutated DNA molecules (Allele Mol Cov) (rs = 0.59; p = 0.001); (c) the LoD of Thr790Met variant (rs = −0.66; p = 0.0002). The arrow indicates a peculiar case of a 0.05% LoD reached with only 12 ng of cfDNA input.
Finally, the LoDs reached by Thr790Met variant through the 27 samples (range 0.05–0.15%, median 0.10%) were correlated with the initial cfDNA amount and, as expected, a moderate negative association was found (rs = −0.66; p = 0.0002) (Figure 6c). It is intriguing that in general, the Oncomine NGS panel outperformed, reaching lower LoDs than expected.
3.5. Manual Review of Variant Caller Format (VCF) on Integrative Genomics Viewer (IGV) Evidenced Accuracy and TVC Plugin Gaps
In order to determine if variants found, including single nucleotide variants (SNVs), small insertions and deletions (InDels), were real events or artifacts as well as to exclude the presence of false negatives or misclassification of variant/s, VCF files were manually reviewed by using IGV software.
In our experience, this visual inspection increased the confidence in SNV calls, and in all small canonical InDels in exons 19 and 20 of EGFR gene reported in TSS by TVC. Again, importantly, no variant misclassification was pointed out.
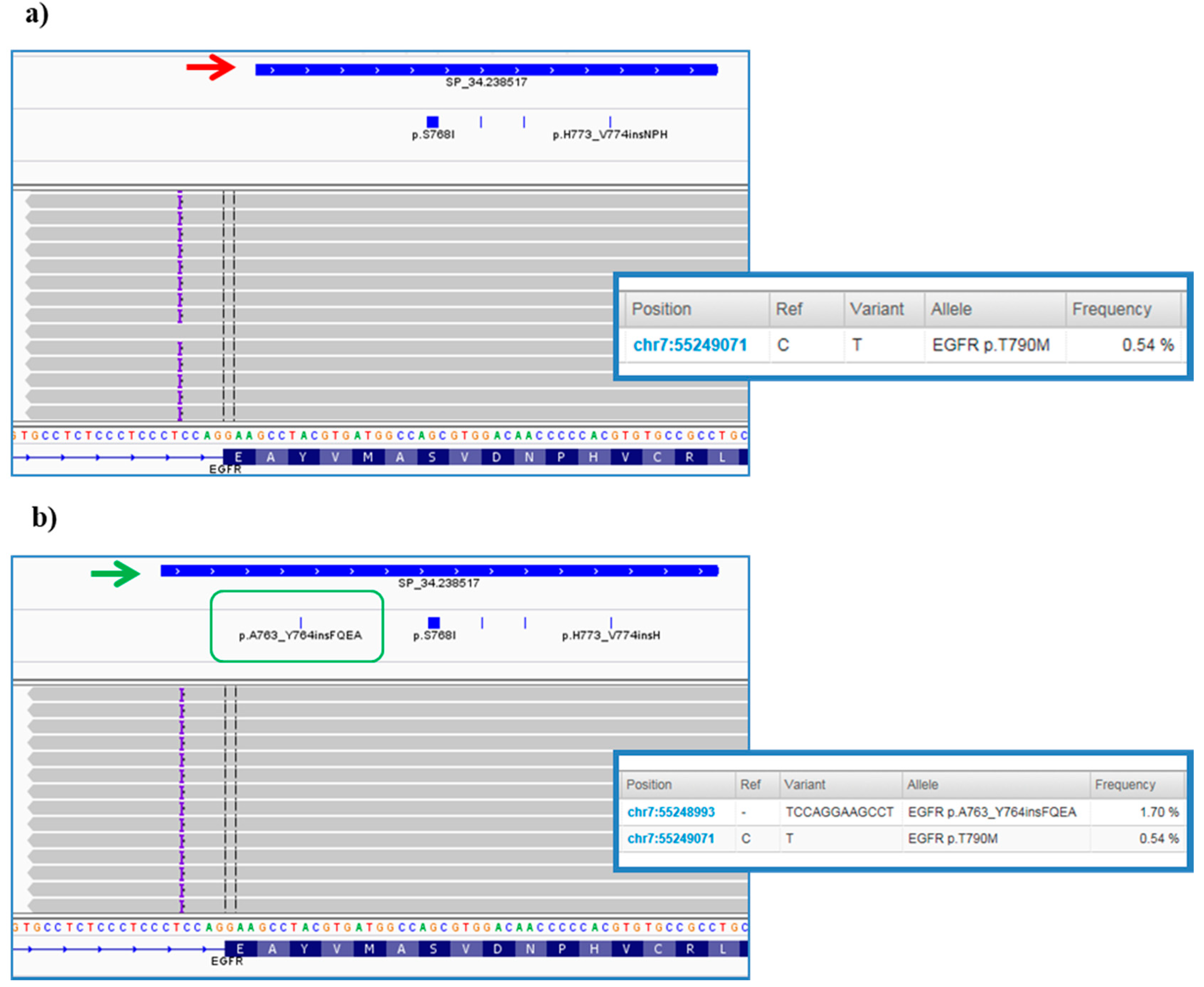
However, further investigation of VCFs allowed us to highlight some critical points of the bioinformatics pipeline predefined by ThermoFisher. In particular, it was noted that the TVC plugin failed to call the EGFR complex mutations Lys745_Glu746insIleProValAlaIleLys (exon 19) and Ala763_Tyr764insPheGlnGluAla (exon 20) (Figure 7a), occurring in pt.25 and pt.35, respectively, and previously validated on tissue samples by Sanger sequencing (data not shown). In both cases, the variant falls outside the DNA target regions covered by the Browser Extensible Data (BED) file of the panel, but, only in the second case we were able to obtain the correct variant call, redoing the analysis after an adjustment in analysis configuration of TVC parameters JSON file (version 5.2), specifically within the TMAP string (Figure 7b). This update was made available to all users by manufacturer in the following versions of the TVC plugin.
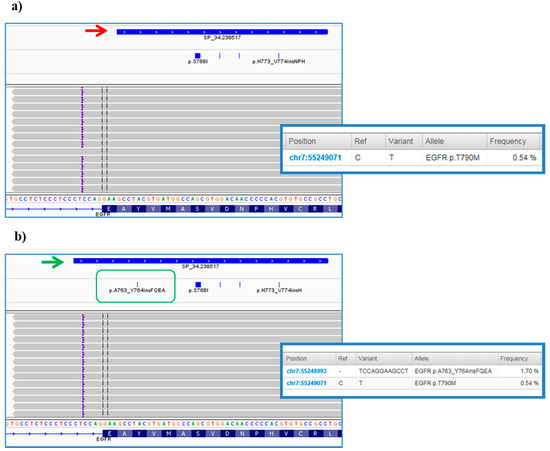
Figure 7.
