Advanced Statistical Analysis of 3D Kinect Data: A Comparison of the Classification Methods
Abstract
:1. Introduction
1.1. Biomedical Background
1.1.1. Electrodiagnostics of Facial Nerve Palsy
1.1.2. Facial Rehabilitation
- 1
- Patient education to explain the pathologic condition and set realistic goals;
- 2
- Soft tissue mobilisation to address facial muscle tightness and edema;
- 3
- Functional retraining to improve oral competence;
- 4
- Facial expression retraining, including stretching exercises;
- 5
- Synkinesis management [22].
1.1.3. Evaluation of Facial Nerve Function
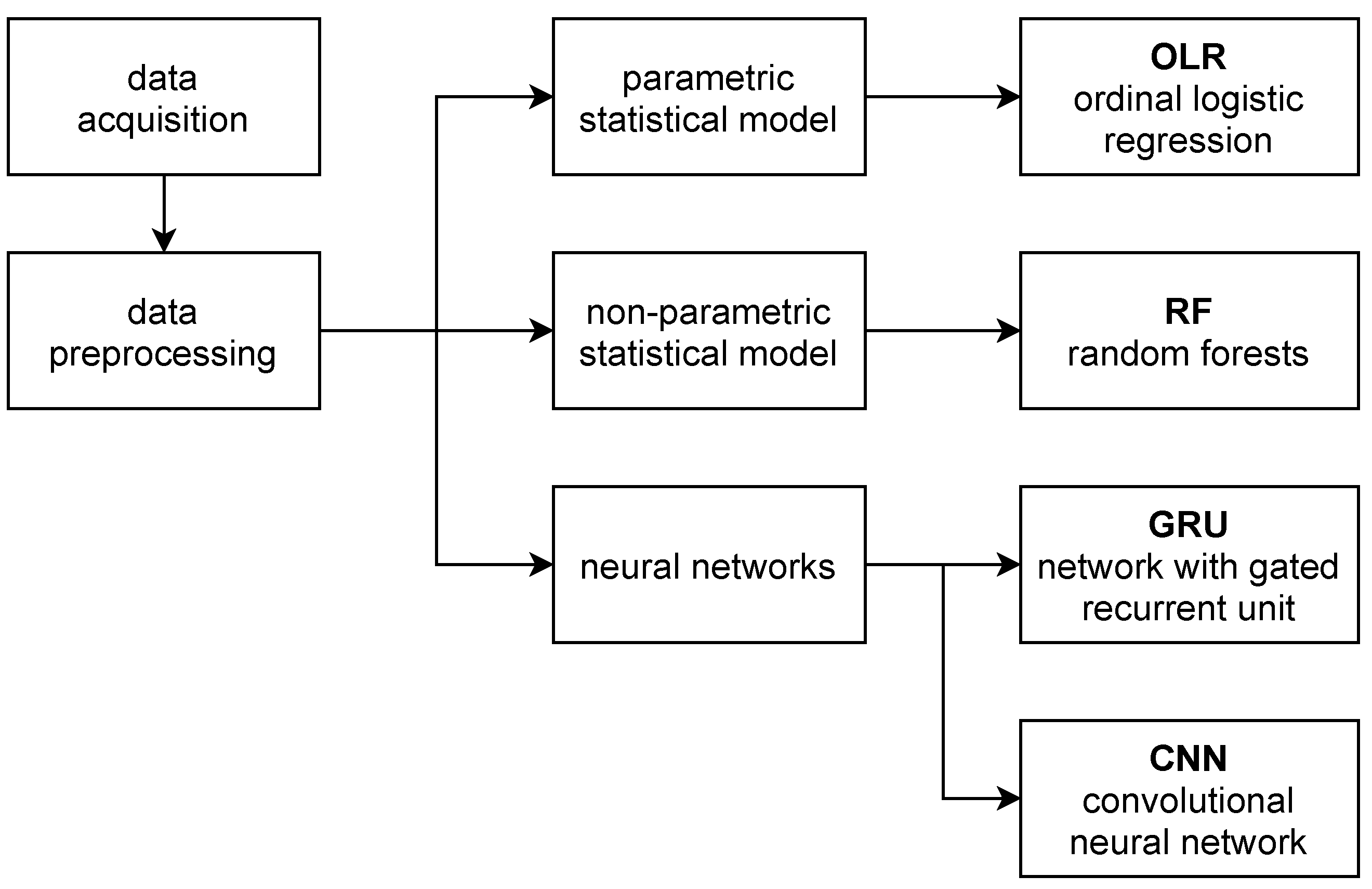
2. Materials and Methods
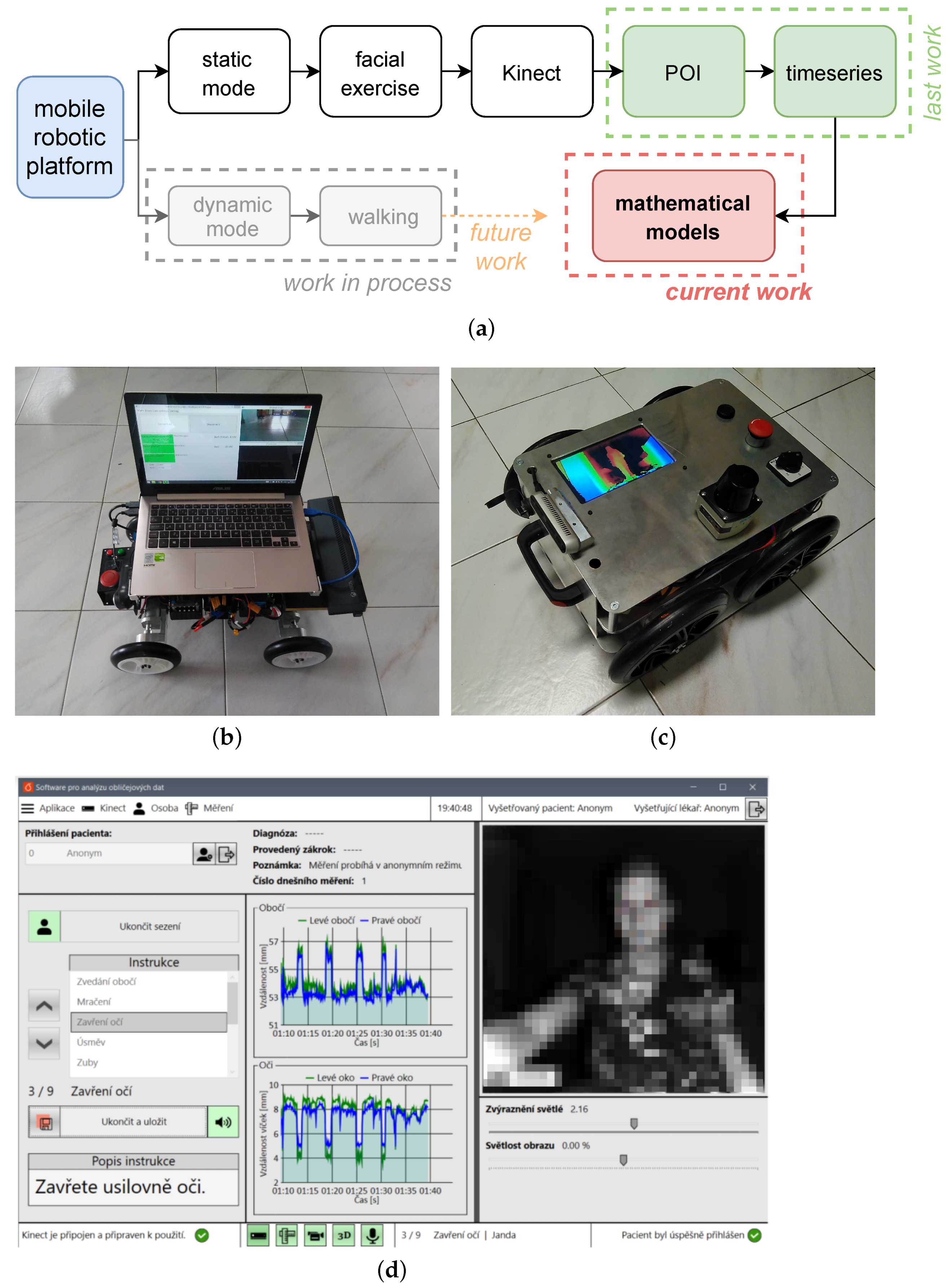
2.1. Data Acquisition
2.2. Data Preprocessing
2.3. Compared Statistics
2.4. Parametric Statistical Model
- The first step consists of application of functional logistic regression (FLR) to indicator curves (understood as functional data) separately for each indicator type (exercise). This turns the indicator curves (functional data) into health scores (real-valued data between 0 and 1).
