Automatic Diagnosis of Coronary Artery Disease in SPECT Myocardial Perfusion Imaging Employing Deep Learning
Abstract
:1. Introduction
1.1. Deep Learning in Image Analysis
1.2. Machine Learning and Deep Learning in SPECT Nuclear Cardiology Imaging
1.3. Contribution of This Research Work
2. Materials and Methods
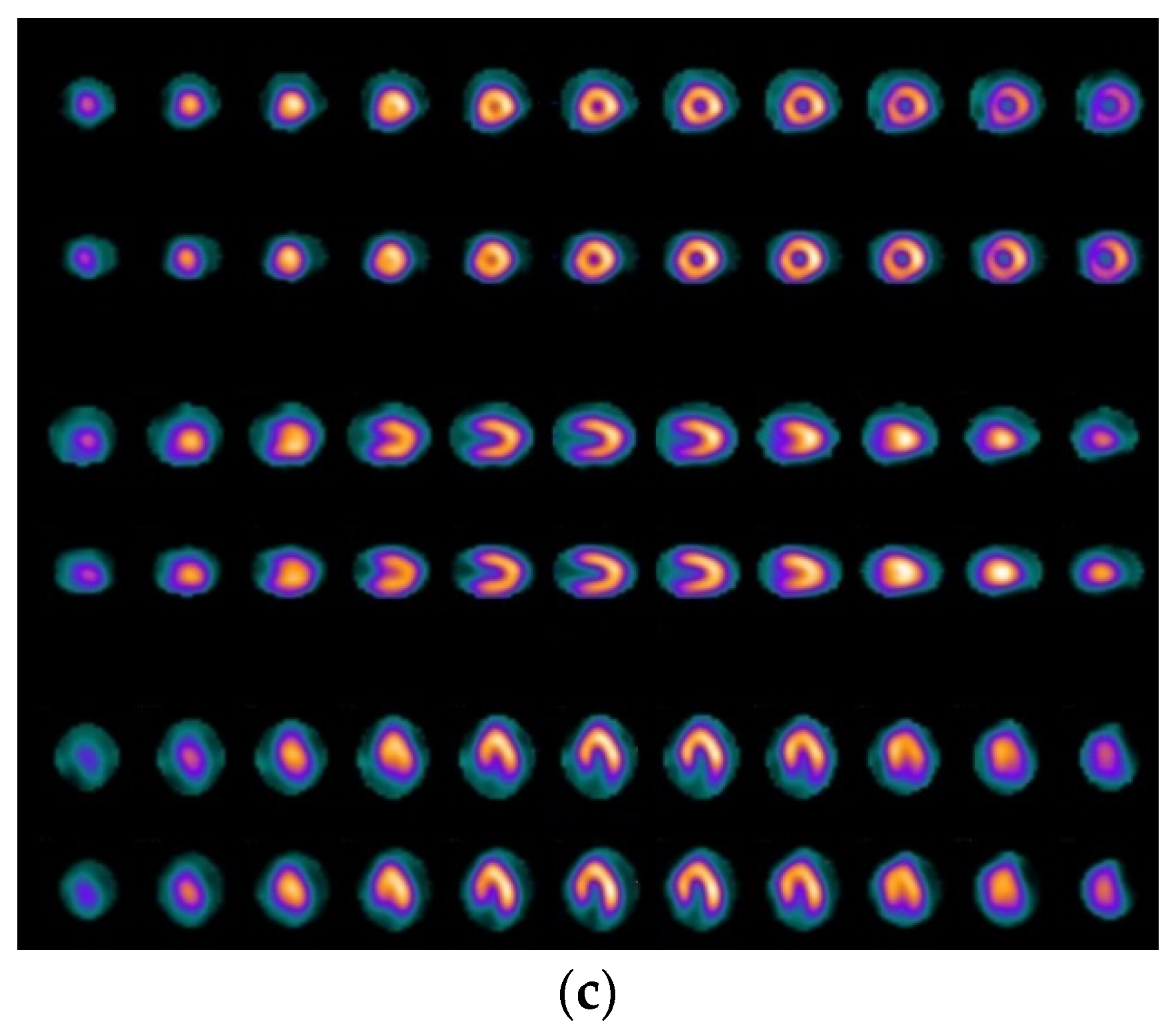
2.1. Patients and Imaging Protocol
2.2. Visual Assessment
2.3. Overview of Convolutional Neural Networks
2.4. Methodology
3. Methodology
3.1. RGB-Based CNN Architecture for Classification in Nuclear Medical Imaging
3.2. Deep Learning Models, Including Transfer Learning for CAD Classification in Medical Imaging
4. Results
4.1. Results from RGB-CNN
4.2. Results from Deep Learning Architectures Applying Transfer Learning and Comparative Analysis
- VGG16: pixel size (300 × 300 × 3), batch size = 32, dropout = 0.2, Global Average Poolong2D, dense nodes 64 × 64, epochs = 400, (average run time = 1853 s),
- DenseNet: pixel size (250 × 250 × 3), batch size = 8, dropout = 0.2, Global Average Poolong2D, dense nodes 16 × 16, epochs = 400, (average run time = 2074 s),
- MobileNet: pixel size (250 × 250 × 3), batch size = 8, dropout = 0.2, Global Average Poolong2D, dense nodes 32 × 32, epochs = 400, (average run time = 3070 s),
- InceptionV3: pixel size (300 × 300 × 3), batch size = 8, dropout = 0.2, Global Average Poolong2D, dense nodes 256 × 256, epochs = 400, (average run time = 1538 s).