Integrative Genomics Viewer (IGV) screenshots of Ala763_Tyr764insPheGlnGluAla variant in EGFR gene. (a) The figure shows the complex InDel in exon 20 of the EGFR gene found at first by visualization with the IGV software. This is a rare insertion of 12 bp (purple braces) mapped outside the of the exon 20 not initially covered by the designed amplicon and consequently not called by the Torrent Variant Caller (TVC, red arrow). (b) The panel below depicts the results of the analysis after some bioinformatics changes to TVC parameters. As it can be noted, the blue bar of the amplicon was elongated to also cover the genomic position at the end of the intron where the mutation occurs (green arrow). Consequently, the hotspot Browser Extensible Data (BED) file was modified (green box) and the specific insertion was correctly reported in the TVC section of the TSS (screens on the right).
Another case of false negative occurred in pt.60, who harbored a complex mutation in EGFR exon 19, i.e., Arg748_Ala755delinsThr. Even in this particular case, the variant was discovered in tissue by Sanger sequencing but found in the plasma only through the IGV visualization.
However, despite fitting the amplicon, the variant was not called and thus a different modification of the TVC parameters, was required. In detail, the parameter "allow_complex" was changed from “0” to “1” and then the variant was found listed into Allele Source “Novel” of TVC plugin section.
4. Discussion
Liquid biopsy has emerged as a promising solution to overcome current limitations associated with tissue biopsies in precision medicine laboratories [18].
CfDNA represents the most stable genetic material and accessible source in the liquid biopsy field and its assessment allows the detection and monitoring of cancer-specific genomic alterations in blood [19]. Even if ctDNA analysis is not a replacement for histologic confirmation, it results in being clinically useful in the diagnostic approach. This molecular testing, due to its high specificity, non-invasive nature and fast turnaround time, can represent a valid tool at diagnosis or progression, even preceding tissue genotyping and can be used to guide treatment [20,21].
Nevertheless, the analysis of ctDNA is challenging due to the mainly low representation of mutated tumor-derived DNA molecules in the plasma. Furthermore, although plasma of cancer patients contains more cfDNA compared to healthy individuals [17,22], the proportion of ctDNA originating from the tumor is highly variable, ranging from <0.1% to >50% of total cfDNA [23,24]. Therefore, methods which are able to detect small number of mutated molecules in an abundance of unmutated DNA fragments with high sensitivity and specificity are required.