- In the second step, classification of the set of health scores for each patient (multivariate data) into HB grades is performed using multivariate ordinal logistic regression (OLR).
2.4.1. Functional Logistic Regression
2.4.2. Multivariate Ordinal Logistic Regression
2.5. Non-Parametric Statistical Model
- In the first step, we apply kernel functional classification to turn curves of individual indicators into real-valued health scores.
- In the second step, HB grades are predicted from the lists (vectors) of health scores using the ordinal forests method.
2.5.1. Kernel Functional Classification
2.5.2. Ordinal Forests
2.6. Neural Networks
3. Results
4. Discussion
5. Conclusions
- Parametric statistics (based on OLR)
- Non-parametric statistics (based on ordinal random forests)
- GRU
- Convolutional neural network
Future Work
- Better data acquisition: As part of data collection and pre-processing, it is expected that the operator will be immediately informed about the quality and usability of the record;
- Unsupervised learning: In the next step, the data will be processed independently of clinical practice, which is heavily burdened by the subjective opinion of the physician;
- Fusion with the project devoted to gait analysis: The data from these experiments are fused with data from the analysis of gait of the same patients (these patients often suffer from balance problems in addition to mimic problems);
- Mobile application: In addition to a full-fledged application in a hospital environment, it is also planned to create a mobile application (using the camera system of a smartphone), which will be available to patients for home rehabilitation.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIC | Akaike information criterion |
| CNN | Neural network with convolutional layers |
| DNN | Deep neural network |
| FLR | Functional logistic regression |
| GLM | Generalised linear models |
| GRU | Neural network with gated recurrent unit |
| HB | House–Brackmann facial nerve grading system |
| MATLAB | a proprietary multi-paradigm programming language and numerical |
| computing environment | |
| OLR | Ordinal logistic regression |
| ORL | Otorhinolaryngology |
| POI | Point of interest |
| R | a free software environment for statistical computing and graphics |
| RF | Random forests |
References
- Procházka, A.; Vyšata, O.; Charvátová, H.; Vališ, M. Motion Symmetry Evaluation Using Accelerometers and Energy Distribution. Symmetry 2019, 11, 871. [Google Scholar] [CrossRef] [Green Version]
- Oudre, L.; Barrois-Müller, R.; Moreau, T.; Truong, C.; Vienne-Jumeau, A.; Ricard, D.; Vayatis, N.; Vidal, P.P. Template-based Step Detection with Inertial Measurement Units. Sensors 2018, 18, 4033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schätz, M.; Procházka, A.; Kuchyňka, J.; Vyšata, O. Sleep Apnea Detection with Polysomnography and Depth Sensors. Sensors 2020, 20, 1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schätz, M.; Centonze, F.; Kuchyňka, J.; Ťupa, O.; Vyšata, O.; Geman, O.; Procházka, A. Statistical Recognition of Breathing by MS Kinect Depth Sensor. In Proceedings of the 2015 International Workshop on Computational Intelligence for Multimedia Understanding (IWCIM), Prague, Czech Republic, 29–30 October 2015; pp. 1–4. [Google Scholar]
