Abstract
Recently, digital pathology is an essential application for clinical practice and medical research. Due to the lack of large annotated datasets, the deep transfer learning technique is often used to classify histopathology images. A softmax classifier is often used to perform classification tasks. Besides, a Support Vector Machine (SVM) classifier is also popularly employed, especially for binary classification problems. Accurately determining the category of the histopathology images is vital for the diagnosis of diseases. In this paper, the conventional softmax classifier and the SVM classifier-based transfer learning approach are evaluated to classify histopathology cancer images in a binary breast cancer dataset and a multiclass lung and colon cancer dataset. In order to achieve better classification accuracy, a methodology that attaches SVM classifier to the fully-connected (FC) layer of the softmax-based transfer learning model is proposed. The proposed architecture involves a first step training the newly added FC layer on the target dataset using the softmax-based model and a second step training the SVM classifier with the newly trained FC layer. Cross-validation is used to ensure no bias for the evaluation of the performance of the models. Experimental results reveal that the conventional SVM classifier-based model is the least accurate on either binary or multiclass cancer datasets. The conventional softmax-based model shows moderate classification accuracy, while the proposed synthetic architecture achieves the best classification accuracy.
1. Introduction
In medical practices, digital pathology is gaining momentum, which focuses on management and analysis of the information generated by the digitalized specimen slides, due to the rapid progress in scanning technologies [1]. Its applications are spreading across diagnostic medicine and disease prediction. By incorporating AI and machine learning, digital pathology retains great power for clinical application and biomedical research [2,3,4,5].
Accurately classifying histopathology images is an important task in clinical practice to gain a reliable diagnosis of diseases. With the help of machine learning, in particular transfer learning, this kind of task can be automated to replace the tedious and expensive labor work of human experts and suit the demands for high accuracy, large data scales, and efficient computation. Due to the lack of large publicly available, annotated digitized slides, transfer learning is commonly used. Transfer learning addresses the cross-domain learning problems by transferring helpful information from the source domain to the task domain, and it is actively applied in visual categorization [6]. Different transfer learning techniques are classified in [6] into feature representation transfer, including cross-domain knowledge transfer and cross-view knowledge transfer, and classifier-based knowledge transfer, including SVM-based, TrAdaboost, and generative models. The employment of deep transfer learning is prevalent due to its superior performance and flexibility [7,8,9,10,11].
Support Vector Machine (SVM) is a supervised learning model initially designed for binary classification tasks [12,13] and has high-quality generalization ability for binary classification problems [14,15]. SVM can be extended to solve multinomial classification problems employing Error-Correcting Output Code (ECOC) [16,17,18]. The outcome is, however, not as good compared with binary problems. SVM is also actively used as the classifier in deep transfer learning. In [2], different deep transfer learning strategies for digital pathology are comprehensively compared on eight classification datasets, where SVM shows promising results when used as the classifier. However, some other researchers claim the advantage of SVM on binary problems is not always evident. The study in [19] tests three backend deep learning architectures, namely VGG, ResNet, and Inception, as feature extractors and three different classifiers, namely FC multilayer, SVM, and Random Forests, on four datasets in digital pathology, and evaluates reproducibility by examining the issue of evaluating the accuracy of predictive models. The emerging AI-based computational pathology has shown great promise in increasing the accuracy of high-quality health care, and contributes insights to the diagnosis and treatment of diseases [20]. Deep learning has been used for analysis of histology images, tumor detection, grading and subtyping, prediction of mutations, survival and response from histology, and largely automates clinical workflows [21].
In this paper, a transfer learning methodology combining the FC layer trained by a softmax classifier on the target dataset with the SVM classifier is proposed to classify histopathology images. Due to the shortages of the specific publicly available annotated histopathology dataset, some previous researches use transfer learning based on off-the-shelf deep CNN architectures, pre-trained on ImageNet as the backbone for classifying histopathology images. A softmax classifier is commonly used in transfer learning which is effective for binary or multinomial classification. An SVM classifier is, however, usually confined to binary classification. The proposed methodology contains a two-step procedure. It first uses AlexNet pre-trained on ImageNet as the backbone in the transferred layers and softmax classifier, and trained on the target dataset, then the SVM classifier is attached to the already trained FC layer in the first step, and the network is trained a second time. Experiments are performed using four-fold cross-validation for the softmax-based model, the SVM-based model and the proposed model, on a binary breast cancer image dataset and a multiclass lung and colon cancer dataset, and the obtained results show evident improvement from the proposed synthetic architecture in classification accuracy compared with the softmax classifier and the SVM classifier used individually.
2. Introduction of Background Knowledge
2.1. Transfer Learning
Transfer learning is a popular machine learning method where a model developed for one task is reused as the starting point for the model on a different task. It can transfer already obtained knowledge to new conditions. In deep learning, transfer learning means using the networks pre-trained on one large dataset as the starting point to construct a new network architecture that can be used on a new dataset with fine-tuning. This significantly reduces the effort for training and is usually much faster and easier than constructing and training a network from scratch, given the vast resources required to train the deep CNN.
