Abstract
Skin lesion segmentation is one of the pivotal stages in the diagnosis of melanoma. Many methods have been proposed but, to date, this is still a challenging task. Variations in size and color, the fuzzy boundary and the low contrast between lesion and normal skin are the adverse factors for deficient or excessive delineation of lesions, or even inaccurate lesion location detection. In this paper, to counter these problems, we introduce a deep learning method based on U-Net architecture, which performs three tasks, namely lesion segmentation, boundary distance map regression and contour detection. The two auxiliary tasks provide an awareness of boundary and shape to the main encoder, which improves the object localization and pixel-wise classification in the transition region from lesion tissues to healthy tissues. Moreover, concerning the large variation in size, the Selective Kernel modules, which are placed in the skip connections, transfer the multi-receptive field features from the encoder to the decoder. Our methods are evaluated on three publicly available datasets: ISBI2016, ISBI 2017 and PH2. The extensive experimental results show the effectiveness of the proposed method in the task of skin lesion segmentation.
1. Introduction
Skin lesions could be recognized as patches with darkened pigment or redness compared to the overall skin appearance. In most cases, they are harmless, but they may be indications of melanoma, which is a type of skin cancer. Although melanoma is among the most common cancers, the survival rate is high, at 92% [1], as long as it does not reach the late stage of cancer. The severity of melanoma is proportional to the detection time from the emergence of skin lesion or its metastasis to other organs, such as the lung or brain. Therefore, early-stage diagnosis of melanoma is critical for success in cancer treatment. The conventional method of removing the malignant lesions from the skin is surgical excision. Before the main treatment, skin lesion’s condition is recorded as dermoscopic images, which visualize the impaired skin area in high resolution and detail. Subsequently, this image is scrutinized in order to locate the accurate area of lesion and diagnose whether it is dangerous. The preparation step was carried out manually in the past by experienced dermatologists, but this was time-consuming and the clinical inspections from individuals were subjective and inconsistent. There are requirements for an automatic inspection system that has the knowledge and follows the standard rules of skin evaluation, and could deal with an increasing number of dermoscopic images. Recently, computer-aided diagnosis (CAD) systems have been built up as human assistance and shown a promising ability in both skin lesion segmentation and classification.
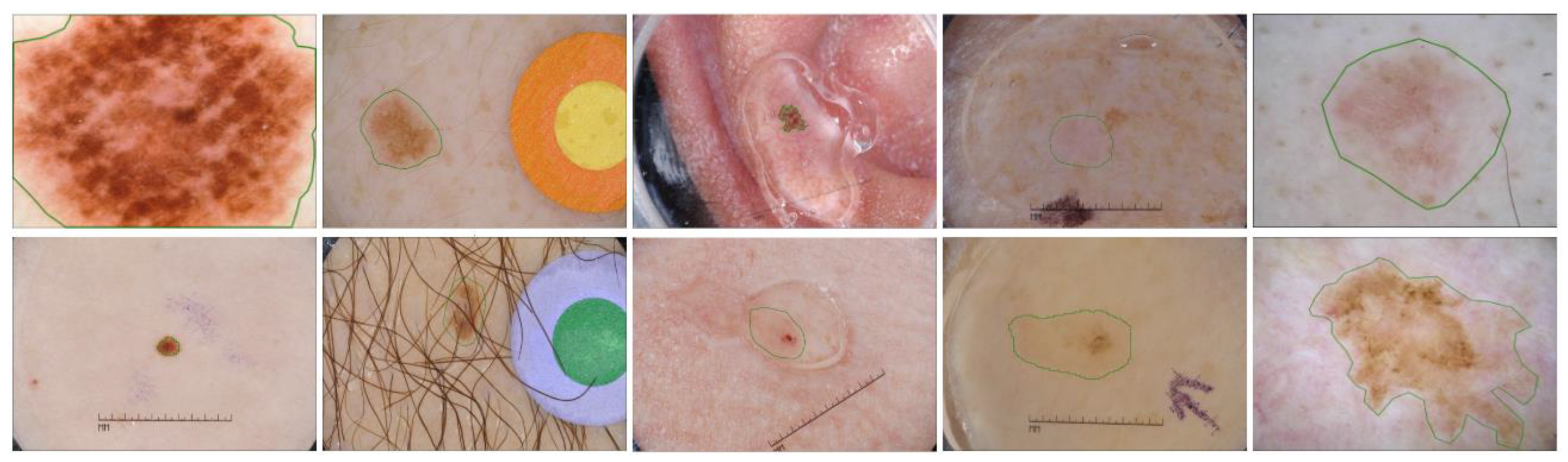
Skin lesion segmentation is a key step, prior to the skin problem categorization. In the past, researchers came up with solutions based on classical techniques like thresholding, clustering, edge and region detection [2,3,4,5,6,7,8]. They are able to deal with many apparent cases of different types of skin, but there are existing impediments that these methods could not overcome. Hence, for the past decade, CAD systems for this task have favored the efficiency of deep learning methods, contributing a lot of novel approaches and positive results [9,10,11,12,13,14,15,16,17]. The architecture of the deep-learning methods used to separate the concerned area from healthy skin generally consists of an encoder and a decoder. The former extracts both the low-level and high-level features, while the latter exploits the latent representation and interprets it as a desirable outcome. Despite the undisputed capacity, deep learning methods still encounter some of the intrinsic and artificial problems of skin lesion segmentation, as briefly shown in Figure 1. The first one is the variability in the size of lesions in dermoscopic images. Some skin lesions are too small compared to the whole image and some occupy the entire image, with little unaffected skin remaining. Another problem is the heterogeneity of the shape and color. Skin pigmentation ordinarily differs from one person to another, and so do the pigments of lesions. In some cases, the contrast between lesion and healthy skin is insignificant and suffers from artifacts and obstructions such as hair, artificial marks and air bubbles. Furthermore, lesion detection becomes entangled with the capricious annotation from experts. There is no agreement among dermatologists, so the lesion delineation could either be loose or excessively detailed, causing deep learning methods to achieve the same performance over the dataset. The aforementioned characteristics of dermoscopic images make this task challenging, even with the impressive discriminative abilities of deep neural networks.
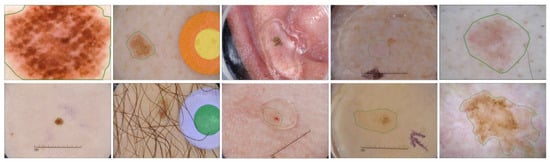
Figure 1.