- Nussbaum, R.; Kelly, C.; Quinby, E.; Mac, A.; Parmanto, B.; Dicianno, B.E. Systematic Review of Mobile Health Applications in Rehabilitation. Arch. Phys. Med. Rehabil. 2019, 100, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Mirniaharikandehei, S.; Heidari, M.; Danala, G.; Lakshmivarahan, S.; Zheng, B. Applying a Random Projection Algorithm to Optimise Machine Learning Model for Predicting Peritoneal Metastasis in Gastric Cancer Patients Using CT Images. Comput. Methods Progr. Biomed. 2021, 200, 105937. [Google Scholar] [CrossRef] [PubMed]
- Leary, O.P.; Crozier, J.; Liu, D.D.; Niu, T.; Pertsch, N.J.; Camara-Quintana, J.Q.; Svokos, K.A.; Syed, S.; Telfeian, A.E.; Oyelese, A.A.; et al. Three-dimensional Printed Anatomic Modeling for Surgical Planning and Real-time Operative Guidance in Complex Primary Spinal Column Tumors: Single-center Experience and Case Series. World Neurosurg. 2021, 145, e116–e126. [Google Scholar] [CrossRef]
- Von Arx, T.; Nakashima, M.J.; Lozanoff, S. The Face—A Musculoskeletal Perspective. A literature review. Swiss Dent. J. 2018, 128, 678–688. [Google Scholar]
- Ullah, S.; Finch, C.F. Applications of Functional Data Analysis: A Systematic Review. BMC Med. Res. Methodol. 2013, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.L.; Chiou, J.M.; Müller, H.G. Functional Data Analysis. Annu. Rev. Stat. Its Appl. 2016, 3, 257–295. [Google Scholar] [CrossRef] [Green Version]
- Wongvibulsin, S.; Wu, K.C.; Zeger, S.L. Clinical Risk Prediction with Random Forests for Survival, Longitudinal, and Multivariate (RF-SLAM) Data Analysis. BMC Med. Res. Methodol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, B.G.; Fradet, G.; Nedzelski, J.M. Development of a Sensitive Clinical Facial Grading System. Otolaryngol. Neck Surg. 1996, 114, 380–386. [Google Scholar] [CrossRef]
- Kohout, J.; Verešpejová, L.; Kříž, P.; Červená, L.; Štícha, K.; Crha, J.; Trnková, K.; Chovanec, M.; Mareš, J. Advanced Statistical Analysis of 3D Kinect Data: Mimetic Muscle Rehabilitation Following Head and Neck Surgeries Causing Facial Paresis. Sensors 2021, 21, 103. [Google Scholar] [CrossRef] [PubMed]
- Owusu, J.A.; Stewart, C.M.; Boahene, K. Facial Nerve Paralysis. Med. Clin. N. Am. 2018, 102, 1135–1143. [Google Scholar] [CrossRef]
- Cockerham, K.; Aro, S.; Liu, W.; Pantchenko, O.; Olmos, A.; Oehlberg, M.; Sivaprakasam, M.; Crow, L. Application of MEMS Technology and Engineering in Medicine: A New Paradigm for Facial Muscle Reanimation. Expert Rev. Med. Devices 2008, 5, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Gordin, E.; Lee, T.S.; Ducic, Y.; Arnaoutakis, D. Facial nerve trauma: Evaluation and considerations in management. Craniomaxillofac. Trauma Reconstr. 2015, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Thielker, J.; Grosheva, M.; Ihrler, S.; Wittig, A.; Guntinas-Lichius, O. Contemporary management of benign and malignant parotid tumors. Front. Surg. 2018, 5, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guntinas-Lichius, O.; Volk, G.F.; Olsen, K.D.; Mäkitie, A.A.; Silver, C.E.; Zafereo, M.E.; Rinaldo, A.; Randolph, G.W.; Simo, R.; Shaha, A.R.; et al. Facial nerve electrodiagnostics for patients with facial palsy: A clinical practice guideline. Eur. Arch. Oto Rhino Laryngol. 