5. Discussion of Results and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cassar, A.; Holmes, D.R.; Rihal, C.S.; Gersh, B.J. Chronic Coronary Artery Disease: Diagnosis and Management. Mayo Clin. Proc. 2009, 84, 1130–1146. [Google Scholar] [CrossRef] [Green Version]
- Ross, R. Atherosclerosis—An Inflammatory Disease. N. Engl. J. Med. 1999, 340, 115–126. [Google Scholar] [CrossRef]
- Girelli, M.; Martinelli, N.; Peyvandi, F.; Olivieri, O. Genetic Architecture of Coronary Artery Disease in the Genome-Wide Era: Implications for the Emerging “Golden Dozen” Loci. Semin. Thromb. Hemost. 2009, 35, 671–682. [Google Scholar] [CrossRef]
- Álvarez-Álvarez, M.M.; Zanetti, D.; Carreras-Torres, R.; Moral, P.; Athanasiadis, G. A survey of sub-Saharan gene flow into the Mediterranean at risk loci for coronary artery disease. Eur. J. Hum. Genet. 2017, 25, 472–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Łukaszewski, B.; Nazar, J.; Goch, M.; Łukaszewska, M.; Stępiński, A.; Jurczyk, M. Diagnostic methods for detection of bone metastases. Współczesna Onkol. 2017, 21, 98–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartor, O. Radium and targeted alpha therapy in prostate cancer: New data and concepts. Ann. Oncol. 2020, 31, 165–166. [Google Scholar] [CrossRef] [PubMed]
- Underwood, S.R.; Anagnostopoulos, C.; Cerqueira, M.; Ell, P.J.; Flint, E.J.; Harbinson, M.; Kelion, A.D.; Al-Mohammad, A.; Prvulovich, E.M.; Shaw, L.J.; et al. Myocardial perfusion scintigraphy: The evidence. Eur. J. Nucl. Med. Mol. Imaging 2004, 31, 261–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuijf, J.D.; Poldermans, D.; Shaw, L.J.; Jukema, J.W.; Lamb, H.J.; De Roos, A.; Wijns, W.; Van Der Wall, E.E.; Bax, J.J. Diagnostic and prognostic value of non-invasive imaging in known or suspected coronary artery disease. Eur. J. Nucl. Med. Mol. Imaging 2005, 33, 93–104. [Google Scholar] [CrossRef]
- Talbot, J.N.; Paycha, F.; Balogova, S. Diagnosis of bone metastasis: Recent comparative studies of imaging modalities. Q. J. Nucl. Med. Mol. Imaging 2011, 55, 374–410. [Google Scholar] [PubMed]
- Doi, K. Computer-Aided Diagnosis in Medical Imaging: Historical Review, Current Status and Future Poten-tial. Computerized medical imaging and graphics. Off. J. Comput. Med. Imaging Soc. 2007, 31, 198–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, G.J.; Carty, F.L.; Cronin, C.G. Imaging of Bone Metastasis: An Update. World J. Radiol. 2015, 7, 202–211. [Google Scholar] [CrossRef]
- Chang, C.Y.; Gill, C.M.; Simeone, F.J.; Taneja, A.K.; Huang, A.J.; Torriani, M.; A Bredella, M. Comparison of the diagnostic accuracy of 99 m-Tc-MDP bone scintigraphy and 18 F-FDG PET/CT for the detection of skeletal metastases. Acta Radiol. 2016, 57, 58–65. [Google Scholar] [CrossRef]
- Wyngaert, T.V.D.; On behalf of the EANM Bone & Joint Committee and the Oncology Committee; Strobel, K.; Kampen, W.U.; Kuwert, T.; Van Der Bruggen, W.; Mohan, H.K.; Gnanasegaran, G.; Bolton, R.D.; Weber, W.A.; et al. The EANM practice guidelines for bone scintigraphy. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 1723–1738. [Google Scholar] [CrossRef] [Green Version]
- Coleman, R. Metastatic bone disease: Clinical features, pathophysiology and treatment strategies. Cancer Treat. Rev. 2001, 27, 165–176. [Google Scholar] [CrossRef]
