Degradation of Brominated Organic Compounds (Flame Retardants) by a Four-Strain Consortium Isolated from Contaminated Groundwater
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Solvents
2.2. Isolates
2.3. Degradation
2.3.1. TBNPA and DBNPG Degradation Kinetics with Yeast Extract
2.3.2. Simultaneous Biodegradation of TBNPA and DBNPG with Yeast Extract
2.3.3. Individual and Simultaneous Degradation of TBNPA and DBNPG with Vitamin Mix or Glucose Amendment
2.4. Isotopic Fractionation
2.5. Analytical Methods
2.6. Calculations
3. Results and Discussion
3.1. Microbial Isolates
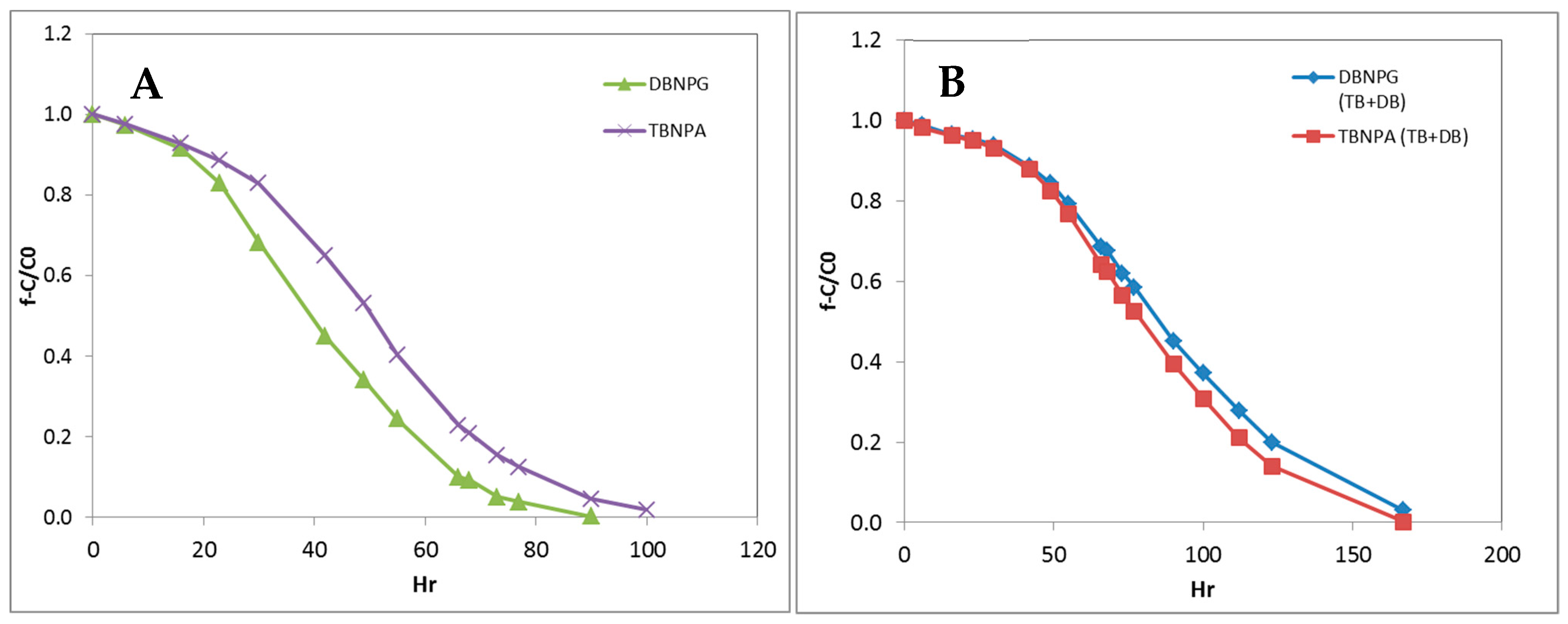
3.2. DBNPG and TBNPA Degradation
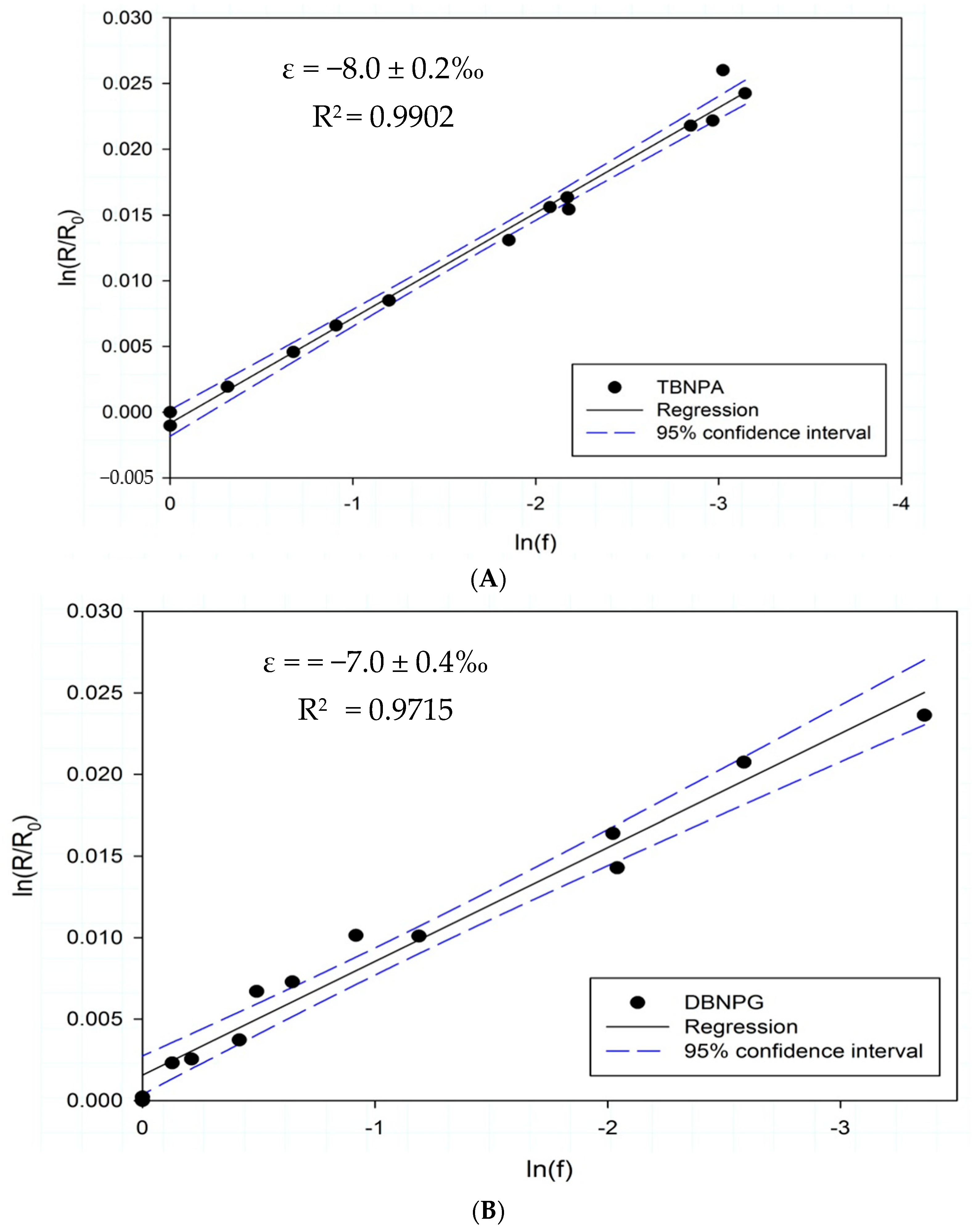
3.3. Isotope Fractionation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tyagi, M.; da Fonseca, M.M.R.; de Carvalho, C.C.C.R. Bioaugmentation and Biostimulation Strategies to Improve the Effectiveness of Bioremediation Processes. Biodegradation 2011, 22, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Haggblom, M.M.; Bossert, I.D. Halogenated organic compounds-A global perspective. In Dehalogenation: Microbial Processes and Environmental Applications; Haggblom, M.M., Bossert, I.D., Eds.; Kluwer Academic Publishers: Norwell, MA, USA, 2003. [Google Scholar]
- Ezra, S.; Feinstein, S.; Bilkis, I.; Adar, E.; Ganor, J. Chemical Transformation of 3-Bromo-2,2-Bis(Bromomethyl)-Propanol under Basic Conditions. Environ. Sci. Technol. 2005, 39, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Yankelzon, I.; Adar, E.; Gelman, F.; Ronen, Z.; Bernstein, A. The Spatial Distribution of the Microbial Community in a Contaminated Aquitard below an Industrial Zone. Water 2019, 11, 2128. [Google Scholar] [CrossRef] [Green Version]
- U.S Environmental Protection Agency (EPA). Furniture Flame Retardancy Partnership: Environmental Profiles of Chemical Flame-Retardant Alternatives for Low-Density Polyurethane Foam. Chemical Hazard Review; U.S Environmental Protection Agency (EPA): Washington, DC, USA, 2005. [Google Scholar]
- Dunnick, J.K.; Heath, J.E.; Farnell, D.R.; Prejean, J.D.; Haseman, J.K.; Elwell, M.R. Carcinogenic Activity of the Flame Retardant, 2,2-Bis(Bromomethyl)-1,3-Propanediol in Rodents, and Comparison with the Carcinogenicity of Other NTP Brominated Chemicals. Toxicol. Pathol. 1997, 25, 541–548. [Google Scholar] [CrossRef]
- Sharma, B.; Shukla, P. Designing Synthetic Microbial Communities for Effectual Bioremediation: A Review. Biocatal. Biotransformation 2020, 38. [Google Scholar] [CrossRef]