Challenging skin lesion samples. First column: Large variation in size. Second and third columns: Lesions with artifacts (mark, hair, bubbles). Fourth column: Low contrast compared to background. Fifth column: loose and detailed annotation.
To tackle those problems, CAD systems must comprehend not only the pigmented areas, but also the adjacent regions of healthy tissues. They must grasp the discriminative features that could convey the foreground and background and the contextual information that could reproduce the structure of the lesion. In this paper, we propose a new deep neural network based on the U-net architecture [15] for the skin lesion segmentation. Our model contains three outputs: lesion detection as the main task, and signed distance map regression and contour delineation as the auxiliary tasks. The auxiliary tasks are operated with self-generated ground truth maps from the provided lesion ground truth, conveying the information of the boundary and the pattern of the lesion to the encoder. The structural information brings spatial awareness to the backbone model, broadening the predicted areas if the anticipated boundary covers bigger areas and vice versa. This also improves the localization of skin lesions since the spatial information is lost after sequences of pooling layers in the downsampling path. Considering the variation in size, we integrate Selective Kernel (SK) modules [18] into the skip connections to locate the receptive fields correlated with the scale of the lesions. The application of this module is based on its original concept, which dynamically adjusts the receptive field in harmony with the size of the target objects. While the atrous convolutional layer [19] enlarges the receptive field and then aggregates multi-scale features from different kernels, the Selective Kernel module evaluates the combination of information from multiple kernels and selects effective spatial scales. Furthermore, we attach the deep supervision module to each layer of the decoder with the purpose of multi-scale feature fusion, improving the decision of the last score map.
Our architecture is trained and used for end-to-end prediction. The contributions of our paper are described as follows:
- We evaluate the effectiveness of the two auxiliary tasks that are integrated into the decoder of U-Net architecture [15] for skin lesion segmentation. By feeding the information of the boundary and shape constraints of the lesions to the backbone model, we improve the pixel-wise classification and localization ability of the network;
- We develop a new skip connection which contains a Selective Kernel module [18] for learning and adopting multi-scale features from the encoder. This module accumulates information from different kernel sizes and yields only beneficial features from the global and comprehensive representation.
The remainder of the paper is organized as follows. In Section 2, we provide an overview of the related literature in the field of skin lesion segmentation. The details of our neural network are analyzed and discussed in Section 3. Section 4 displays the achievements of our model through experiments on some public datasets. The conclusion is presented in Section 5.
2. Related Works
Skin lesion segmentation has been an intriguing matter of research for a long time. Traditional algorithms in this field revolved around three concepts: thresholding [3,4], clustering [5,6] and deformable contour model [7,8]. Thresholding methods discriminate lesions and background by estimating the pixel intensity through image histogram. Clustering methods separate the two classes by learning the differences in the extrinsic characteristics. Deformable models initiate a curve surrounding the lesion and the curve evolves into the boundary of the object by mapping the chromatic changes. These methods extract low-level features like pixel values, color or contextual structure, which face the weakness of the variation in the appearance of skin lesions. It is arduous for clustering methods to deal with non-skin related noises or for thresholding, and for deformable methods to not be affected by the inconspicuous transition from pigmented region to the healthy skin tissues. The fact that they could not derive the semantic information from dermoscopic images restricts the skin lesion localization ability and the generalization on larger or partially dissimilar datasets.
With the latest surge in deep neural networks, methods based on deep learning have been widely preferred thanks to the of semantic and latent feature acquisition ability and superior performance in myriad types of projects. Great success has been ascertained in medical image analysis such as optic disc segmentation [20], blood vessel segmentation [21], lung segmentation [22] and brain segmentation [23], and so is the task of skin lesion segmentation [9,10,11,12,13,14,15,16,17]. Bi et al. [9] applied a fully convolutional network (FCN) [10] with a multistage learning method that, in the early-stage FCN, extracted the coarse low-level information and, in the late stage, learned the subtle characteristic of lesion boundaries. Yuan et al. [11] optimized their FCN with Jaccard distance loss. To obtain high-resolution predictions, Li et al. [12] proposed a dense deconvolutional network to learn rich features from local and global contextual information. Yu et al. [13] extracted multiscale features from the layers of a residual network and aggregated them. Lin et al. [14] compared the skin lesion segmentation performance between U-Net [15] and clustering, which observed the advantage of the former. Considering the global context feature extraction, SkinNet [16] integrated the dilated convolutions into the encoder branch of U-Net. In SLSDeep [17], the author introduced an encoder–decoder network with dilated residual and pyramid pooling networks for the coarse and fine representation of dermoscopic images.
There are several networks that have been proposed in terms of enlarging the receptive field. Zhao et al. [24] proposed the pyramid pooling module in the Pyramid Scene Parsing network to obtain the global contextual prior along with the sub-region context and then concatenate different levels of features as the final global feature. In DeepLab [25], atrous convolution pyramid pooling analyzed the convolutional layers with filter at multiple sampling rates and effective fields-of-view, encoding the target object and the context of the image at multiple scales. In later years, the authors of [25] upgraded their paper to DeepLabv3+ [26], integrating the depthwise separable convolution into the atrous spatial pyramid pooling for faster and stronger network and reconsidering the decoder module for better object boundary recovery. The Dynamic Filter network [27] produced filters based on the input, and output the generated parameters to the next input, learning both spatial and photometric changes. To model the geometric transformation, Deformable Convolutional network [28] included deformable convolution which enables free-form deformation of the 2D sampling grid by the knowledge of preceding feature maps, and deformable ROI pooling, which enables the adaptation of object localization to different shapes. Different from the above methods, to obtain denser information from multiple kernel sizes, we capitalize on the SK Network from [18]. The light weight of the module and its great capacity to enlarge the receptive field support the adaptation of the main model to different sizes of lesion. Moreover, we built the model with the multitasking approach, which uses the label information of the distance map and the contour of skin lesion. Utilizing auxiliary tasks helps to guide the process of feature extraction and make the model aware of the boundary constraint and the shape of the unsettled appearance of the lesion.