- Savvopoulos, C.A.; Spyridonidis, T.; Papandrianos, N.; Vassilakos, P.J.; Alexopoulos, D.; Apostolopoulos, D.J. CT-based attenuation correction in Tl-201 myocardial perfusion scintigraphy is less effective than non-corrected SPECT for risk stratification. J. Nucl. Cardiol. 2014, 21, 519–531. [Google Scholar] [CrossRef]
- Malek, H. Nuclear Cardiology. In Practical Cardiology; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 167–172. [Google Scholar]
- American Heart Association Editorial Staff. Myocardial Perfusion Imaging (MPI) Test. Available online: https://www.heart.org/en/health-topics/heart-attack/diagnosing-a-heart-attack/myocardial-perfusion-imaging-mpi-test (accessed on 15 March 2021).
- Alexanderson, E.; Better, N.; Bouyoucef, S.-E.; Dondi, M.; Dorbala, S.; Einstein, A.J.; El-Haj, N.; Giubbini, R.; Keng, F.; Kumar, A.; et al. Nuclear Cardiology: Guidance on the Implementation of SPECT Myocardial Perfusion Imaging; Human Health Series; International Atomic Energy Agency: Vienna, Australia, 2016. [Google Scholar]
- Slart, R.H.J.A.; Williams, M.C.; Juarez-Orozco, L.E.; Rischpler, C.; Dweck, M.R.; Glaudemans, A.W.J.M.; Gimelli, A.; Georgoulias, P.; Gheysens, O.; Gaemperli, O.; et al. Position paper of the EACVI and EANM on artificial intelligence applications in multimodality cardiovascular imaging using SPECT/CT, PET/CT, and cardiac CT. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Sartor, A.O.; DiBiase, S.J. Bone Metastases in Advanced Prostate Cancer: Management. 2018. Available online: https://www.uptodate.com/contents/bone-metastases-in-advanced-prostate-cancer-management (accessed on 7 July 2021).
- Johansson, L.; Edenbrandt, L.; Nakajima, K.; Lomsky, M.; Svensson, S.-E.; Trägårdh, E. Computer-aided diagnosis system outperforms scoring analysis in myocardial perfusion imaging. J. Nucl. Cardiol. 2014, 21, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Slomka, P.J.; Betancur, J.; Liang, J.X.; Otaki, Y.; Hu, L.-H.; Sharir, T.; Dorbala, S.; Di Carli, M.; Fish, M.B.; Ruddy, T.D.; et al. Rationale and design of the REgistry of Fast Myocardial Perfusion Imaging with NExt generation SPECT (REFINE SPECT). J. Nucl. Cardiol. 2018, 27, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Orozco, L.E.; Martinez-Manzanera, O.; Storti, A.E.; Knuuti, J. Machine Learning in the Evaluation of Myocardial Ischemia Through Nuclear Cardiology. Curr. Cardiovasc. Imaging Rep. 2019, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Suri, J.S. State-of-the-art review on deep learning in medical imaging. Front. Biosci. 2019, 24, 392–426. [Google Scholar] [CrossRef]
- Lundervold, A.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Abdelhafiz, D.; Yang, C.; Ammar, R.; Nabavi, S. Deep convolutional neural networks for mammography: Advances, challenges and applications. BMC Bioinform. 2019, 20, 281. [Google Scholar] [CrossRef] [Green Version]
- Sadik, M.; Hamadeh, I.; Nordblom, P.; Suurkula, M.; Höglund, P.; Ohlsson, M.; Edenbrandt, L. Computer-Assisted Interpretation of Planar Whole-Body Bone Scans. J. Nucl. Med. 2008, 49, 1958–1965. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, H.; Kikuchi, A.; Onoguchi, M.; Sjöstrand, K.; Edenbrandt, L. Computer-aided diagnosis system for bone scintigrams from Japanese patients: Importance of training database. Ann. Nucl. Med. 2012, 26, 622–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koizumi, M.; Miyaji, N.; Murata, T.; Motegi, K.; Miwa, K.; Koyama, M.; Terauchi, T.; Wagatsuma, K.; Kawakami, K.; Richter, J. Evaluation of a revised version of computer-assisted diagnosis system, BONENAVI version 2.1.7, for bone scintigraphy in cancer patients. Ann. Nucl. Med. 2015, 29, 659–665. [Google Scholar] [CrossRef]
