The Isolation of a Novel Streptomyces sp. CJ13 from a Traditional Irish Folk Medicine Alkaline Grassland Soil that Inhibits Multiresistant Pathogens and Yeasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Samples
2.2. Soil pH Measurement
2.3. Microorganisms
2.4. Soil Sample Pretreatment
2.5. Media
2.6. Antibiotic Tests
2.7. DNA Extraction
2.8. Sequencing Protocol
2.9. Phylogenetic Tree
2.10. In Silico Secondary Metabolite Analysis
2.11. Accession Numbers
3. Results
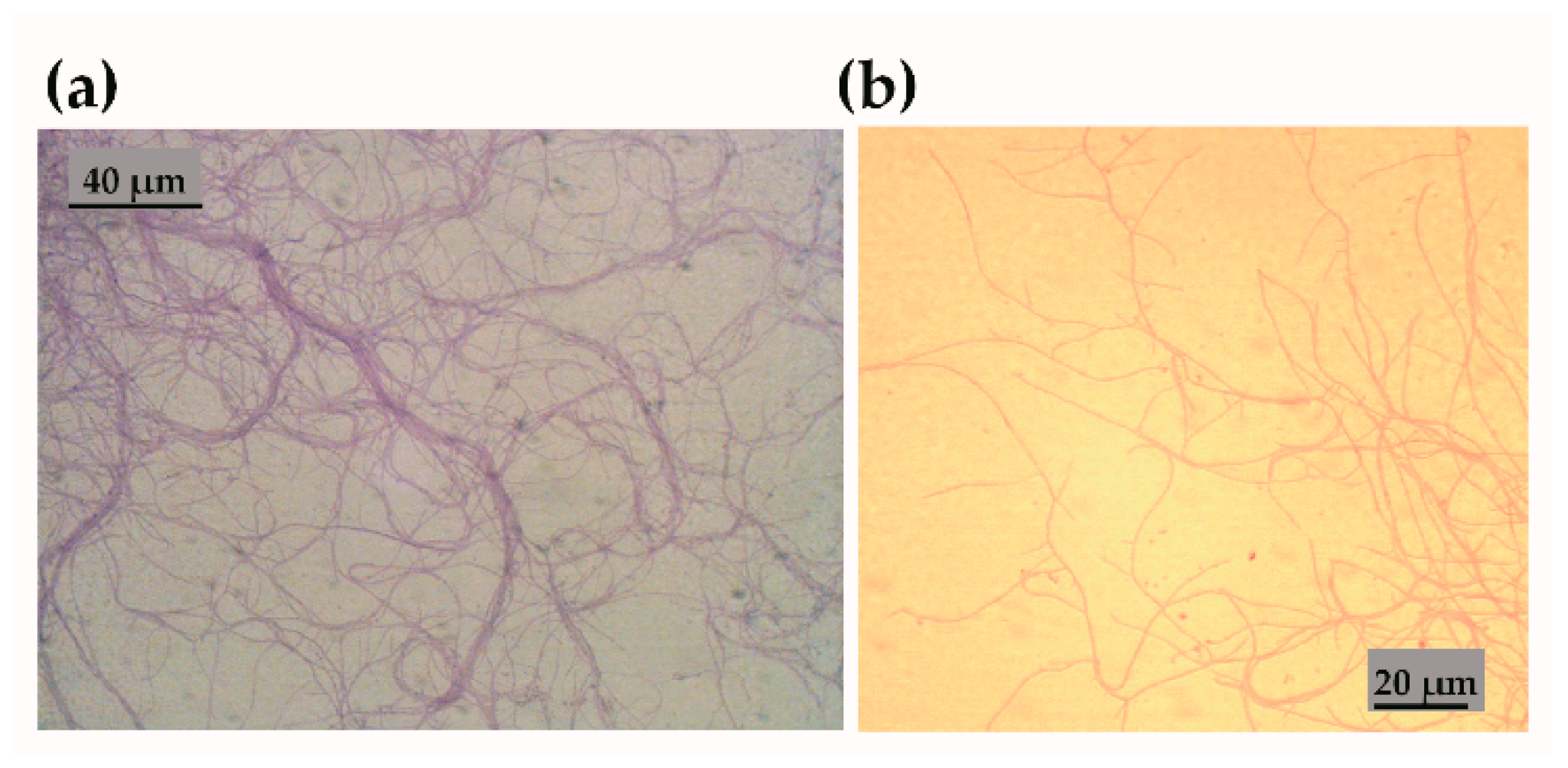
3.1. Characterisation of Streptomyces sp. Isolate CJ13
3.2. Antibiotic Tests
3.2.1. Growth Inhibition by Streptomyces sp. CJ13
3.2.2. Stability of Inhibitory Factor from Streptomyces sp. CJ13
3.3. Whole Genome Sequencing of Streptomyces CJ13
3.4. Phylogeny of Streptomyces sp. CJ13
3.5. Streptomyces sp. CJ13 Secondary Metabolite Prediction
3.6. In Silico Prediction of Streptomyces sp. CJ13 Resistance Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capozzi, C.; Maurici, M.; Pana, A. Antimicrobial resistance: It is a global crisis, “a slow tsunami”. Igiene e Sanita Pubblica 2019, 75, 429–450. [Google Scholar]
- Paulin, S.; Beyer, P. 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Liu, R.; Li, X.; Lam, K.S. Combinatorial chemistry in drug discovery. Curr. Opin. Chem. Biol. 2017, 38, 117–126. [Google Scholar] [CrossRef]
- Ribeiro da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Antibiotic Discovery: Where Have We Come from, Where Do We Go? Antibiotics 2019, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-H.; Hsieh, Y.-H.; Powers, Z.M.; Kao, C.-Y. Defeating Antibiotic-Resistant Bacteria: Exploring Alternative Therapies for a Post-Antibiotic Era. Int. J. Mol. Sci. 2020, 21, 1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rateb, M.E.; Ebel, R.; Jaspars, M. Natural product diversity of actinobacteria in the Atacama Desert. Antonie Van Leeuwenhoek 2018, 111, 1467–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, U.; Jacobo-Herrera, N.; Altemimi, A.; Lakhssassi, N. A Comprehensive Review on Medicinal Plants as Antimicrobial Therapeutics: Potential Avenues of Biocompatible Drug Discovery. Metabolites 2019, 9, 258. [Google Scholar] [CrossRef] [Green Version]
- Dettweiler, M.; Lyles, J.T.; Nelson, K.; Dale, B.; Reddinger, R.M.; Zurawski, D.V.; Quave, C.L. American Civil War plant medicines inhibit growth, biofilm formation, and quorum sensing by multidrug-resistant bacteria. Sci. Rep. 2019, 9, 7692. [Google Scholar] [CrossRef] [Green Version]