- Varjani, S.J. Microbial Degradation of Petroleum Hydrocarbons. Bioresour. Technol. 2017, 223. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Thavamani, P.; Venkateswarlu, K.; Lee, Y.B.; Naidu, R.; Megharaj, M. Remediation Approaches for Polycyclic Aromatic Hydrocarbons (PAHs) Contaminated Soils: Technological Constraints, Emerging Trends and Future Directions. Chemosphere 2017, 168. [Google Scholar] [CrossRef]
- Dell’Anno, A.; Beolchini, F.; Rocchetti, L.; Luna, G.M.; Danovaro, R. High Bacterial Biodiversity Increases Degradation Performance of Hydrocarbons during Bioremediation of Contaminated Harbor Marine Sediments. Environ. Pollut. 2012, 167, 85–92. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Bhatia, R.K.; Choi, Y.-K.; Kan, E.; Kim, Y.-G.; Yang, Y.-H. Biotechnological Potential of Microbial Consortia and Future Perspectives. Crit. Rev. Biotechnol. 2018, 38. [Google Scholar] [CrossRef]
- Maphosa, F.; de Vos, W.M.; Smidt, H. Exploiting the Ecogenomics Toolbox for Environmental Diagnostics of Organohalide-Respiring Bacteria. Trends Biotechnol. 2010, 28, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Mee, M.T.; Collins, J.J.; Church, G.M.; Wang, H.H. Syntrophic Exchange in Synthetic Microbial Communities. Proc. Natl. Acad. Sci. USA 2014, 111, E2149–E2156. [Google Scholar] [CrossRef] [Green Version]
- Morris, B.E.L.; Henneberger, R.; Huber, H.; Moissl-Eichinger, C. Microbial Syntrophy: Interaction for the Common Good. FEMS Microbiol. Rev. 2013, 37, 384–406. [Google Scholar] [CrossRef]
- Zelezniak, A.; Andrejev, S.; Ponomarova, O.; Mende, D.R.; Bork, P.; Patil, K.R. Metabolic Dependencies Drive Species Co-Occurrence in Diverse Microbial Communities. Proc. Natl. Acad. Sci. USA 2015, 112, 201522642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronen, Z.; Visnovsky, S.; Nejidat, A. Soil Extracts and Co-Culture Assist Biodegradation of 2,4,6-Tribromophenol in Culture and Soil by an Auxotrophic Achromobacter Piechaudii Strain TBPZ. Soil Biol. Biochem. 2005, 37, 1640–1647. [Google Scholar] [CrossRef]
- Tao, K.; Liu, X.; Chen, X.; Hu, X.; Cao, L.; Yuan, X. Biodegradation of Crude Oil by a Defined Co-Culture of Indigenous Bacterial Consortium and Exogenous Bacillus Subtilis. Bioresour. Technol. 2017, 224, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Abuhamed, T.; Bayraktar, E.; Mehmetoǧlu, T.; Mehmetoǧlu, Ü. Kinetics Model for Growth of Pseudomonas Putida F1 during Benzene, Toluene and Phenol Biodegradation. Process. Biochem. 2004, 39, 983–988. [Google Scholar] [CrossRef]