2020, 277, 1855–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heckmann, J.G.; Urban, P.P.; Pitz, S.; Guntinas-Lichius, O.; Gágyor, I. The diagnosis and treatment of idiopathic facial paresis (bell’s palsy). Dtsch. Ärzteblatt Int. 2019, 116, 692. [Google Scholar]
- Kennelly, K.D. Electrodiagnostic approach to cranial neuropathies. Neurol. Clin. 2012, 30, 661–684. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Kühn, D.; Jungheim, M.; Schwemmle, C.; Ptok, M. Neuromuskuläre Elektrostimulationsverfahren in der HNO-Heilkunde. HNO 2014, 62, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.W.; Baiungo, J. Facial Rehabilitation: Evaluation and Treatment Strategies for the Patient with Facial Palsy. Otolaryngol. Clin. N. Am. 2018, 51, 1151–1167. [Google Scholar] [CrossRef] [PubMed]
- House, W. Facial Nerve Grading System. Otolaryngol. Head Neck Surg. 1985, 93, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Scheller, C.; Wienke, A.; Tatagiba, M.; Gharabaghi, A.; Ramina, K.F.; Scheller, K.; Prell, J.; Zenk, J.; Ganslandt, O.; Bischoff, B.; et al. Interobserver Variability of the House-Brackmann Facial Nerve Grading System for the Analysis of a Randomised Multi-center Phase III Trial. Acta Neurochir. 2017, 159, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.O.; Silverman, B.W. Applied Functional Data Analysis: Methods and Case Studies; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Febrero Bande, M.; Oviedo de la Fuente, M. Statistical Computing in Functional Data Analysis: The R Package fda.usc. J. Stat. Softw. 2012, 51, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Agresti, A. Categorical Data Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2003; Volume 482. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S; Springer Publishing Company, Incorporated: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Ferraty, F.; Vieu, P. Non-parametric Functional Data Analysis: Theory and Practice; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Hornung, R. Ordinal Forests. J. Classif. 2019, 37, 4–17. [Google Scholar] [CrossRef]
- Cho, K.; van Merrienboer, B.; Bahdanau, D.; Bengio, Y. On the Properties of Neural Machine Translation: Encoder-Decoder Approaches. arXiv 2014, arXiv:1409.1259. Available online: http://xxx.lanl.gov/abs/1409.1259 (accessed on 30 April 2021).
- Rawat, W.; Wang, Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017, 29, 2352–2449. Available online: http://xxx.lanl.gov/abs/https://direct.mit.edu/neco/article-pdf/29/9/2352/1017965/neco_a_00990.pdf (accessed on 30 April 2021). [CrossRef]
- Qayyum, A.; Anwar, S.M.; Majid, M.; Awais, M.; Alnowami, M.R. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar]
- Naser, M.Z.; Alavi, A. Insights into Performance Fitness and Error Metrics for Machine Learning. arXiv 2020, arXiv:2006.00887. Available online: http://xxx.lanl.gov/abs/2006.00887 (accessed on 30 April 2021).