- Cordell, G.A.; Farnsworth, N.R.; Beecher, C.W.W.; Doel Soejarto, D.; Kinghorn, A.D.; Pezzuto, J.M.; Wall, M.E.; Wani, M.C.; Cobb, R.R.; O’Neill, M.J.; et al. Novel strategies for the discovery of plant-derived anticancer agents. In Anticancer Drug Discovery and Development: Natural Products and New Molecular Models. Developments in Oncology; Valeriote, F.A., Baker, L.H., Eds.; Springer: Boston, MA, USA, 1994; ISBN 978-1-4613-6118-3. [Google Scholar]
- Liu, M.; Abdel-Mageed, W.M.; Ren, B.; He, W.; Huang, P.; Li, X.; Bolla, K.; Guo, H.; Chen, C.; Song, F.; et al. Endophytic Streptomyces sp. Y3111 from traditional Chinese medicine produced antitubercular pluramycins. Appl. Microbiol. Biotechnol. 2014, 98, 1077–1085. [Google Scholar] [CrossRef]
- Quinn, G.A.; Banat, A.M.; Abdelhameed, A.M.; Banat, I.M. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J. Med. Microbiol. 2020, 69, 1040–1048. [Google Scholar] [CrossRef]
- Falkinham, J.O., 3rd; Wall, T.E.; Tanner, J.R.; Tawaha, K.; Alali, F.Q.; Li, C.; Oberlies, N.H. Proliferation of antibiotic-producing bacteria and concomitant antibiotic production as the basis for the antibiotic activity of Jordan’s red soils. Appl. Environ. Microbiol. 2009, 75, 2735–2741. [Google Scholar] [CrossRef] [Green Version]
- Behroozian, S.; Svensson, S.L.; Davies, J. Kisameet Clay Exhibits Potent Antibacterial Activity against the ESKAPE Pathogens. mBio 2016, 7, e01842-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svensson, S.L.; Behroozian, S.; Xu, W.; Surette, M.G.; Li, L.; Davies, J. Kisameet Glacial Clay: An Unexpected Source of Bacterial Diversity. mBio 2017, 8, e00590-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foley, R. Indigenous narratives of health: (re)placing folk-medicine within Irish health histories. J. Med. Humanit. 2015, 36, 5–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, F.; Sasse, A.; Sheridan, H.; Heinrich, M. Are identities oral? Understanding ethnobotanical knowledge after Irish independence (1937–1939). J. Ethnobiol. Ethnomed. 2017, 13, 65. [Google Scholar] [CrossRef] [Green Version]
- Koay, A.; Shannon, F.; Sasse, A.; Heinrich, M.; Sheridan, H. Exploring the Irish National Folklore Ethnography Database (Dúchas) for Open Data Research on Traditional Medicine Use in Post-Famine Ireland: An Early Example of Citizen Science. Front. Pharmacol. 2020, 11, 1670. [Google Scholar] [CrossRef]
- Terra, L.; Dyson, P.J.; Hitchings, M.D.; Thomas, L.; Abdelhameed, A.; Banat, I.M.; Gazze, S.A.; Vujaklija, D.; Facey, P.D.; Francis, L.W.; et al. A novel alkaliphilic Streptomyces inhibits ESKAPE pathogens. Front. Microbiol. 2018, 9, 2458. [Google Scholar] [CrossRef]
- McLean, E.O. Soil pH and line requirements. In Methods of Soil Analysis, 2nd ed.; Part 2. Chemical and Microbiological Properties; American Society of Agronomy and Soil Sciences Society of America: Madison, WI, USA, 1982. [Google Scholar]