- Alvarez, P.J.J.; Vogel, T.M. Substrate Interactions of Benzene, Toluene, and p-Xylene During Microbial-Degradation by Pure Cultures and Mixed Culture Aquifer Slurries. Appl. Environ. Microbiol. 1991, 57, 2981–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arvin, E.; Jensen, B.K.; Gundersen, A.T. Substrate Interactions during Aerobic Biodegradation of Benzene. Appl. Environ. Microbiol. 1989, 55, 3221–3225. [Google Scholar] [CrossRef] [Green Version]
- Freedman, D.L.; Danko, A.S.; Verce, M.F. Substrate Interactions during Aerobic Biodegradation of Methane, Ethene, Vinyl Chloride and 1,2-Dichloroethenes. Water Sci. Technol. 2001, 43, 333–340. [Google Scholar] [CrossRef] [Green Version]
- McNally, D.L.; Mihelcic, J.R.; Lueking, D.R. Biodegradation of Mixtures of Polycyclic Aromatic Hydrocarbons under Aerobic and Nitrate-Reducing Conditions. Chemosphere 1999, 38, 1313–1321. [Google Scholar] [CrossRef]
- Millette, D.; Barker, J.F.; Comeau, Y.; Butler, B.J.; Frind, E.O.; Clément, B.; Samson, R. Substrate Interaction during Aerobic Biodegradation of Creosote Related Compounds: A Factorial Batch Experiment. Environ. Sci. Technol. 1995, 29, 1944–1952. [Google Scholar] [CrossRef]
- Rogers, J.B.; Reardon, K.F. Modeling Substrate Interactions during the Biodegradation of Mixtures of Toluene and Phenol by Burkholderia Species JS150. Biotechnol. Bioeng. 2000, 70, 428–435. [Google Scholar] [CrossRef]
- van Pée, K.H.; Unversucht, S. Biological Dehalogenation and Halogenation Reactions. Chemosphere 2003, 52, 299–312. [Google Scholar] [CrossRef]
- Fetzner, S.; Lingens, F. Bacterial Dehalogenases: Biochemistry, Genetics, and Biotechnological Applications. Microbiol. Rev. 1994, 58, 641–685. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Bacterial Dehalogenation. Appl. Microbiol. Biotechnol. 1998, 50, 633–657. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.K.; Bae, H. Role of Dehalogenases in Aerobic Bacterial Degradation of Chlorinated Aromatic Compounds. J. Chem. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Cincinelli, A.; Pieri, F.; Zhang, Y.; Seed, M.; Jones, K.C. Compound Specific Isotope Analysis (CSIA) for Chlorine and Bromine: A Review of Techniques and Applications to Elucidate Environmental Sources and Processes. Environ. Pollut. 2012, 169, 112–127. [Google Scholar] [CrossRef]
- Elsner, M. Stable Isotope Fractionation to Investigate Natural Transformation Mechanisms of Organic Contaminants: Principles, Prospects and Limitations. J. Environ. Monit. 2010, 12, 2005–2031. [Google Scholar] [CrossRef]