3. Proposed Method
Even though deep neural networks produce a superior quality of object segmentation to classical and machine learning methods in skin lesion segmentation, there are some problems regarding the inhomogeneous appearance that the recent methods still suffer from. In this paper, we tackle the two problems of skin lesion segmentation, which are the variation in sizes and the fuzzy boundary of the target object. For the first problem, we propose the application of the SK Network [18] in the skip connections of the U-Net model [15]. The lightweight module captures and delivers the features from a larger receptive field and adaptively selects relevant features from different sizes of the lesions. When analyzing the contextual structure of the human skin, we notice that the lesions, in most cases, converge into a unified mass and in one dermoscopic image, there is only one lesion. Hence, provided that we comprehend the exterior constraint of the lesion, the desired object should be laid inside the surround mark. Based on this observation, we attach an additional decoder for both distance map regression and contour generation. The multitask learning approach feeds the model on the awareness of the boundary of the lesion and the gradient shift in skin pigments from impaired to healthy tissues. Details of our model are demonstrated in the following sections.
3.1. Baseline Model
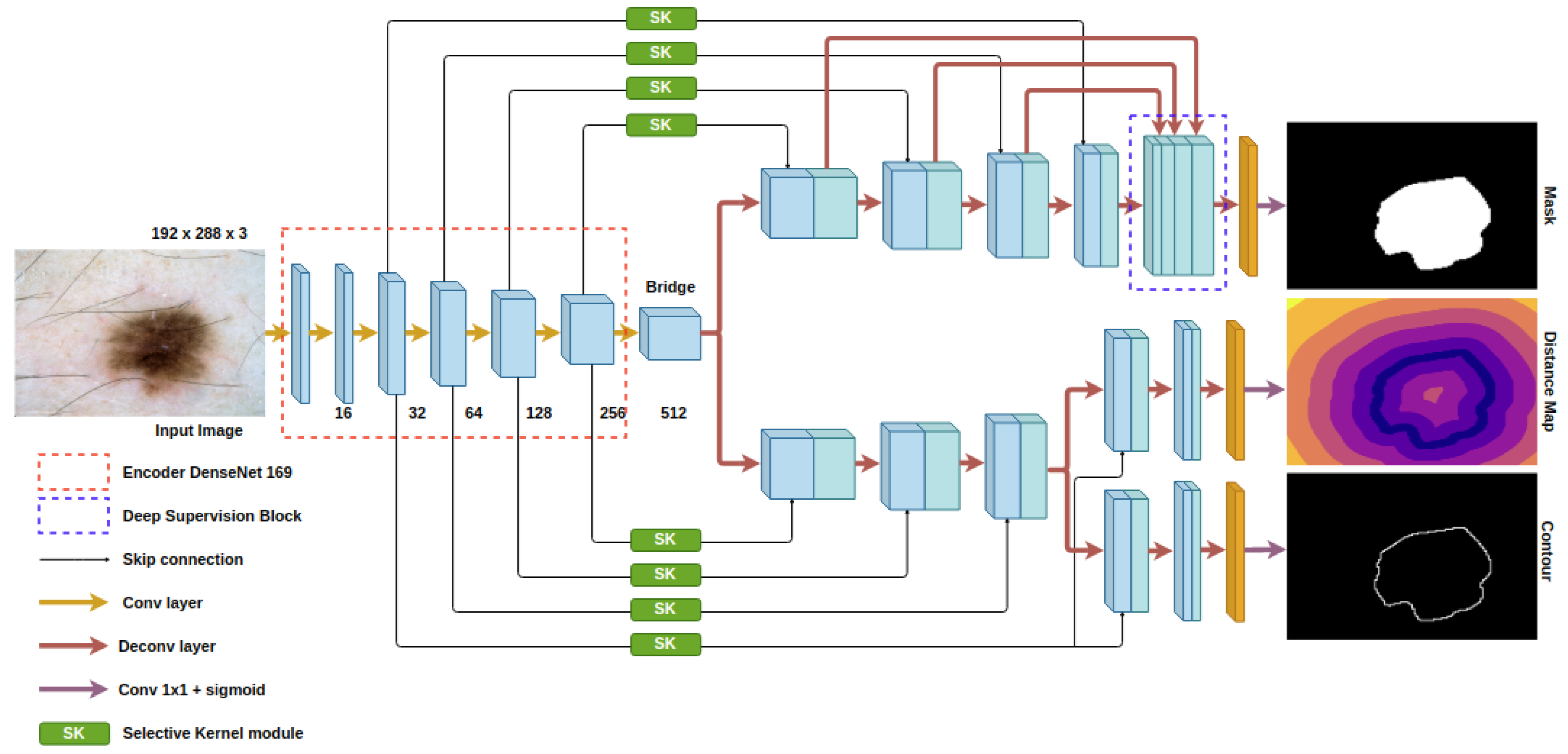
The proposed model is demonstrated in Figure 2. The architecture of our auto encoder–decoder is based on the U-Net model [15]. For skin lesion segmentation, we choose DenseNet [29] as the backbone of the U-Net in order to counter the diverse skin appearance and the low contrast between the lesion and the background. The down-sampling and pooling operations in the conventional encoder cause the reduction in spatial information and affect the efficiency of object localization. The loss of detailed features also impedes the ability to discriminate the ambiguity of the obscure target inside the background. In DenseNet, the connectivity among low and high layers reserves more features from the last layers and has richer patterns so that the top layers gain features from all complexity levels. To further improve the convergence rate and the regularization, we employ the deep supervision block at the decoder. The outputs of the decoder’s layers are fused with the last prediction. Since our whole network is pretty heavy in terms of the number of parameters, while we only provide the cues at the last output to make the model learn the semantics in skin lesion, the model may miss meaningful information during the gradient propagation from the head layer to all of the lower layer. Deep supervision helps the gradients, not only from the last block but from every block, flow straight back to the low-level layers, avoiding the effect of gradient vanishing. This technique not only cuts down the training time but also increases the accuracy.
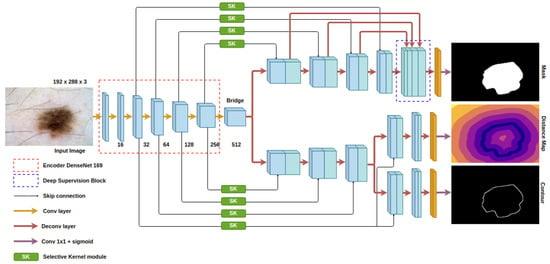
Figure 2.
Architecture of the proposed model.