The deep learning models pre-trained for a large and challenging image classification task, ImageNet competition, are commonly used to perform transfer learning. In this paper, pre-trained AlexNet architecture is used. In order to transfer the richly learned features from AlexNet to a new image classification task, the tail part of the network is cut off and replaced with a new classifier, softmax, for example, to suit the need for the new task. The rest of the newly formed transfer network structure is the same as the pre-trained network except for the last couple of layers. The weights in the transferred layers are kept frozen while training the newly constructed network. Thus, the knowledge learned from the pre-trained dataset images can be transferred to the new task.
2.2. AlexNet Architecture
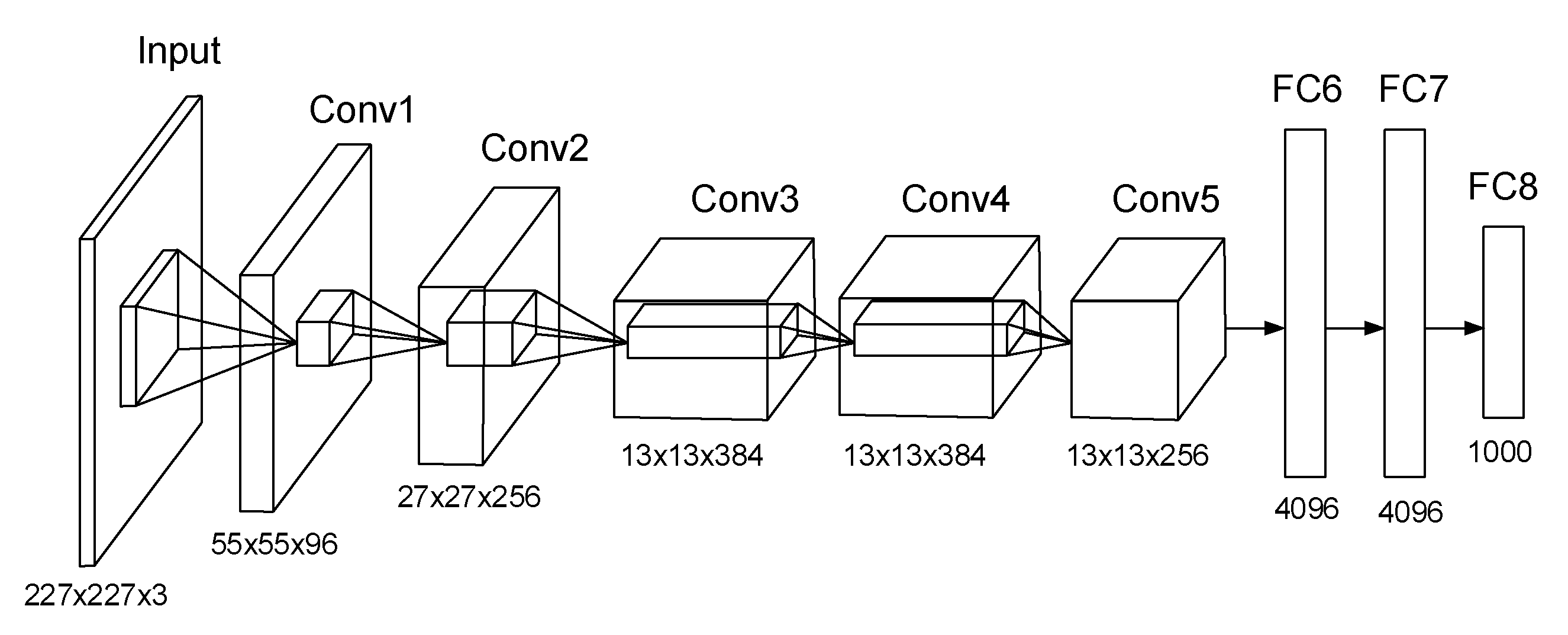
AlexNet is a well-known fast GPU implantation of CNN developed by Alex Krizhevsky, which won the ImageNet contest in 2012. The capability of AlexNet to achieve high accuracy on very challenging datasets is incredible. It has been trained to classify more than a million images on the ImageNet dataset into a thousand different classes and learned rich features from those images; the results on test data are 37.5% top-1 error rates and 17.0% top-5 error rates. The network has over 60 million parameters and 65,000 neurons, and it took around a week to train on two GTX 580 GPUs. The ImageNet is a large image database designed for visual object recognition research. It contains over 14 million annotated images with more than 20,000 categories.
The architecture of AlexNet is illustrated in Figure 1. It consists of five convolutional layers and three fully connected (FC) layers. In the first convolutional layer, there are 96 kernels of size 11 × 11 × 3. Likewise, in other convolutional layers, there are many kernels of the same size. Max pooling layers are appended to the first, the second, and the fifth convolutional layers. The last FC layer feeds into a final 1000-way softmax.
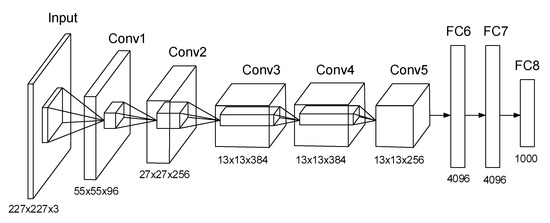
Figure 1.