- Elsner, M.; Zwank, L. A New Concept Linking Observable Stable Isotope Fractionation to Transformation Pathways of Organic Pollutants. Environ. Sci. Technol. 2005, 39, 6896–6916. [Google Scholar] [CrossRef] [Green Version]
- Hofstetter, T.B.; Schwarzenbach, R.P.; Bernasconi, S.M. Assessing Transformation Processes of Organic Compounds Using Stable Isotope Fractionation. Environ. Sci. Technol. 2008, 42, 7737–7743. [Google Scholar] [CrossRef]
- Meckenstock, R.U.; Morasch, B.; Griebler, C.; Richnow, H.H. Stable Isotope Fractionation Analysis as a Tool to Monitor Biodegradation in Contaminated Acquifers. J. Contam. Hydrol. 2004, 75, 215–255. [Google Scholar] [CrossRef]
- Thullner, M.; Centler, F.; Richnow, H.H.; Fischer, A. Quantification of Organic Pollutant Degradation in Contaminated Aquifers Using Compound Specific Stable Isotope Analysis—Review of Recent Developments. Org. Geochem. 2012, 42, 1440–1460. [Google Scholar] [CrossRef]
- Elsner, M.; Jochmann, M.A.; Hofstetter, T.B.; Hunkeler, D.; Bernstein, A.; Schmidt, T.C.; Schimmelmann, A. Current Challenges in Compound-Specific Stable Isotope Analysis of Environmental Organic Contaminants. Anal. Bioanal. Chem. 2012, 403, 2471–2491. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Bernstein, A.; Gelman, F.; Ronen, Z. Microbial Degradation of the Brominated Flame Retardant TBNPA by Groundwater Bacteria: Laboratory and Field Study. Chemosphere 2016, 156, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Beal, J.; Farny, N.G.; Haddock-Angelli, T.; Selvarajah, V.; Baldwin, G.S.; Buckley-Taylor, R.; Gershater, M.; Kiga, D.; Marken, J.; Sanchania, V.; et al. Robust Estimation of Bacterial Cell Count from Optical Density. Commun. Biol. 2020, 3. [Google Scholar] [CrossRef]
- Kozell, A.; Yecheskel, Y.; Balaban, N.; Dror, I.; Halicz, L.; Ronen, Z.; Gelman, F. Application of Dual Carbon-Bromine Isotope Analysis for Investigating Abiotic Transformations of Tribromoneopentyl Alcohol (TBNPA). Environ. Sci. Technol. 2015, 49. [Google Scholar] [CrossRef]
- Gelman, F.; Halicz, L. High Precision Determination of Bromine Isotope Ratio by GC-MC-ICPMS. Int. J. Mass Spectrom. 2010, 289, 167–169. [Google Scholar] [CrossRef]
- Hage, J.C.; Hartmans, S. Monooxygenase-Mediated 1,2-Dichloroetbane Degradation by Pseudomonas Sp Strain DCA1. Appl. Environ. Microbiol. 1999, 65, 2466–2470. [Google Scholar] [CrossRef] [Green Version]