3.2. Selective Kernel Module
Skin lesions come in a wide range of sizes. In some cases, they occupy less than one percent of the total area of the image and, in other cases, the remaining background is so small that the algorithm could not find enough reference points to discern the two classes. Furthermore, the convolutional kernel of DenseNet [29] or other convolutional neural networks are built with a stable size (3 × 3). This makes conventional deep learning networks unable to extract features from larger receptive fields. Nonetheless, to some extent, large receptive fields do not equal adequate outputs. The appropriate receptive field should reconcile the object localization with the contextual information extraction. The accuracy of localization decreases in relation to the excessive size of the receptive field, while the ability to capture the contextual information is confined to the undersized one. Based on that observation as well as the variation in size of the skin lesions, we opt for the SK module [18] which is exceptional at incorporating local and global information into the receptive field.
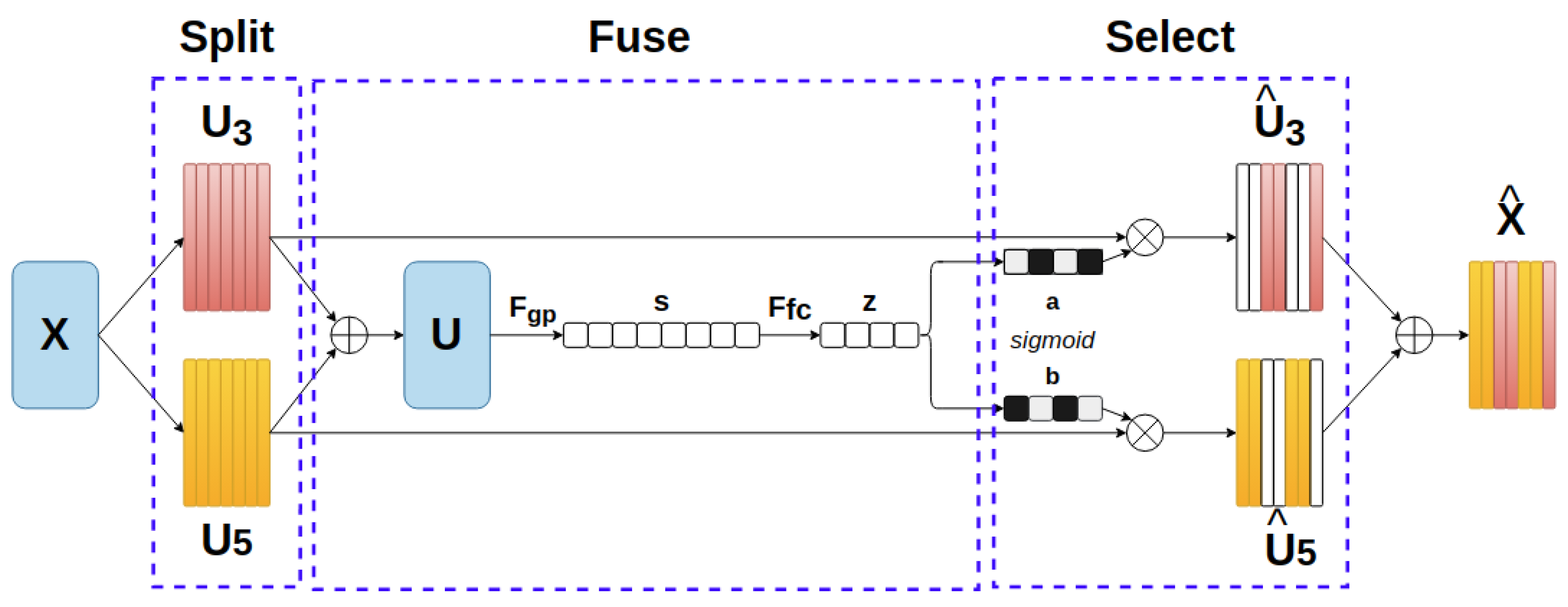
This module is constructed with the ideas of an automatic selection operation and a lightweight design. The architecture is illustrated in Figure 3 and could be briefly described with three steps, in the following order: Split, Fuse and Select. At first, a group of two or three depthwise convolutional blocks (with Batch Normalization [30] and ReLU Activation [31]) with different dilation sizes are applied to the prior feature map X . This can extend to multiple branches with bigger kernel sizes to cover a larger receptive field. In our case, the combination of kernel 3 × 3 and kernel 5 × 5 achieves the best result. Subsequently, in the Fuse process, an element-wise sum function assembles information from U3 and U5 (U = U3 + U5). To embed global spatial information into the channel, a global average pooling function Fgp is placed, which generates channel-wise statistics for s, followed by is a fully- onnected layer Ffc which performs the guidance for the precise and adaptive selections. The feature map z contains the encoded attention weights for features in different scales. At the Select stage, the channel selection weights are computed by a classification layer based on their correspondence with the split feature maps in the beginning. We choose sigmoid as the classification function on an exchange with the softmax function in the original paper, since the task is binary classification in our method. The softmax function will be in charge when more than two data branches are generated in the Split stage. The learned parameters are applied to each feature map that is derived from the beginning, and all of them are then aggregated to be a new feature map. The new feature map is
where and are the selected feature maps of the kernel size 3 and 5, respectively. a and b, in the following order, are the attention weight vectors for each feature map in which the sum of the elements with the same order from two vectors is 1.
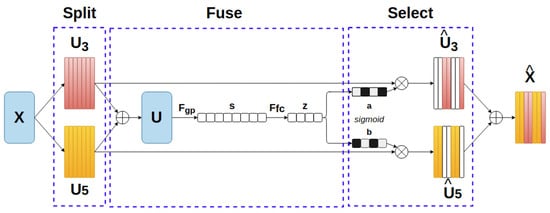
Figure 3.
Selective Kernel module.
SK modules are embedded into the skip connections of U-Net, as shown in Figure 2. The corresponding level features from the encoder are adaptively selected before passing to decoder. In recent papers about object segmentation, researchers focused on the spatial attention and channel attention [32,33,34] to guide the learning process towards only relevant local representation, reducing the distraction from unnecessary features and improving the representation power. However, in the skin lesion task, where objects have great fluctuations in appearance, there are limitations to the attention mechanism. The global contextual information is often disregarded, as the mechanism is biased towards the target object. In addition, the attention mechanism is not constructed with the involvement and process of multi-scale features. According to the predefined problem, we prefer the advantage of the SK module for our network.