NGS methods have been revealing a promising approach to detect more sensitive and specific biomarkers for clinical utility. Considering NSCLC management, targeted therapies, like TKIs, require a sensitive and precise mutation profiling of the tumor to ensure identification of every patient who could potentially benefit from therapy. In particular, acquired resistance mutations of the EGFR gene, for example Thr790Met occurring in up to 60% of NSCLC patients, represent a clinical challenge in the treatment [25,26,27,28]. In this context the analysis of ctDNA has become a hopeful tool to verify the Thr790Met presence in NSCLC patients, which developed a resistance to TKI therapy [29,30]. Therefore, the most relevant requirements for an NGS panel to be used for liquid biopsy purposes are high levels of sensitivity and specificity to ensure true variant calling at very low frequencies. Technically, to overcome the drawbacks of PCR-based NGS (e.g., DNA polymerase errors, etc.), the addition of UMI, i.e., random nucleotide sequences barcoding each DNA molecule prior to PCR amplification, was introduced [5,6,7,8,9,10]. This artifice allows us to distinguish reads amplified from the same original DNA molecule and to identify molecules containing true variants from false positives.
In this study, we tested the feasibility of the OLcfA NGS panel UMI-based on a cohort of NSCLC patients (n = 76) and, mostly, we focused on pre- and analytical details from 92 plasma samples. We validated the assay on both wet bench and bioinformatics processes across a broad spectrum of validation parameters including sequencing performance, analytical sensitivity and specificity and LoD reached. The approach was firstly validated with commercially available cfDNA reference material providing EGFR hotspot variants at MAFs down to 0.1% and confident levels of sensitivity and specificity achieved (92.5% and 100%, respectively).
From an overall view of the sequencing performance of the panel, it emerged that not all amplicons performed similarly. This was likely due to a different PCR amplification efficiency, expected when dealing with multiple genes/target regions panels where DNA library generation occurs in single-tube reactions. According to literature, it is verifiable in both small (as in our case) as well as large NGS panel [31].
Regarding the pre-analytical parameters, in our cohort we found a considerable rate of heterogeneity in levels of cfDNA abundance across the plasma samples (2.6–309 ng/mL of plasma), indicating that it is not possible to predict how much cfDNA will be achieved, neither at an inter- or intra-patient level. However, this issue is well known, and in fact that the shedding of cfDNA into the bloodstream is affected by several individual characteristics, such as tumor stage, metastasis, inflammation, treatment, and comorbidities [1,3,18,32].
LoD evaluation through the samples revealed, not surprisingly, that higher quantities of cfDNA input allowed to reach very low levels of LoD (up to 0.02%), as also similarly reported by others [17], with the advantage to preserve specificity of the NGS workflow. It is interesting to note that at least 0.1% LoD was reached in 37/92 cases (40.22%). In order to achieve such LoD value, the manufacturer recommends to employ at least 20 ng of cfDNA input for NGS library preparation. In our data, we demonstrated that even when <20 ng of cfDNA were available, it was still possible a 0.1% LoD for almost the half (16/37, 43.20%) of these samples. This further suggests that predetermined cfDNA input for LoD can, therefore, only be a vague reference of what would be expected.
Bearing in mind what is reported by manufacturer to reach the main goal of a 0.1% LoD, we showed a fulfillment of the median value of molecular families (MedMolCov >2500) and the median values of read coverage (MedReadCov >25,000) in more than half of sequenced samples (60.9%). Not surprisingly, a strong correlation between the cfDNA input and MedMolCov was found, since it is expected that the more cfDNA that is introduced for NGS libraries, the higher will be the number of molecular families sequenced, as already reported [17]. By contrast, no significant association was found between DNA quantity (both cfDNA input and cfDNA abundance) and the EGFR mutational status (p > 0.15). However, this finding is not surprising, indeed it is known as the ctDNA portion can widely vary of total cfDNA, also in NSCLC [33,34]. Anyways, our data are encouraging since we showed that it is possible to carry out mutations even in plasma samples with low amounts of cfDNA, with the huge advantage of a reduced number of “non-informative” blood draws in routine clinical practice.
These data together demonstrate, first of all, the high translational strength of the assay that is shown to be reliable and sensitive. Secondly, the automatic bioinformatics pipeline allows us to directly have the output files (BAM and VCF) for each sequenced sample and execute a graphical and statistical overall evaluation through predefined parameters available in TSS.