- McClay, K.; Fox, B.G.; Steffan, R.J. Chloroform Mineralization by Toluene-Oxidizing Bacteria. Appl. Environ. Microbiol. 1996, 62, 2716–2722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryoo, D.; Shim, H.; Canada, K.; Barbieri, P.; Wood, T.K. Aerobic Degradation of Tetrachloroethylene by Toluene-o-Xylene Monooxygenase of Pseudomonas Stutzeri OX1. Nat. Biotechnol. 2000, 18, 775–778. [Google Scholar] [CrossRef]
- Hartmans, S.; de Bont, J.A.M. Aerobic Vinyl Chloride Metabolism in Aerobic Vinyl Chloride Metabolism in Mycobacterium. Appl. Environ. Microbiol. 1992, 58, 1220–1226. [Google Scholar] [CrossRef] [Green Version]
- Vanderberg, L.A.; Perry, J.J. Dehalogenation by Mycobacterium Vaccae JOB-5: Role of the Propane Monooxygenase. Can. J. Microbiol. 1994, 40, 169–172. [Google Scholar] [CrossRef]
- Brown, L.M.; Gunasekera, T.S.; Striebich, R.C.; Ruiz, N. Draft Genome Sequence of Gordonia Sihwensis Strain 9, a Branched Alkane-Degrading Bacterium. Genome Announc. 2016, 4, 9–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Shen, J.; Wu, R.; Sun, X.; Li, J.; Han, W.; Wang, L. Biodegradation Mechanism of 1H-1,2,4-Triazole by a Newly Isolated Strain Shinella Sp. NJUST26. Sci. Rep. 2016, 6, 29675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.A.; Hyman, M.R. Oxidation of Methyl Tert-Butyl Ether by Alkane Hydroxylase in Dicyclopropylketone-Induced and n-Octane-Grown Pseudomonas Putida GPo1. Appl. Environ. Microbiol. 2004, 70. [Google Scholar] [CrossRef] [Green Version]
- Dejonghe, W.; Berteloot, E.; Goris, J.; Boon, N.; Crul, K.; Maertens, S.; Höfte, M.; de Vos, P.; Verstraete, W.; Top, E.M. Synergistic Degradation of Linuron by a Bacterial Consortium and Isolation of a Single Linuron-Degrading Variovorax Strain. Appl. Environ. Microbiol. 2003, 69, 1532–1541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzeszcz, J.; Kaszycki, P. Aerobic Bacteria Degrading Both N-Alkanes and Aromatic Hydrocarbons: An Undervalued Strategy for Metabolic Diversity and Flexibility. Biodegradation 2018, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.P.; Pan, J.C.; Ma, Y.L. Elucidation of Multiple Alkane Hydroxylase Systems in Biodegradation of Crude Oil n -alkane Pollution by Pseudomonas Aeruginosa DN1. J. Appl. Microbiol. 2020, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hennessee, C.T.; Li, Q.X. Effects of Polycyclic Aromatic Hydrocarbon Mixtures on Degradation, Gene Expression, and Metabolite Production in Four Mycobacterium Species. Appl. Environ. Microbiol. 2016, 82, 3357–3369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, N.B.; Coleman, N.v. Biodegradation of Vinyl Chloride, Cis-Dichloroethene and 1,2-Dichloroethane in the Alkene/Alkane-Oxidising Mycobacterium Strain NBB4. Biodegradation 2011, 22, 1095–1108. [Google Scholar] [CrossRef]