| Grade | Description | Characteristic |
|---|---|---|
| I | Normal function | normal facial function in all areas |
| II | Mild dysfunction | Gross: slight weakness on close inspection; very slight synkinesis |
| At rest: normal tone and symmetry | ||
| Motion Forehead: moderate to good function | ||
| Eye: complete closure with minimum effort | ||
| Mouth: slight asymmetry | ||
| III | Moderate dysfunction | Gross: obvious but not disfiguring |
| difference between two sides; noticeable synkinesis | ||
| At rest: normal tone and symmetry | ||
| Motion Forehead: slight to moderate movement | ||
| Eye: complete closure with effort | ||
| Mouth: slightly weak with maximum effort | ||
| IV | Moderately severe dysfunction | Gross: obvious weakness and disfiguring asymmetry |
| At rest: normal tone and symmetry | ||
| Motion Forehead: none | ||
| Eye: incomplete closure | ||
| Mouth: asymmetric with maximum effort | ||
| V | Severe dysfunction | Gross: only barely perceptible motion |
| At rest: asymmetry | ||
| Motion Forehead: none | ||
| Eye: incomplete closure | ||
| Mouth: slight movement | ||
| VI | Total paralysis | no movement |
| HB by a Clinician | HB by OLR | HB by RF | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 6 | 1 | 2 | 3 | 6 | |
| 1 | 28 | 17 | 10 | 3 | 48 | 1 | 9 | 0 |
| 2 | 6 | 13 | 2 | 0 | 17 | 2 | 2 | 0 |
| 3 | 2 | 3 | 15 | 3 | 13 | 1 | 8 | 1 |
| 6 | 0 | 1 | 5 | 14 | 3 | 0 | 12 | 5 |
| HB by a Clinician | HB by | HB by | ||||||
|---|---|---|---|---|---|---|---|---|
| GRU-tp100k | GRU-tp1.6M | |||||||
| 1 | 2 | 3 | 6 | 1 | 2 | 3 | 6 | |
| 1 | 38 | 7 | 7 | 6 | 37 | 7 | 7 | 7 |
| 2 | 9 | 7 | 2 | 3 | 6 | 11 | 2 | 2 |
| 3 | 7 | 2 | 9 | 5 | 10 | 4 | 5 | 4 |
| 6 | 5 | 2 | 5 | 8 | 9 | 4 | 4 | 3 |
| HB by a Clinician | HB by | HB by | ||||||
|---|---|---|---|---|---|---|---|---|
| CNN-tp160k | CNN-tp4M | |||||||
| 1 | 2 | 3 | 6 | 1 | 2 | 3 | 6 | |
| 1 | 43 | 1 | 8 | 6 | 43 | 4 | 11 | 0 |
| 2 | 8 | 6 | 4 | 3 | 9 | 9 | 3 | 0 |
| 3 | 7 | 4 | 12 | 0 | 8 | 2 | 12 | 1 |
| 6 | 5 | 1 | 3 | 11 | 4 | 1 | 1 | 14 |
| Correct Classification | ||||||
|---|---|---|---|---|---|---|
| HB by a Clinician | OLR | RF | GRU-tp100k | GRU-tp1.6M | CNN-tp160k | CNN-tp4M |
| 1 | 48% | 83% | 66% | 64% | 74% | 74% |
| 2 | 62% | 10% | 33% | 52% | 29% | 43% |
| 3 | 65% | 35% | 39% | 22% | 52% | 52% |
| 6 | 70% | 25% | 40% | 15% | 55% | 70% |
| Overall accuracy | 57% | 52% | 51% | 46% | 59% | 64% |
| Approximate classification | ||||||
| HB by a clinician | OLR | RF | GRU-tp100k | GRU-tp1.6M | CNN-tp160k | CNN-tp4M |
| 1 | 78% | 84% | 78% | 76% | 76% | 81% |
| 2 | 100% | 100% | 86% | 90% | 86% | 100% |
| 3 | 78% | 39% | 48% | 39% | 70% | 61% |
| 6 | 70% | 25% | 40% | 15% | 55% | 70% |
| Overall accuracy | 80% | 69% | 67% | 61% | 73% | 79% |
| HB by a Model | OLR | RF | GRU-tp100k | GRU-tp1.6M | CNN-tp160k | CNN-tp4M |
|---|---|---|---|---|---|---|
| 1 | 78% | 59% | 64% | 60% | 68% | 67% |
| 2 | 38% | 50% | 39% | 42% | 50% | 56% |
| 3 | 47% | 26% | 39% | 28% | 44% | 44% |
| 6 | 70% | 83% | 36% | 19% | 55% | 93% |
| OLR | RF | GRU-tp100k | GRU-tp1.6M | CNN-tp160k | CNN-tp4M | |
|---|---|---|---|---|---|---|
| Test set | 57% | 52% | 51% | 46% | 59% | 64% |