Nevertheless, despite the previous promising results, this cannot still be considered an optimal analysis workflow in all instances; indeed, if on the one hand molecular tagging lowered drastically the rate of false positives, on the other false negatives can still occur. In particular, in our cohort, we reported the variant calling failure of three complex mutations. It is important to note that in these cases the mutations were filtered out by the TVC plugin despite reaching the values of MAF and coverage to enable a call. These were in particular two insertions on exon 19 and 20 and a deletion on exon 19 of EGFR, only displayable through the inspection of the genomic positions of interest on IGV. For two out of three InDels (exon 20 insertion and exon 19 deletion) an adjustment of current default setting, consisting in a lower stringency of specific TVC parameter JSON file, enabled the subsequent successful variant detection. In these cases, the variant interception was facilitated by retrospective molecular analysis on tissues. However, it is not always possible to know a priori the EGFR driver mutation and so it is necessary to obtain help from software visualization (e.g., IGV) and/or a validated external bioinformatics pipeline for variant calling.
5. Conclusions
By now, ctDNA testing for the determination of the driver and resistance mutations have entered the daily practice of clinical laboratories. Here we tried to focus on the main advantages as well as potential drawbacks of the OLcfA application on a routine NSCLC clinical setting. We also highlighted the fundamental bioinformatics elements to conduct an analysis consistent with the parameters predefined by the TSS local analysis software. From analytical evaluation of the OLcfA, we were able to demonstrate an accurate detection of low-frequency variants in cfDNA from plasma samples of post-TKI NSCLC patients. The assay performed well also when sub-optimal amounts of cfDNA input were available for NGS library preparation, without affecting the variant detection rate. The assay also revealed to be effective for patients with a known EGFR primary mutation receiving TKI therapy to detect upcoming EGFR Thr790Met mutation, even present below 0.1% MAF in the blood, with no lowering of the specificity. These data, together with those presented in our previous study, support clinical implementation of the OLcfA panel in medicine precision laboratories for NGS liquid biopsy of advanced NSCLC patients.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/8/2895/s1. Table S1: EGFR mutations found in all 61 mutated plasma samples.
Author Contributions
Conceptualization, G.D.L. and M.D.; methodology, G.D.L., S.L., R.C. and M.D.; validation, G.D.L. and M.D.; formal analysis, G.D.L.; investigation, G.D.L. and M.D.; resources, M.M., C.G., G.R., M.T. and M.G.D.B.; data curation, G.D.L.; writing—original draft preparation, G.D.L.; writing—review and editing, M.D. and S.C.; visualization, G.D.L.; supervision, S.Z.; project administration, S.Z. and M.D.; funding acquisition, S.C. and S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Italian Ministry of Health (GR2011-12; 02350922) and partially by the Compagnia San Paolo (2017-0529) to S. Coco, by the Italian Ministry of Health 5 × 1000 funds 2013, 2014 and 2015 to S. Zupo.
Acknowledgments
Andrea Luchetti and Alessandra Gasparini for bioinformatics support (ThermoFisher Scientific).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547. [Google Scholar] [CrossRef] [PubMed]
- Stahlberg, A.; Krzyzanowski, P.M.; Jackson, J.B.; Egyud, M.; Stein, L.; Godfrey, T.E. Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res. 2016, 44, e105. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.W.; Kennedy, S.R.; Salk, J.J.; Fox, E.J.; Hiatt, J.B.; Loeb, L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 14508–14513. [Google Scholar] [CrossRef]
- Narayan, A.; Carriero, N.J.; Gettinger, S.N.; Kluytenaar, J.; Kozak, K.R.; Yock, T.I.; Muscato, N.E.; Ugarelli, P.; Decker, R.H.; Patel, A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012, 72, 3492. [Google Scholar] [CrossRef]
- Kivioja, T.; Vaharautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2011, 9, 72–74. [Google Scholar] [CrossRef]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530. [Google Scholar] [CrossRef]