- Lehrer, R.I.; Rosenman, M.; Harwig, S.S.; Jackson, R.; Eisenhauer, P. Ultrasensitive Assays for Endogenous Antimicrobial Polypeptides. J. Immunol. Methods 1991, 137, 167–173. [Google Scholar] [CrossRef]
- Nkanga, E.; Hagedorn, C. Detection of antibiotic-producing Streptomyces inhabiting forest soils. Antimicrob. Agents Chemother. 1978, 14, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinforma. Oxf. Engl. 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Börnigen, D.; Morgan, X.C.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 2304. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Paine, E.; Wall, R.; Kam, G.; Lauriente, T.; Sa-ngarmangkang, P.-C.; Horne, D.; Cheeptham, N. In Situ Cultured Bacterial Diversity from Iron Curtain Cave, Chilliwack, British Columbia, Canada. Diversity 2017, 9, 36. [Google Scholar] [CrossRef] [Green Version]
- Gosse, J.T.; Ghosh, S.; Sproule, A.; Overy, D.; Cheeptham, N.; Boddy, C.N. Whole Genome Sequencing and Metabolomic Study of Cave Streptomyces Isolates ICC1 and ICC4. Front. Microbiol. 2019, 10, 1020. [Google Scholar] [CrossRef]
- Waksman, S.A.; Harris, D.; Lechevalier, M. Studies on Streptomyces lavendulae. J. Bacteriol. 1951, 62, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Nicault, M.; Tidjani, A.-R.; Gauthier, A.; Dumarcay, S.; Gelhaye, E.; Bontemps, C.; Leblond, P. Mining the Biosynthetic Potential for Specialized Metabolism of a Streptomyces Soil Community. Antibiotics 2020, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Santana-Molina, C.; Rivas-Marin, E.; Rojas, A.M.; Devos, D.P. Origin and Evolution of Polycyclic Triterpene Synthesis. Mol. Biol. Evol. 2020, 37, 1925–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terra, L.; Dyson, P.; Ratcliffe, N.; Castro, H.C.; Vicente, A.C.P. Biotechnological Potential of Streptomyces Siderophores as New Antibiotics. Curr. Med. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Letzel, A.-C.; Li, J.; Amos, G.C.A.; Millán-Aguiñaga, N.; Ginigini, J.; Abdelmohsen, U.R.; Gaudêncio, S.P.; Ziemert, N.; Moore, B.S.; Jensen, P.R. Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Environ. Microbiol. 2017, 19, 3660–3673. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zaleta-Rivera, K.; Zhu, X.; Huffman, J.; Millet, J.C.; Harris, S.D.; Yuen, G.; Li, X.-C.; Du, L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 2007, 51, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.; Samborskyy, M.; Usachova, K.; Schnatz, K.; Leadlay, P.F. Sulfation and amidinohydrolysis in the biosynthesis of giant linear polyenes. Beilstein J. Org. Chem. 2017, 13, 2408–2415. [Google Scholar] [CrossRef] [Green Version]