- Komeda, Y.; Handa, H.; Watanabe, T.; Nomura, T.; Kitahashi, M.; Sakurai, T.; Okamoto, A.; Minami, T.; Kono, M.; Arizumi, T.; et al. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology 2017, 93, 30–34. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [Green Version]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Feleki, A. A Deep-Learning Approach for Diagnosis of Metastatic Breast Cancer in Bones from Whole-Body Scans. Appl. Sci. 2020, 10, 997. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Chen, S.; Qin, J.; Liu, Y.; Huang, B.; Chen, H. Application of Deep Learning in Automated Analysis of Molecular Images in Cancer: A Survey. Contrast Media Mol. Imaging 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Jackel, L.; Bottou, L.; Cortes, C.; Denker, J.; Drucker, H.; Guyon, I.; Muller, U.; Sackinger, E.; Simard, P.; et al. Learning Algorithms for Classification: A Comparison on Handwritten Digit Recognition. 1995. Available online: http://yann.lecun.com/exdb/publis/pdf/lecun-95a.pdf (accessed on 7 July 2021).
- Qian, N. On the momentum term in gradient descent learning algorithms. Neural Netw. 1999, 12, 145–151. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks BT—Computer Vision–ECCV 2014. In Proceedings of the European Conference on Computer Vision (ECCV); Springer: Berlin/Heidelberg, Germany, 2014; pp. 818–833. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Guerra, E.; de Lara, J.; Malizia, A.; Díaz, P. Supporting user-oriented analysis for multi-view domain-specific visual languages. Inf. Softw. Technol. 2009, 51, 769–784. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Arsanjani, R.; Xu, Y.; Dey, D.; Vahistha, V.; Shalev, A.; Nakanishi, R.; Hayes, S.; Fish, M.; Berman, D.; Germano, G.; et al. Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J. Nucl. Cardiol. 2013, 20, 553–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.-H.; Betancur, J.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; A Kaufmann, P.; Sinusas, A.J.; Miller, E.J.; et al. Machine learning predicts per-vessel early coronary revascularization after fast myocardial perfusion SPECT: Results from multicentre REFINE SPECT registry. Eur. Hear. J. Cardiovasc. Imaging 2020, 21, 549–559. [Google Scholar] [CrossRef]
- Lomsky, M.; Gjertsson, P.; Johansson, L.; Richter, J.; Ohlsson, M.; Tout, D.; Van Aswegen, A.; Underwood, S.R.; Edenbrandt, L. Evaluation of a decision support system for interpretation of myocardial perfusion gated SPECT. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 1523–1529. [Google Scholar] [CrossRef]
- Rahmani, R.; Niazi, P.; Naseri, M.; Neishabouri, M.; Farzanefar, S.; Eftekhari, M.; Derakhshan, F.; Mollazadeh, R.; Meysami, A.; Abbasi, M. Precisión diagnóstica mejorada para la imagen de perfusión miocárdica usando redes neuronales artificiales en diferentes variables de entrada incluyendo datos clínicos y de cuantificación. Rev. Española Med. Nucl. Imagen Mol. 2019, 38, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Arsanjani, R.; Xu, Y.; Dey, D.; Fish, M.; Dorbala, S.; Hayes, S.; Berman, D.; Germano, G.; Slomka, P. Improved Accuracy of Myocardial Perfusion SPECT for the Detection of Coronary Artery Disease Using a Support Vector Machine Algorithm. J. Nucl. Med. 2013, 54, 549–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Güner, L.A.; Karabacak, N.I.; Akdemir, O.U.; Karagoz, P.S.; Kocaman, S.A.; Cengel, A.; Ünlü, M. An open-source framework of neural networks for diagnosis of coronary artery disease from myocardial perfusion SPECT. J. Nucl. Cardiol. 2010, 17, 405–413. [Google Scholar] [CrossRef]