3.3. Structural Awareness Module
In the architecture of convolutional neural networks, low-level layers extract the features such as edges and curves and are then accumulated to generate high-level features in order to perceive the context in image. Through this process, the spatial information is dissipated and partially transformed, which causes a lack of confidence in predicting the thin edge around the target object. Skin lesions which have unclear borders may degrade the performance of pattern classification, while conventional segmentation methods and loss functions designed for this task do not regard the shape information of the target objects. In this paper, we equip our model with boundary distance map regression task and contour regression task to better distinguish the lesion and non-lesion pixels at the boundary area, enhance the localization accuracy and further improve the whole segmentation performance. The maps for these two tasks are inferred from the masks of skin lesions. Since the contour map regression task could easily suffer from the imbalance problem, the boundary distance map task is used at the same time to deliver more insightful appearance constraints to the encoder. Both types of map are generated by an additional decoder. They share the information at half of the decoder and split into separate heads at the other half. Features in the adaptive skip connections are still transferred to the additional decoder to strengthen the regression performance. When predicting, the second decoder could be detached to reduce the total parameters.
3.4. Loss Function
For the main task, we use the combination of weighted binary cross entropy loss LWBCE and Dice loss LDice due to the skewed distribution of lesion and non-lesion pixels. The function for LWBCE, LDice, Lmask can be represented as below, respectively
where indicates the ground truth pixel label and indicates the predicted pixel label. β is a self-deduced coefficient related to the ratio of the object and background in the whole dataset. The weighted binary cross entropy loss deals with the pixel-wise classification while the Dice loss takes care of the intersection of area. Both of them can reduce the imbalance problem.
For the axillary tasks, we use Log-Cosh loss to regress the distance map and the contour map. Log-Cosh loss is smoother than the mean squared error loss for regression tasks and less influenced by irregular incorrect prediction. The Log-Cosh loss function is written as
After all, we have to optimize the total loss LTotal for our model
λ1, λ2 and λ3 are the coefficients for lesion segmentation loss, distance map loss and contour map loss, respectively. The equal contribution of three tasks to the knowledge of the encoder is prone to the lack of attention to the main task, leading to the inefficient lesion segmentation and waste of supplementary data. Hence, we empirically set λ1, λ2 and λ3 as 1, 0.01 and 0.001 so the network can fully exploit the provided information.
4. Experiments and Results
4.1. Datasets
We test our method on three public benchmark datasets, namely ISBI 2016 [35], ISBI 2017 [36] and PH2 [37]. The ISBI 2016 and ISBI 2017 datasets are from the annual challenges organized by the International Skin Image Collaboration (ISIC) in 2016 and 2017, respectively. The ISBI 2016 consists of 900 training images and 379 test images, while the ISBI 2017 comprises 2000 training images, 600 test images and a validation set of 150 images. The test dataset of each of the above datasets is evaluated with results from other existing methods to prove the efficiency of our method. There are 200 dermoscopic images with annotation in the PH2 dataset and we only use this dataset for evaluation. All of the provided dermoscopy images are in RGB format and their corresponding ground truths are binary masks to delineate lesion and non-lesion areas.
4.2. Evaluation Criterion
The metrics to evaluate the proposed method are selected from the suggestion of the ISBI 2017 challenge, which includes Jaccard index (JA), Dice coefficient (DI), Accuracy (AC). They are defined in the Equations (7)–(9)
where TP, TN, FP, and FN correspond to true positive, true negative, false positive, and false negative, respectively. JA and DI estimate the overlap and similarity between the prediction area and the ground truth. AC indicates the ratio of correctly segmented area over the ground truth. All of the metrics follow the rule that the higher, the better.
4.3. Implementation Details
The proposed method is implemented in Python and Keras framework. We employ the DenseNet169 pre-trained on ImageNet [38] as the backbone of the auto encoder-decoder. The images in the aforementioned datasets have different resolutions, so the input sizes for training and testing are uniformly set as 288 in width and 192 in height. To enhance the generalization ability, we use online data augmentation, which comprises horizontal and vertical flips, random rotation with low degrees, image scaling and distortion. The annotations for boundary distance map and contour map are not provided, so we self-generate them based on the ground truth maps of lesions. We adopt the Adam algorithm [39] with a batch size of 8 to optimize the whole network. The initial learning rate is set at 0.00003 and is dropped by 10 percent after each 20 epochs. The total number of epochs used for training is 120. During the inference phase, only the main decoder is retained to generate the lesion mask. We also use Test Time Augmentation (TTA), which includes image flip and rotation, as a post processing step to improve the prediction accuracy. We do not train the PH2 dataset. It is evaluated using the model trained on ISBI 2016 and ISBI 2017.
4.4. Results
4.4.1. Ablation Studies
To evaluate the effectiveness of the proposed method on skin lesion segmentation, we conduct the ablation studies on the ISBI 2016 and ISBI 2017 datasets. The DenseNet169 model is set as the baseline benchmark and we subsequently append other configurations to it, to verify their supportive impact on the baseline model. The detailed quantitative experimental results for the ISBI 2016 and ISBI 2017 datasets are displayed in Table 1 with the accompanying methods, namely, the SK module in the skip connections, structural awareness module as the additional decoder, deep supervision module at the end of the main decoder and TTA as the post-processing step. By incorporating the new modules, we observe the increase in JA, DI and AC in both datasets. The baseline model has a greater boost with the assistance of the structural awareness than with the addition of the SK module. The former enhances the DI by 0.67 and 1.18, while the latter enhances the DI score by 0.4 and 0.92 on the ISBI 2016 and ISBI 2017, respectively. The combination of two aforementioned methods and the deep supervision module achieves the superior performance to the baseline model, with the DI increase of 0.84 and 1.82 on each dataset. The post-processing step TTA further improves the performance of our model, obtaining the final DI results of 92.61 for the ISBI 2016 dataset and 87.61 for the ISBI 2017. In conclusion, Table 1 proves that the additional modules along with their abilities to extract the contextual and multi-scale information obtain better segmentation results and the coordination among auxiliary modules does not conflict each other.

Table 1.
Ablation studies on the ISBI 2016 and ISBI 2017 datasets.