- Pan, G.; Xu, Z.; Guo, Z.; Ming Ma, H.; Yang, D.; Zhou, H.; Gansemans, Y.; Zhu, X.; Huang, Y.; Zhao, L.-X.; et al. Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc. Natl. Acad. Sci. USA 2017, 114, E11131–E11140. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Blodgett, J.A.V.; Clardy, J. Targeted Discovery of Polycyclic Tetramate Macrolactams from an Environmental Streptomyces Strain. Org. Lett. 2010, 12, 4652–4654. [Google Scholar] [CrossRef] [Green Version]
- Kersten, R.D.; Yang, Y.-L.; Xu, Y.; Cimermancic, P.; Nam, S.-J.; Fenical, W.; Fischbach, M.A.; Moore, B.S.; Dorrestein, P.C. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011, 7, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Waglechner, N.; McArthur, A.G.; Wright, G.D. Phylogenetic reconciliation reveals the natural history of glycopeptide antibiotic biosynthesis and resistance. Nat. Microbiol. 2019, 4, 1862–1871. [Google Scholar] [CrossRef]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial Heavy-Metal and Antibiotic Resistance Genes in a Copper Tailing Dam Area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.-Y.; Logue, M.; Robinson, N. Antimicrobial resistance is a global problem—A UK perspective. Eur. J. Integr. Med. 2020, 36, 101136. [Google Scholar] [CrossRef] [PubMed]
- Yucel, S.; Yamac, M. Selection of Streptomyces isolates from Turkish karstic caves against antibiotic resistant microorganisms. Pak. J. Pharm. Sci. 2010, 23, 1–6. [Google Scholar] [PubMed]
- Maciejewska, M.; Adam, D.; Martinet, L.; Naome, A.; Calusinska, M.; Delfosse, P.; Carnol, M.; Barton, H.A.; Hayette, M.-P.; Smargiasso, N.; et al. A Phenotypic and Genotypic Analysis of the Antimicrobial Potential of Cultivable Streptomyces Isolated from Cave Moonmilk Deposits. Front. Microbiol. 2016, 7, 1455. [Google Scholar] [CrossRef] [Green Version]
- Rangseekaew, P.; Pathom-aree, W. Cave Actinobacteria as Producers of Bioactive Metabolites. Front. Microbiol. 2019, 10, 387. [Google Scholar] [CrossRef] [Green Version]
- Butler, M.S.; Paterson, D.L. Antibiotics in the clinical pipeline in October 2019. J. Antibiot. 2020, 73, 329–364. [Google Scholar] [CrossRef]
- Chiang, Y.-M.; Chang, S.-L.; Oakley, B.R.; Wang, C.C. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Omics 2011, 15, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [Green Version]
- Okada, B.K.; Seyedsayamdost, M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2017, 41, 19–33. [Google Scholar] [CrossRef] [Green Version]
- Pishchany, G.; Mevers, E.; Ndousse-Fetter, S.; Horvath, D.J.; Paludo, C.R.; Silva-Junior, E.A.; Koren, S.; Skaar, E.P.; Clardy, J.; Kolter, R. Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. Proc. Natl. Acad. Sci. USA 2018, 115, 10124. [Google Scholar] [CrossRef] [Green Version]