- Betancur, J.; Hu, L.-H.; Commandeur, F.; Sharir, T.; Einstein, A.J.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.A.; Sinusas, A.J.; Miller, E.J.; et al. Deep Learning Analysis of Upright-Supine High-Efficiency SPECT Myocardial Perfusion Imaging for Prediction of Obstructive Coronary Artery Disease: A Multicenter Study. J. Nucl. Med. 2018, 60, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Betancur, J.; Commandeur, F.; Motlagh, M.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.; Sinusas, A.J.; et al. Deep Learning for Prediction of Obstructive Disease From Fast Myocardial Perfusion SPECT. JACC: Cardiovasc. Imaging 2018, 11, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Spier, N.; Nekolla, S.G.; Rupprecht, C.; Mustafa, M.; Navab, N.; Baust, M. Classification of Polar Maps from Cardiac Perfusion Imaging with Graph-Convolutional Neural Networks. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berkaya, S.K.; Sivrikoz, I.A.; Gunal, S. Classification models for SPECT myocardial perfusion imaging. Comput. Biol. Med. 2020, 123, 103893. [Google Scholar] [CrossRef]
- Verberne, H.J.; Acampa, W.; Anagnostopoulos, C.D.; Ballinger, J.R.; Bengel, F.; De Bondt, P.; Buechel, R.R.; Cuocolo, A.; Van Eck-Smit, B.L.F.; Flotats, A.; et al. EANM procedural guidelines for radionuclide myocardial perfusion imaging with SPECT and SPECT/CT: 2015 revision. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 1929–1940. [Google Scholar] [CrossRef] [Green Version]
- Arsanjani, R.; Dey, D.; Khachatryan, T.; Shalev, A.; Hayes, S.W.; Fish, M.; Nakanishi, R.; Germano, G.; Berman, D.S.; Slomka, P. Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population. J. Nucl. Cardiol. 2014, 22, 877–884. [Google Scholar] [CrossRef]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Papageorgiou, K. Bone metastasis classification using whole body images from prostate cancer patients based on convolutional neural networks application. PLoS ONE 2020, 15, e0237213. [Google Scholar] [CrossRef]
- Papandrianos, N.; Papageorgiou, E.I.; Anagnostis, A. Development of Convolutional Neural Networks to identify bone metastasis for prostate cancer patients in bone scintigraphy. Ann. Nucl. Med. 2020, 34, 824–832. [Google Scholar] [CrossRef]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Papageorgiou, K. Efficient Bone Metastasis Diagnosis in Bone Scintigraphy Using a Fast Convolutional Neural Network Architecture. Diagnostics 2020, 10, 532. [Google Scholar] [CrossRef]
- Papandrianos, N.; Alexiou, S.; Xouria, X.; Apostolopoulos, D.J. Atypical Bilateral Stress Fractures of the Femoral Shaft Diagnosed by Bone Scintigraphy in a Woman With Osteoporosis. Clin. Nucl. Med. 2013, 38, 910–912. [Google Scholar] [CrossRef]
- O’Shea, K.T.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net. arXiv 2014, arXiv:1412.6806. [Google Scholar]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Colaboratory Cloud Environment Supported by Google. Available online: https://colab.research.google.com/ (accessed on 7 July 2021).