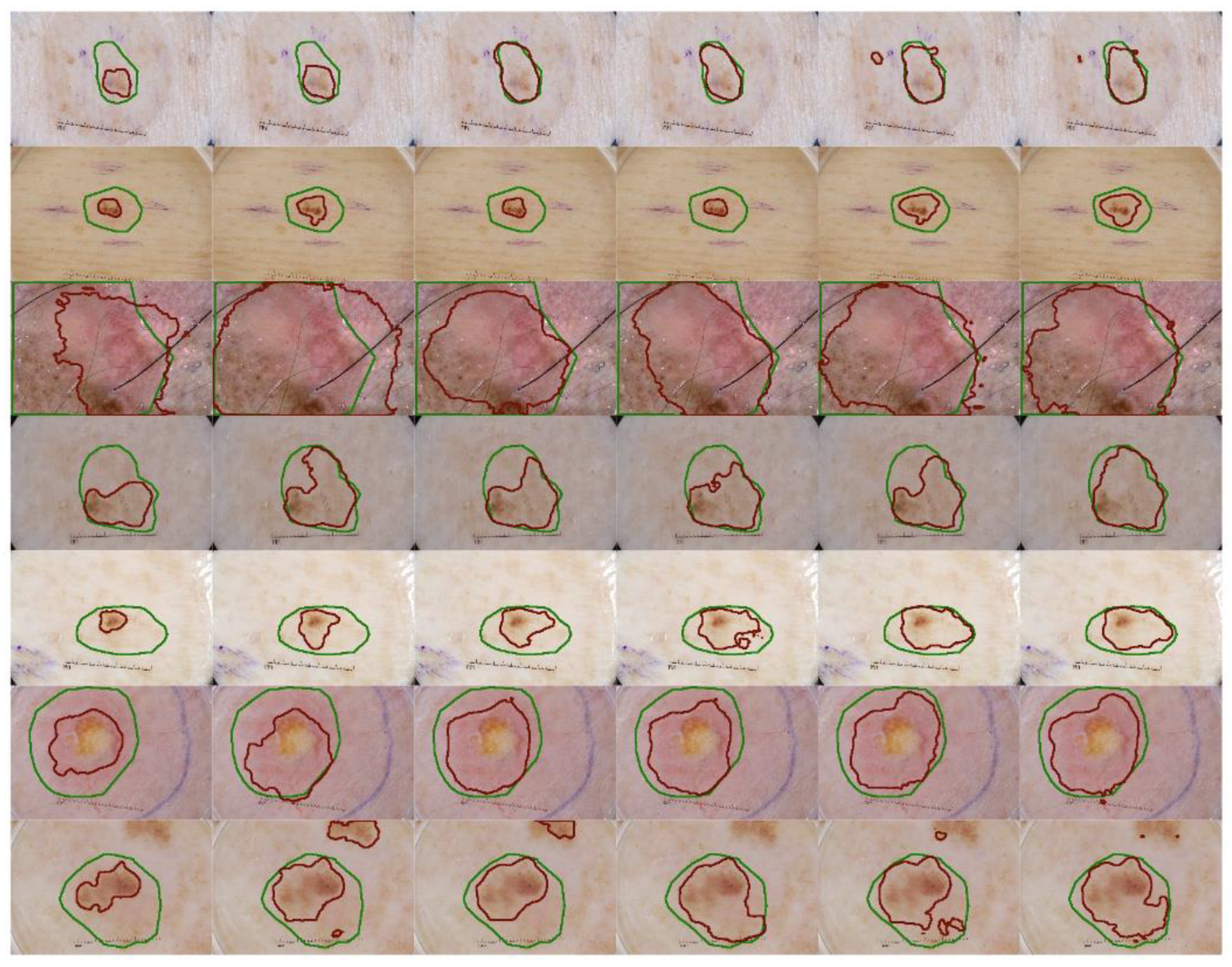
We also implement a qualitative analysis on the effectiveness of the proposed methods on skin lesion segmentation. Figure 4 shows the experimental results of several challenging cases. Compared to the baseline segmentation results, the SK module clearly expands the receptive field, since the prediction area is broader while the structure module delineates the boundary that is closer to boundary of the ground truth. The baseline model could discern the noticeable discrepancy between the impaired and healthy tissues but the proposed method gains a deeper understanding of the texture of the lesions. The SK module may produce the lesion map that excessively spread over a larger region than expected (Figure 4, second image in third row) but when it is associated with the structural module, its influence is appropriately weakened to satisfy the two mentioned problems. In the cases of low contrast, we see that our model performs well in keeping track of the lesion pattern and produce a satisfactory lesion map (Figure 4, fourth row). The object localization capability is obviously enhanced in the later implementations, where the prediction areas outside the lesions are eliminated (Figure 4, last row).
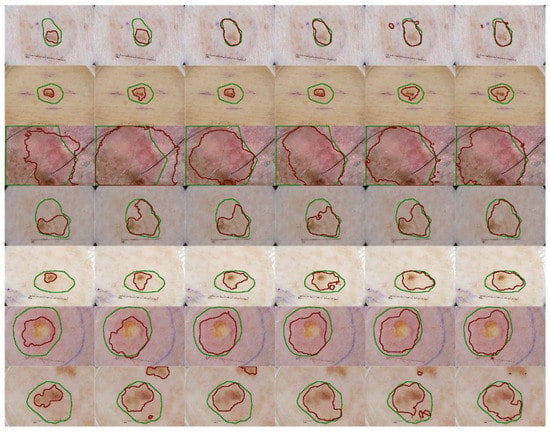
Figure 4.
Qualitative segmentation result comparison with different configurations. The green lines indicate the location and the area of the ground truth of the skin lesion and the red lines demonstrate the prediction of our method on the same input. From the first column to the sixth columns: the baseline method, baseline + SK module, baseline + structural awareness, baseline + SK + structural awareness, the proposed method and itself after TTA as post-processing step.
To examine the effectiveness of applying different kernel sizes in the SK module, we conduct the experiments with four types of kernel combination on the ISBI 2017 dataset, as shown in Table 2. As we stated in the above section, a large receptive field does not always promise best performance. Among the kernel size configurations, the combination of kernel size 3 × 3 and kernel size 5 × 5 produces the best segmentation performance, which is 87.61 in DI. Surprisingly, the worst segmentation performance is from the combination of triple kernel size, which obtains a DI result of only 86.71.

Table 2.
Performance comparison of different kernel sizes in the SK module.
4.4.2. Comparison to Other Published Methods
In this section, we analyze the performance of our method on the ISBI 2016, ISBI 2017 and PH2 datasets. Our results are compared to the teams participating in the ISBI challenges and other published methods. The results are taken from their publications. Table 3, Table 4 and Table 5 show the segmentation performance on the ISBI 2016, ISBI 2017 and PH2 datasets, respectively.