- Groupe, V.; Frankel, J.W.; Leche-Valier, M.P.; Waksman, S.A. Antiviral properties of ehrlichin, an antibiotic produced by Streptomyces lavendulae. J. Immunol. 1951, 67, 471–482. [Google Scholar] [PubMed]
- Arai, T.; Yazawa, K.; Mikami, Y. Isolation and characterization of satellite antibiotics, mimosamycin and chlorocarcins from Streptomyces lavendulae, streptothricin source. J. Antibiot. 1976, 29, 398–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balitz, D.M.; Bush, J.A.; Bradner, W.T.; Doyle, T.W.; O’Herron, F.A.; Nettleton, D.E. Isolation of lavendamycin, a new antibiotic from Streptomyces lavendulae. J. Antibiot. 1982, 35, 259–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matulova, M.; Feckova, L.; Novakova, R.; Mingyar, E.; Csolleiova, D.; Zduriencikova, M.; Sedlak, J.; Patoprsty, V.; Sasinkova, V.; Uhliarikova, I.; et al. A Structural Analysis of the Angucycline-Like Antibiotic Auricin from Streptomyces lavendulae Subsp. Lavendulae CCM 3239 Revealed Its High Similarity to Griseusins. Antibiotics 2019, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Classification | |
|---|---|
| Domain: | Bacteria |
| Phylum: | Actinobacteria |
| Class: | Actinobacteria |
| Order: | Actinomycetales |
| Family: | Streptomycetaceae |
| Genus: | Streptomyces |
| Species: | Streptomyces sp. isolate CJ_13 |
| Gram stain: | Positive |
| Motility: | Negative |
| Pigmentation: | Brown |
| Cell shape: | Branched hyphae |
| Oxygen: | Aerobic, anaerobic tolerant |
| Spores: | Purple/white |
| pH in ISP2 broth | 6.85 to 7.85 |
| Antimicrobial | Microorganisms | |||||
|---|---|---|---|---|---|---|
| MSSA | S. bombicola | B. subtilis | E. coli | P. aeruginosa | MRSA | |
| CJ13 Dark | 13 | 18 | 11 | 0 | 13 | 14 |
| CJ13 Light | 12 | 6 | 0 | 0 | 10 | 13 |
| CEF 20 | 33 | - | 0 | - | 0 | 18 |
| TE 20 | 18 | - | - | - | - | 15 |
| TE 10 | - | - | - | - | 18 | - |
| TE 1 | - | - | 24 | - | - | - |
| CN 10 | na | - | - | 17 | - | - |
| E 10 | na | - | - | 0 | - | - |
| CHL 50 | na | - | - | - | - | - |
| Scheme 20. | ZOI at 20 °C | ZOI at 100 °C | CEP | CHL | TE |
|---|---|---|---|---|---|
| MRSA | 16 | 9 | 18 | - | 17 |
| P. aeruginosa | 12 | 9 | - | - | 16 |
| S. bombicola | 22 | 16 | - | 18 | - |
| Attribute. | Streptomyces sp. CJ13 |
|---|---|
| Genome size | 9,230,829 bp |
| GC Content | 71.9% |
| N50 | 22,457 |
| L50 | 114 |
| Number of Contigs | 901 |
| Number of Subsystems | 314 |
| Total genes | 9260 |
| Number of RNAs | 86 |
| Type | Antibiotic Gene Cluster | Similarity |
|---|---|---|
| NRPS | Salinichelins | 53% |
| NRPS | Antifungal factor | 50% |
| T1PKS | Mediomycin A | 46% |