| Pixels | Dense Nodes | Val. Acc. | Val. Loss | Test Acc. | Test Loss | AUC | Time (s) |
|---|---|---|---|---|---|---|---|
| 200 × 200 × 3 | 32–32 | 90.12 | 0.25 | 89.7 | 0.31 | 0.916 | 890 |
| 64–64 | 93.41 | 0.2 | 92.64 | 0.21 | 0.935 | 831 | |
| 128–128 | 89.05 | 0.36 | 89.21 | 0.33 | 0.871 | 860 | |
| 250 × 250 × 3 | 32–32 | 94.72 | 0.16 | 93.62 | 0.12 | 0.926 | 1125 |
| 64–64 | 92.53 | 0.25 | 93.47 | 0.13 | 0.921 | 1116 | |
| 128–128 | 82.89 | 0.44 | 80.87 | 0.18 | 0.845 | 1043 | |
| 300 × 300 × 3 | 32–32 | 90.78 | 0.17 | 88.23 | 0.28 | 0.9025 | 1736 |
| 64–64 | 85.3 | 0.36 | 86.47 | 0.29 | 0.893 | 1641 | |
| 128–128 | 86.84 | 0.45 | 73.52 | 0.69 | 0.716 | 1469 | |
| 350 × 350 × 3 | 32–32 | 78.94 | 0.51 | 70.58 | 0.61 | 0.78 | 2200 |
| 64–64 | 80.52 | 0.47 | 68.35 | 0.62 | 0.765 | 2221 | |
| 128–128 | 74.21 | 0.57 | 65.43 | 0.71 | 0.711 | 2185 |
| Pixels | Dense Nodes | Val. Acc. | Val. Loss | Test Acc. | Test Loss | AUC | Time (s) |
|---|---|---|---|---|---|---|---|
| 200 × 200 × 3 | 32–32 | 92.73 | 0.183 | 92.15 | 0.24 | 0.885 | 748 |
| 64–64 | 92.53 | 0.24 | 93.13 | 0.265 | 0.948 | 679 | |
| 128–128 | 91.72 | 0.23 | 89.91 | 0.22 | 0.885 | 674 | |
| 250 × 250 × 3 | 32–32 | 94.73 | 0.12 | 91.17 | 0.22 | 0.769 | 990 |
| 64–64 | 91.21 | 0.235 | 90.36 | 0.26 | 0.873 | 971 | |
| 128–128 | 92.1 | 0.195 | 90.3 | 0.21 | 0.898 | 1089 | |
| 300 × 300 × 3 | 32–32 | 94.73 | 0.158 | 91.66 | 0.21 | 0.920 | 1547 |
| 64–64 | 91.42 | 0.24 | 91.905 | 0.183 | 0.93 | 1387 | |
| 128–128 | 92.03 | 0.19 | 92.01 | 0.218 | 0.915 | 1409 | |
| 350 × 350 × 3 | 32–32 | 88.46 | 0.31 | 87.74 | 0.29 | 0.871 | 1854 |
| 64–64 | 89.47 | 0.325 | 91.17 | 0.198 | 0.887 | 1856 | |
| 128–128 | 92.1 | 0.185 | 92.01 | 0.205 | 0.914 | 1910 |
| Pixels | Dense Nodes | Val. Acc. | Val. Loss | Test Acc. | Test Loss | AUC | Time (s) |
|---|---|---|---|---|---|---|---|
| 200 × 200 × 3 | 32–32 | 87.71 | 0.323 | 92.15 | 0.31 | 0.935 | 630 |
| 64–64 | 90.34 | 0.33 | 91.9 | 0.3 | 0.931 | 790 | |
| 128–128 | 93.82 | 0.18 | 92.04 | 0.245 | 0.923 | 707 | |
| 250 × 250 × 3 | 32–32 | 90.78 | 0.2 | 91.17 | 0.21 | 0.855 | 1110 |
| 64–64 | 89.91 | 0.253 | 93.12 | 0.187 | 0.921 | 1065 | |
| 128–128 | 89.47 | 0.263 | 89.69 | 0.305 | 0.909 | 1039 | |
| 300 × 300 × 3 | 32–32 | 88.59 | 0.29 | 90.19 | 0.276 | 0.917 | 1440 |
| 64–64 | 89.46 | 0.224 | 91.15 | 0.24 | 0.907 | 1569 | |
| 128–128 | 92.1 | 0.21 | 91.37 | 0.2 | 0.914 | 1573 | |
| 350 × 350 × 3 | 32–32 | 87.28 | 0.26 | 88.57 | 0.23 | 0.898 | 1650 |
| 64–64 | 90.78 | 0.38 | 86.77 | 0.42 | 0.854 | 2077 | |
| 128–128 | 89.47 | 0.295 | 91.17 | 0.245 | 0.898 | 1980 |
| 2-Classes | Abnormal | Normal |
|---|---|---|
| Abnormal | 26 | 0 |
| Normal | 1 | 7 |
| Reference | Input Data | ML/DL Methods | Classification Problem | Reference Standard | Results |
|---|---|---|---|---|---|
| Quantitative Data and Clinical Data from Polar Maps | |||||
| Arsanjani et al. [48] | Quantitative data | Boosted ensemble learning | Normal/Abnormal | Coronary Angiography | Stress TPD: Accuracy 0.88, AUC 0.94, Specificity 093, Sensitivity 0.81 |
| Hu et al. [45], 2020 | Clinical and quantitative data | Ensemble LogitBoost algorithm (ML) | Normal/Abnormal | Coronary Angiography | Accuracy 0.72, AUC 0.79. |