| Train set 1 | 55% | 98% | 94% | 43% | 99% | 99% |
| Train set 2 | 63% | 100% | 100% | 100% | 100% | 100% |
| Train set 3 | 50% | 99% | 100% | 96% | 100% | 100% |
| Train set 4 | 60% | 98% | 100% | 97% | 100% | 100% |
| Train set 5 | 62% | 100% | 98% | 69% | 100% | 100% |
| Method | Pros | Cons |
|---|---|---|
| statistical models in general |  easily tractable, allow for analysis of underlying drivers easily tractable, allow for analysis of underlying drivers |  require careful choice of models and methods require careful choice of models and methods |
| parametric statistics |  provide explicit dependence formulas (enable in-depth analysis of the studied phenomena) provide explicit dependence formulas (enable in-depth analysis of the studied phenomena) |  sensitive to model misspecifications sensitive to model misspecifications |
 do not require large datasets do not require large datasets | ||
| non-parametric statistics |  good flexibility good flexibility |  prone to overfitting on small datasets prone to overfitting on small datasets |
 strict (distributional and model) assumptions not required strict (distributional and model) assumptions not required | ||
| neural models in general |  highly modular highly modular |  susceptible to sparse training data susceptible to sparse training data |
 black box black box | ||
| GRU based models |  infer long temporal dependencies infer long temporal dependencies |  can not reduce dimensionality, slow training/inference for longer measurement;s can not reduce dimensionality, slow training/inference for longer measurement;s |
 ineffective parallelisation, longer training ineffective parallelisation, longer training | ||
| CNN based models |  infers patterns from data, both in temporal and cross-feature dimensions infers patterns from data, both in temporal and cross-feature dimensions |  more susceptible to overfitting more susceptible to overfitting |
 for dimensionality reduction for dimensionality reduction |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Červená, L.; Kříž, P.; Kohout, J.; Vejvar, M.; Verešpejová, L.; Štícha, K.; Crha, J.; Trnková, K.; Chovanec, M.; Mareš, J. Advanced Statistical Analysis of 3D Kinect Data: A Comparison of the Classification Methods. Appl. Sci. 2021, 11, 4572. https://doi.org/10.3390/app11104572
Červená L, Kříž P, Kohout J, Vejvar M, Verešpejová L, Štícha K, Crha J, Trnková K, Chovanec M, Mareš J. Advanced Statistical Analysis of 3D Kinect Data: A Comparison of the Classification Methods. Applied Sciences. 2021; 11(10):4572. https://doi.org/10.3390/app11104572
Chicago/Turabian StyleČervená, Lenka, Pavel Kříž, Jan Kohout, Martin Vejvar, Ludmila Verešpejová, Karel Štícha, Jan Crha, Kateřina Trnková, Martin Chovanec, and Jan Mareš. 2021. "Advanced Statistical Analysis of 3D Kinect Data: A Comparison of the Classification Methods" Applied Sciences 11, no. 10: 4572. https://doi.org/10.3390/app11104572
APA StyleČervená, L., Kříž, P., Kohout, J., Vejvar, M., Verešpejová, L., Štícha, K., Crha, J., Trnková, K., Chovanec, M., & Mareš, J. (2021). Advanced Statistical Analysis of 3D Kinect Data: A Comparison of the Classification Methods. Applied Sciences, 11(10), 4572. https://doi.org/10.3390/app11104572