| NRPS | Weishanmycin | 45% |
| T1PKS, NRPS | Combamide (macrolactam) | 44% |
| Lanthipeptide | SAL-2242 | 44% |
| Biological Role. | Protein Name |
|---|---|
| Aminoglycoside adenylyltransferases | Aminoglycoside N6’-acetyltransferase (EC 2.3.1.82) |
| Resistance to fluoroquinolones | DNA gyrase subunit B (EC 5.99.1.3) |
| Resistance to fluoroquinolones | DNA gyrase subunit A (EC 5.99.1.3) |
| Beta-lactamase | Beta-lactamase |
| Copper homeostasis | Multicopper oxidase |
| Copper homeostasis | Copper resistance protein |
| Copper homeostasis | Copper-translocating P-type ATPase (EC 3.6.3.4) |
| Copper homeostasis | Copper resistance protein |
| Copper homeostasis | Multidrug resistance transporter, |
| Copper homeostasis: copper tolerance | Cytoplasmic copper homeostasis protein |
| Magnesium and cobalt tolerance | Magnesium and cobalt efflux protein |
| Cobalt–zinc–cadmium resistance | Cobalt–zinc–cadmium resistance protein |
| Cobalt–zinc–cadmium resistance | Transcriptional regulator |
| Mercuric reductase | FAD-dependent NAD(P)-disulphide oxidoreductase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quinn, G.A.; Abdelhameed, A.M.; Alharbi, N.K.; Cobice, D.; Adu, S.A.; Swain, M.T.; Castro, H.C.; Facey, P.D.; Bakshi, H.A.; Tambuwala, M.M.; et al. The Isolation of a Novel Streptomyces sp. CJ13 from a Traditional Irish Folk Medicine Alkaline Grassland Soil that Inhibits Multiresistant Pathogens and Yeasts. Appl. Sci. 2021, 11, 173. https://doi.org/10.3390/app11010173
Quinn GA, Abdelhameed AM, Alharbi NK, Cobice D, Adu SA, Swain MT, Castro HC, Facey PD, Bakshi HA, Tambuwala MM, et al. The Isolation of a Novel Streptomyces sp. CJ13 from a Traditional Irish Folk Medicine Alkaline Grassland Soil that Inhibits Multiresistant Pathogens and Yeasts. Applied Sciences. 2021; 11(1):173. https://doi.org/10.3390/app11010173
Chicago/Turabian StyleQuinn, Gerry A., Alyaa M. Abdelhameed, Nada K. Alharbi, Diego Cobice, Simms A. Adu, Martin T. Swain, Helena Carla Castro, Paul D. Facey, Hamid A. Bakshi, Murtaza M. Tambuwala, and et al. 2021. "The Isolation of a Novel Streptomyces sp. CJ13 from a Traditional Irish Folk Medicine Alkaline Grassland Soil that Inhibits Multiresistant Pathogens and Yeasts" Applied Sciences 11, no. 1: 173. https://doi.org/10.3390/app11010173
APA StyleQuinn, G. A., Abdelhameed, A. M., Alharbi, N. K., Cobice, D., Adu, S. A., Swain, M. T., Castro, H. C., Facey, P. D., Bakshi, H. A., Tambuwala, M. M., & Banat, I. M. (2021). The Isolation of a Novel Streptomyces sp. CJ13 from a Traditional Irish Folk Medicine Alkaline Grassland Soil that Inhibits Multiresistant Pathogens and Yeasts. Applied Sciences, 11(1), 173. https://doi.org/10.3390/app11010173