| Rahmani et al. [47], 2019 | Clinical and Quantitative data | Feed-forward ANN (multi-layer perceptron) | Absence/Presence of coronary artery stenosis >50% in at least a vessel | Coronary Angiography | Accuracy 0.86, Specificity 1.00, Sensitivity 0.82 |
| Feed-forward ANN | (2 classes) Normal/Abnormal angiography result | Coronary Angiography | Accuracy 0.93, Specificity 1.00, Sensitivity 0.92 | ||
| Arsanjani et al. [55] | Quantitative and functional data | SVM | Absence/Presence of coronary artery stenosis ≥70% | Coronary Angiography | Stress TPD: Accuracy 0.86, AUC 0.92, Specificity 0.88, Sensitivity 0.84 |
| Guner et al. [49], 2010 | Quantitative polar maps | ANN: multilayer perceptron (WEKA), input layer of 50 nodes, one hidden layer of 5 nodes and one output node. | Absence/Presence of coronary artery stenosis | Coronary Angiography | AUC 0.74, Specificity 0.68, Sensitivity 0.71 |
| Polar Maps (Images) | |||||
| Betancur et al. [50], 2019 | Upright and supine polar maps | DL: 3 × (Conv.Layers, Relu, Max-Pooling filter) and 3 fully connected layers. | Presence of coronary artery stenosis ≥70% and >50% in left main coronary artery (Normal/Abnormal) | Coronary Angiography | The AUC score, specificity and sensitivity were 81%, 83%, and 66% |
| Betancur et al. [51], 2018 | Raw and quantitative polar maps | Deep CNN: 3 × (Conv.Layers, Relu, Max-Pooling filter), 3 fully connected layers, followed by 3 parallel outputs—1 per coronary territory | Presence of coronary artery stenosis ≥70% (Normal/Abnormal) | Coronary Angiography | The AUC score, specificity and sensitivity were 80%, 58%, and 82% |
| Spier et al. [52], 2019 | Polar maps | Graph-based CNN, Chebyshev | Presence of coronary artery stenosis (Normal/Abnormal) | Expert Reader | The accuracy, specificity and sensitivity were 91%, 96%, and 86% |
| 2D SPECT MPI images | |||||
| Berkaya et al. [53], 2020 | SPECT MPI images | SVM with deep features, VGG-19 | (2 classes) Normal, Abnormal | Expert Reader | The accuracy, specificity and sensitivity were 79%, 57%, and 100% |
| SVM with shallow features, VGG-19 | (2 classes) Normal, Abnormal | Expert Reader | The accuracy, specificity and sensitivity were 94%, 100%, and 88% | ||
| Transfer learning, VGG-16 | (2 classes) Normal, Abnormal | Expert Reader | The accuracy, AUC, specificity and sensitivity were 86%, 92%, 71%, and 100% | ||
| Knowledge-based | (2 classes) Normal, Abnormal | Expert Reader | The accuracy, specificity and sensitivity were 93%, 86%, and 100% | ||
| Proposed work | SPECT MPI images | RGB-CNN batch size = 8, dropout = 0.2, conv 16–32–64–128–256, dense nodes 32.32 | (2 classes) Normal, Abnormal | Coronary Angiography | The accuracy, AUC, specificity and sensitivity were 94%, 93%, 78%, and 94% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papandrianos, N.; Papageorgiou, E. Automatic Diagnosis of Coronary Artery Disease in SPECT Myocardial Perfusion Imaging Employing Deep Learning. Appl. Sci. 2021, 11, 6362. https://doi.org/10.3390/app11146362
Papandrianos N, Papageorgiou E. Automatic Diagnosis of Coronary Artery Disease in SPECT Myocardial Perfusion Imaging Employing Deep Learning. Applied Sciences. 2021; 11(14):6362. https://doi.org/10.3390/app11146362
Chicago/Turabian StylePapandrianos, Nikolaos, and Elpiniki Papageorgiou. 2021. "Automatic Diagnosis of Coronary Artery Disease in SPECT Myocardial Perfusion Imaging Employing Deep Learning" Applied Sciences 11, no. 14: 6362. https://doi.org/10.3390/app11146362