Table 3.
Performance comparison with other methods on the ISBI 2016 dataset.

Table 4.
Performance comparison with other methods on the ISBI 2017 dataset.

Table 5.
Performance comparison with other methods on the PH2 dataset.
In the ISBI 2016 dataset, we achieve the best segmentation performance in JA, DI and AC. Based on the JA and DI results, we see that the proposed method is more capable of locating the lesion and find the neighboring elements than other methods.
In the ISBI 2017 dataset, we achieve the best segmentation performance in JA and AC but not in DI. Our results prove that the proposed method is consistent in both datasets. The ISBI 2017 dataset is more difficult for segmentation, since there are more sophisticated skin lesions in melanoma cases.
In the PH2 dataset, we conduct two experiments on the pretrained model of the ISBI 2016 and the ISBI 2017, which other methods also followed, because this dataset is small. We achieve the best performance on the PH2 dataset in both cases. The result on the trained dataset of ISBI 2017 is expected to acquire a better performance, since the ISBI 2017 dataset is bigger. However, the trained model on the ISBI 2016 dataset generalizes better on the PH2 dataset.
5. Conclusions
In this paper, based on the observation that the sizes of the skin lesion are varied across images and the low contrast between the lesion pigment and the healthy tissues, we propose the method that could learn the multi-scale information and the structural constraint simultaneously. We apply the Selective Kernel module into the skip connections of U-Net to transfer an appropriate and larger receptive field to the decoder, helping the model deal with the variation in lesion sizes. Moreover, we propose the auxiliary decoder of distance map delineation and contour detection with the purposed of acknowledging the skin lesion structure. The qualitative and quantitative results of our method prove its effectiveness in three public datasets. It can deal with the inconsistent appearance of skin lesions and solve several image segmentation problems. For future research, the segmentation result can be integrated to the lesion classification for further diagnosis of the detailed lesion area or even for prediction of the possible progression of the lesion. Skin lesion could be benign or malignant and, in the case of malignant tumor, early diagnosis is greatly recommended to avoid metastasis or even fatality. We will incorporate the clinical knowledge into the lesion image to automatically diagnose the states of the skin lesion with high precision.
Author Contributions
Conceptualization, T.-D.-T.P.; Methodology, T.-D.-T.P.; Writing—review and editing, T.-D.-T.P. and G.-S.L.; Supervision, G.-S.L., S.-H.K. and H.-J.Y.; Project administration, G.-S.L., S.-H.K. and H.-J.Y.; Funding acquisition, G.-S.L., S.-H.K. and H.-J.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT). (NRF-2020R1A4A1019191) and also by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIT) (NRF-2019M3E5D1A02067961).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Abbas, Q.; Garcia, I.F.; Celebi, M.E.; Ahmad, W.; Mushtaq, Q. A perceptually oriented method for contrast enhancement and segmentation of dermoscopy images. Ski. Res. Technol. 2012, 19, e490–e497. [Google Scholar] [CrossRef] [PubMed]
- Grana, C.; Pellacani, G.; Cucchiara, R.; Seidenari, S. A new algorithm for border description of polarized light surface microscopic images of pigmented skin lesions. IEEE Trans. Med. Imaging 2003, 22, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Emre Celebi, M.; Wen, Q.; Hwang, S.; Iyatomi, H.; Schaefer, G. Lesion Border Detection in Dermoscopy Images Using Ensembles of Thresholding Methods. Skin Res. Technol. 2013, 19, e252–e258. [Google Scholar] [CrossRef]
- Melli, R.; Grana, C.; Cucchiara, R. Comparison of color clustering algorithms for segmentation of dermatological images. Med. Imaging 2006, 6144, 61443. [Google Scholar] [CrossRef]
- Zhou, H.; Schaefer, G.; Sadka, A.H.; Celebi, M.E. Anisotropic Mean Shift Based Fuzzy C-Means Segmentation of Dermoscopy Images. IEEE J. Sel. Top. Signal Process. 2009, 3, 26–34. [Google Scholar] [CrossRef]
- Zhou, H.; Schaefer, G.; Celebi, M.E.; Lin, F.; Liu, T. Gradient Vector Flow ưith Mean Shift for Skin Lesion Segmentation. Comput. Med. Imaging Graph. 2011, 35, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Tavares, J.M.R.S. A Novel Approach to Segment Skin Lesions in Dermoscopic Images Based on a Deformable Model. IEEE J. Biomed. Health Inform. 2016, 20, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Kim, J.; Ahn, E.; Kumar, A.; Fulham, M.; Feng, D. Dermoscopic Image Segmentation via Multistage Fully Convolutional Networks. IEEE Trans. Biomed. Eng. 2017, 64, 2065–2074. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Yuan, Y.; Chao, M.; Lo, Y.-C. Automatic Skin Lesion Segmentation Using Deep Fully Convolutional Networks With Jaccard Distance. IEEE Trans. Med. Imaging 2017, 36, 1876–1886. [Google Scholar] [CrossRef]
- Li, H.; He, X.; Zhou, F.; Yu, Z.; Ni, D.; Chen, S.; Wang, T.; Lei, B. Dense Deconvolutional Network for Skin Lesion Segmentation. IEEE J. Biomed. Health Inform. 2019, 23, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Jiang, X.; Zhou, F.; Qin, J.; Ni, D.; Chen, S.; Lei, B.; Wang, T. Melanoma Recognition in Dermoscopy Images via Aggregated Deep Convolutional Features. IEEE Trans. Biomed. Eng. 2018, 66, 1006–1016. [Google Scholar] [CrossRef]
- Lin, B.S.; Michael, K.; Kalra, S.; Tizhoosh, H. Skin Lesion Segmentation: U-Nets Versus Clustering. In Proceedings of the 2017 IEEE Symposium Series on Computational Intelligence (SSCI), Honolulu, HI, USA, 27 November–1 December 2017; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2017; pp. 1–7. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Cham, Switzedland, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Vesal, S.; Ravikumar, N.; Maier, A. SkinNet. In A Deep Learning Framework for Skin Lesion Segmentation, Proceedings of the 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference Proceedings (NSS/MIC), Sydney, Australia, 10–17 November 2018; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2018; pp. 1–3. [Google Scholar]