| ID | Strain (Highest Similarity, 99%) | Gene Bank Accession Number | OD 600 (Initial Density CFU/mL) |
|---|---|---|---|
| DB2 | Pseudomonas citronellolis | KY229738 | 0.001 (1.10 × 105) |
| DB3 | Gordonia sihwensis | KY229739 | 0.001 (1.70 × 105) |
| DB4 | Shinella zoogloeoides | KY229740 | 0.004 (4.90 × 105) |
| DB5 | Microbacterium oxydans | KY229741 | 0.027 (3.00 × 106) |
| TB1 | Pseudomonas aeruginosa | KY229734 | |
| TB2 | Delftia tsuruhatensis | KY229735 | |
| TB3 | Pseudomonas citronellolis | KY229736 | |
| TB4 | Sphingobacterium siyangense | KY229752 | |
| TB5 | Microbacterium paraoxydans | KY229753 |
| Amendment * | OD600 | Time (Days) | |
|---|---|---|---|
| TBNPA | 1 Yeast extract | 0.066 | 4 |
| 1 Vitamin mix | 0.021 | 30–60 | |
| 2 Glucose | 0.089 ± 0.0049 | 4–5 | |
| DBNPG | 1 Yeast extract | 0.079 | 4 |
| 1 Vitamin mix | 0.020 | 30–60 | |
| 2 Glucose | 0.100 ± 0.0007 | 4–5 | |
| TBNPA and DBNPG | 3 Yeast extract | 0.069 ± 0.0119 | 7 |
| 1 Vitamin mix | 0.023 | 30–60 | |
| 2 Glucose | 0.087 ± 0.0070 | 4–5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balaban, N.; Gelman, F.; Taylor, A.A.; Walker, S.L.; Bernstein, A.; Ronen, Z. Degradation of Brominated Organic Compounds (Flame Retardants) by a Four-Strain Consortium Isolated from Contaminated Groundwater. Appl. Sci. 2021, 11, 6263. https://doi.org/10.3390/app11146263
Balaban N, Gelman F, Taylor AA, Walker SL, Bernstein A, Ronen Z. Degradation of Brominated Organic Compounds (Flame Retardants) by a Four-Strain Consortium Isolated from Contaminated Groundwater. Applied Sciences. 2021; 11(14):6263. https://doi.org/10.3390/app11146263
Chicago/Turabian StyleBalaban, Noa, Faina Gelman, Alicia A. Taylor, Sharon L. Walker, Anat Bernstein, and Zeev Ronen. 2021. "Degradation of Brominated Organic Compounds (Flame Retardants) by a Four-Strain Consortium Isolated from Contaminated Groundwater" Applied Sciences 11, no. 14: 6263. https://doi.org/10.3390/app11146263
APA StyleBalaban, N., Gelman, F., Taylor, A. A., Walker, S. L., Bernstein, A., & Ronen, Z. (2021). Degradation of Brominated Organic Compounds (Flame Retardants) by a Four-Strain Consortium Isolated from Contaminated Groundwater. Applied Sciences, 11(14), 6263. https://doi.org/10.3390/app11146263