- Volckmar, A.L.; Sultmann, H.; Riediger, A.; Fioretos, T.; Schirmacher, P.; Endris, V.; Stenzinger, A.; Dietz, S. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018, 57, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018, 16, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Masunaga, N.; Kagara, N.; Motooka, D.; Nakamura, S.; Miyake, T.; Tanei, T.; Naoi, Y.; Shimoda, M.; Shimazu, K.; Kim, S.J.; et al. Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res. Treat. 2018, 167, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Bartels, S.; Persing, S.; Hasemeier, B.; Schipper, E.; Kreipe, H.; Lehmann, U. Molecular Analysis of Circulating Cell-Free DNA from Lung Cancer Patients in Routine Laboratory Practice: A Cross-Platform Comparison of Three Different Molecular Methods for Mutation Detection. J. Mol. Diagn. 2017, 19, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, C.; Lehmann, A.; Lenze, D.; Hummel, M. Validation and comparison of two NGS assays for the detection of EGFR T790M resistance mutation in liquid biopsies of NSCLC patients. Oncotarget 2018, 9, 18529–18539. [Google Scholar] [CrossRef]
- Dono, M.; De Luca, G.; Lastraioli, S.; Anselmi, G.; Dal Bello, M.G.; Coco, S.; Vanni, I.; Grossi, F.; Vigani, A.; Genova, C.; et al. Tag-based next generation sequencing: A feasible and reliable assay for EGFR T790M mutation detection in circulating tumor DNA of non small cell lung cancer patients. Mol. Med. 2019, 25, 15. [Google Scholar] [CrossRef]
- Alborelli, I.; Generali, D.; Jermann, P.; Cappelletti, M.R.; Ferrero, G.; Scaggiante, B.; Bortul, M.; Zanconati, F.; Nicolet, S.; Haegele, J.; et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: A proof of concept and technical validation study. Cell Death Dis. 2019, 10, 534. [Google Scholar] [CrossRef]
- De Rubis, G.; Rajeev Krishnan, S.; Bebawy, M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Sabari, J.K.; Offin, M.; Stephens, D.; Ni, A.; Lee, A.; Pavlakis, N.; Clarke, S.; Diakos, C.I.; Datta, S.; Tandon, N.; et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. J. Natl. Cancer Inst. 2019, 111, 575–583. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, G.; Conte, D.; Leon, M.; Ciricione, R.; Roz, L.; Ratcliffe, C.; Roz, E.; Cirenei, N.; Bellomi, M.; Pelosi, G.; et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003, 21, 3902. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Janku, F.; Zhan, Q.; Fan, J.B. Accessing genetic information with liquid biopsies. Trends Genet. 2015, 31, 564. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.; Barrett, J.C.; Jänne, P.A. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 2016, 34, 3375. [Google Scholar] [CrossRef]
- Imamura, F.; Uchida, J.; Kukita, Y.; Kumagai, T.; Nishino, K.; Inoue, T.; Kimura, M.; Oba, S.; Kato, K. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 2016, 94, 68. [Google Scholar] [CrossRef]
- Thress, K.S.; Brant, R.; Carr, T.H.; Dearden, S.; Jenkins, S.; Brown, H.; Hammett, T.; Cantarini, M.; Barrett, J.C. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015, 90, 509. [Google Scholar] [CrossRef]
- Sequist, L.V.; Waltman, B.A.; Dias-Santagata, D.; Digumarthy, S.; Turke, A.B.; Fidias, P.; Bergethon, K.; Shaw, A.T.; Gettinger, S.; Cosper, A.K.; et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011, 3, 75ra26. [Google Scholar] [CrossRef]
- Xu, S.; Lou, F.; Wu, Y.; Sun, D.Q.; Zhang, J.B.; Chen, W.; Ye, H.; Liu, J.H.; Wei, S.; Zhao, M.-Y.; et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett. 2016, 370, 324–331. [Google Scholar] [CrossRef]
- Ye, X.; Zhu, Z.Z.; Zhong, L.; Lu, Y.; Sun, Y.; Yin, X.; Yang, Z.; Zhu, G.; Ji, Q. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: Truth or artifact? J. Thorac. Oncol. 2013, 8, 1118–1120. [Google Scholar] [CrossRef]
- Williams, H.L.; Walsh, K.; Diamond, A.; Oniscu, A.; Deans, Z.C. Validation of the Oncomine™ focus panel for next-generation sequencing of clinical tumour samples. Virchows Arch. 2018, 473, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jurchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar] [PubMed]
- Thompson, J.C.; Yee, S.S.; Troxel, A.B.; Savitch, S.L.; Fan, R.; Balli, D.; Lieberman, D.B.; Morrissette, J.D.; Evans, T.L.; Bauml, J.; et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin. Cancer Res. 2016, 22, 5772–5782. [Google Scholar] [CrossRef] [PubMed]
- Sacher, A.G.; Paweletz, C.; Dahlberg, S.E.; Alden, R.S.; O’Connell, A.; Feeney, N.; Mach, S.L.; Jänne, P.A.; Oxnard, G.R. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016, 2, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).