- Sarker, M.K.; Rashwan, H.A.; Akram, F.; Banu, S.F.; Saleh, A.; Singh, V.K.; Chowdhury, F.U.H.; Abdulwahab, S.; Romani, S.; Radeva, P.; et al. SLS Deep: Skin Lesion Segmentation Based on Dilated Residual and Pyramid Pooling Networks. In Transactions on Petri Nets and Other Models of Concurrency XV; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018; pp. 21–29. [Google Scholar]
- Li, X.; Wang, W.; Hu, X.; Yang, J. Selective Kernel Networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–21 June 2019; pp. 510–519. [Google Scholar]
- Yu, F.; Koltun, V. Multi-Scale Context Aggregation by Dilated Convolutions. In Proceedings of the International Conference on Learning Representations (ICLR), San Juan, Puerto Rico, 2–4 May 2015. [Google Scholar]
- Fu, H.; Cheng, J.; Xu, Y.; Wong, D.W.K.; Liu, J.; Cao, X. Joint Optic Disc and Cup Segmentation Based on Multi-Label Deep Network and Polar Transformation. IEEE Trans. Med. Imaging 2018, 37, 1597–1605. [Google Scholar] [CrossRef]
- Kromm, C.; Rohr, K. Inception Capsule Network for Retinal Blood Vessel Segmentation and Centerline Extraction. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2020; pp. 1223–1226. [Google Scholar]
- Jiang, J.; Hu, Y.C.; Liu, C.J.; Halpenny, D.; Hellmann, M.D.; Deasy, J.O.; Mageras, G.; Veeraraghavan, H. Multiple Resolution Residually Connected Feature Streams for Automatic Lung Tumor Segmentation from CT Images. IEEE Trans. Med. Imaging 2018, 38, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Jagan, A. A New Approach for Segmentation and Detection of Brain Tumor in 3D Brain MR Imaging. In Proceedings of the 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, IN, USA, 29–31 March 2018; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2018; pp. 1230–1235. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1063–6919. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Jia, X.; De Brabandere, B.; Tuytelaars, T.; Van Gool, L. Dynamic Filter Networks. NIPS 2016, 29, 667–675. [Google Scholar]
- Dai, J.; Qi, H.; Xiong, Y.; Li, Y.; Zhang, G.; Hu, H.; Wei, Y. Deformable convolutional networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22 October 2017; pp. 764–773. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified Linear Units Improve Restricted Boltzmann Machines. ICML 2010, 27, 807–814. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where To Look For The Pancreas. In Proceedings of the Medical Imaging with Deep Learning, Amsterdam, The Netherlands, 4–6 July 2018. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Gutman, D.; Codella, C.F.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2017, arXiv:1710.05006. [Google Scholar]
- Mendonca, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.S.; Rozeira, J. PH2 - A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. In Proceedings of the International Conference Learn. Represent. (ICLR), San Diego, CA, USA, 5–8 May 2015. [Google Scholar]
- Li, Y.; Shen, L. Skin Lesion Analysis towards Melanoma Detection Using Deep Learning Network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Alpaslan, N.; Bhattacharya, P. Developing A Retrieval Based Diagnostic Aid for Automated Melanoma Recognition of Dermoscopic Images. In Proceedings of the 2016 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 18–20 October 2016; IEEE: New York, NY, USA, 2016; pp. 1–7. [Google Scholar]
- Deng, Z.; Fan, H.; Xie, F.; Cui, Y.; Liu, J. Segmentation of dermoscopy images based on fully convolutional neural network. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2017; pp. 1732–1736. [Google Scholar]
- Nasr-Esfahani, E.; Rafiei, S.; Jafari, M.H.; Karimi, N.; Wrobel, J.S.; Samavi, S.; Soroushmehr, S.R. Dense pooling layers in fully convolutional network for skin lesion segmentation. Comput. Med. Imaging Graph. 2019, 78, 101658. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Yang, J.; Liu, J.; Jiang, Z.; Zheng, Y.; Wang, Y. Skin lesion segmentation using high-resolution convolutional neural network. Comput. Methods Programs Biomed. 2020, 186, 105241. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Fang, Z.; Yuan, S.; Zhan, C.; Xing, Y.; Zhou, J.T.; Yang, F. iMSCGnet: Iterative Multi-Scale Context-Guided Segmentation of Skin Lesion in Dermoscopic Images. IEEE Access 2020, 8, 39700–39712. [Google Scholar] [CrossRef]
- Yuan, Y.; Lo, Y.-C. Improving Dermoscopic Image Segmentation With Enhanced Convolutional-Deconvolutional Networks. IEEE J. Biomed. Health Inform. 2019, 23, 519–526. [Google Scholar] [CrossRef]
- Berseth, M. ISIC 2017-Skin Lesion Analysis towards Melanoma Detection. CoRR 2017. abs/1703.00523. [Google Scholar]
- Bi, L.; Kim, J.; Ahn, E.; Feng, D. Automatic Skin Lesion Analysis Using Large-Scale Dermoscopy Images and Deep Residual Networks. arXiv 2017, arXiv:1703.04197. [Google Scholar]
- Wang, H.; Wang, G.; Sheng, Z.; Zhang, S. Automated Segmentation of Skin Lesion Based on Pyramid Attention Network. In Transactions on Petri Nets and Other Models of Concurrency XV; Springer: Berlin/Heidelberg, Germany, 2019; pp. 435–443. [Google Scholar]
- Tu, W.; Liu, X.; Hu, W.; Pan, Z. Dense-Residual Network with Adversarial Learning for Skin Lesion Segmentation. IEEE Access 2019, 7, 77037–77051. [Google Scholar] [CrossRef]
- Al-Masni, M.A.; Al-Antari, M.A.; Choi, M.-T.; Han, S.-M.; Kim, T.-S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef]
- Goyal, M.; Oakley, A.; Bansal, P.; Dancey, D.; Yap, M.H. Skin Lesion Segmentation in Dermoscopic Images with Ensemble Deep Learning Methods. IEEE Access 2020, 8, 4171–4181. [Google Scholar] [CrossRef]
- Baghersalimi, S.; Bozorgtabar, B.; Schmid-Saugeon, P.; Ekenel, H.K.; Thiran, J.-P. DermoNet: Densely linked convolutional neural network for efficient skin lesion segmentation. EURASIP J. Image Video Process. 2019, 2019, 71. [Google Scholar] [CrossRef]
- Bi, L.; Kim, J.; Ahn, E.; Kumar, A.; Feng, D.; Fulham, M. Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recognit. 2019, 85, 78–89. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).