Architecture of AlexNet.
Rectified Linear Unit (ReLU) is a critical feature of the AlexNet instead of using the tanh function. By using ReLU nonlinearity, AlexNet could be trained much faster than using the saturating activation functions. Besides dropout, data augmentation, including image translation and reflection, altering the intensities of the RGB channels are employed to prevent overfitting.
2.3. Support Vector Machine
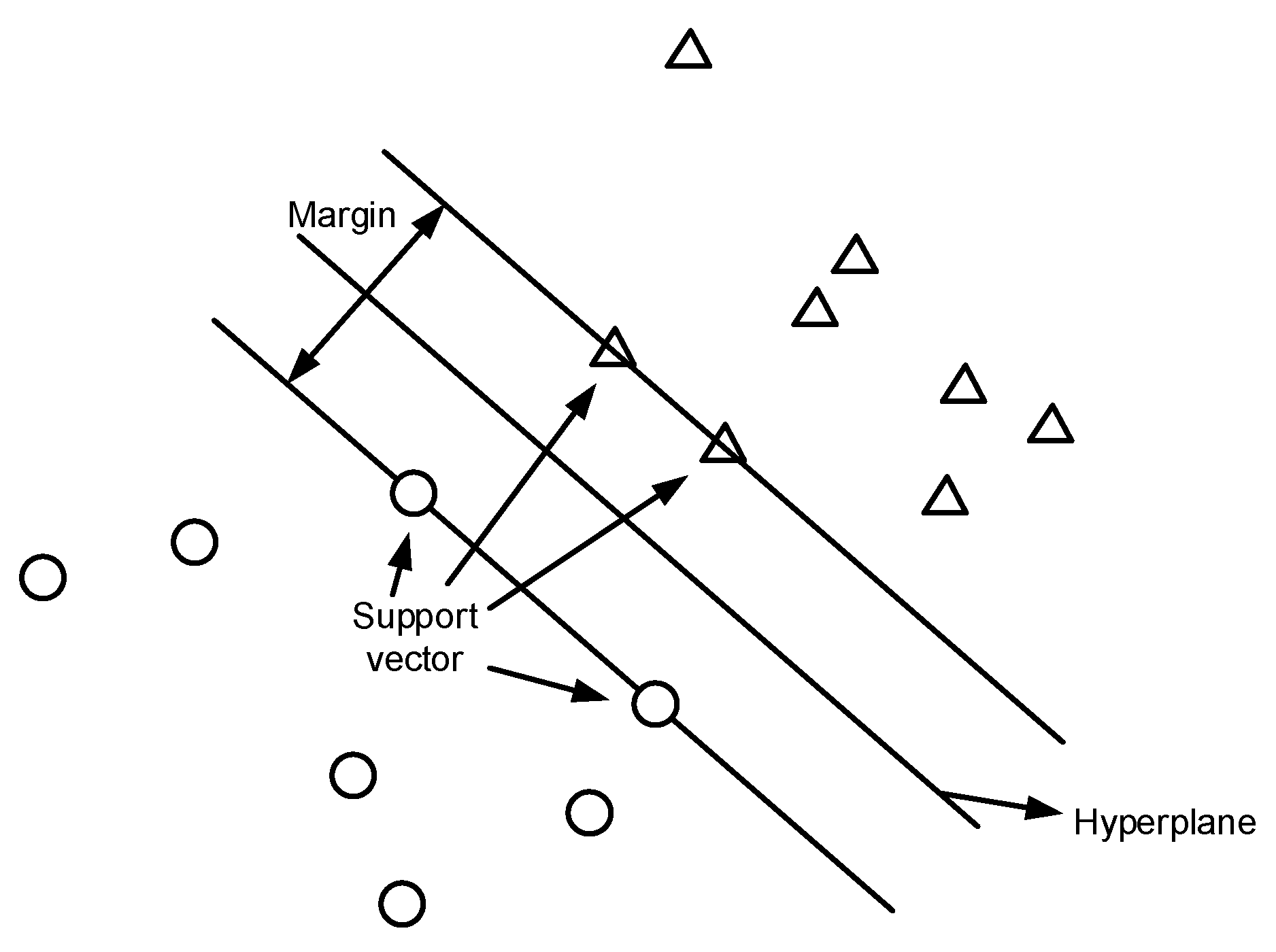
SVM is one of the widely used supervised learning models in machine learning that can assign new data points to different categories when given a set of training examples. It maps data points in space and tries to separate all data points of different classes by finding the best hyperplane in n-dimensional space with the most significant margin between classes, where n is the number of features. The samples fall into one of the sections separated by the hyperplane. Thus, the category to which the sample belongs can be determined.
An illustration of SVM for binary data classification is shown in Figure 2. The two classes of data points are labeled as circles and triangles in the figure. SVM constructs a hyperplane that separates different classes of data points. The support vectors are the points that are closest to the hyperplane. Typically, many possible hyperplanes can be used; the one with the maximum distance or margin between data points of different classes is chosen, such that the new data points can be classified with more confidence.
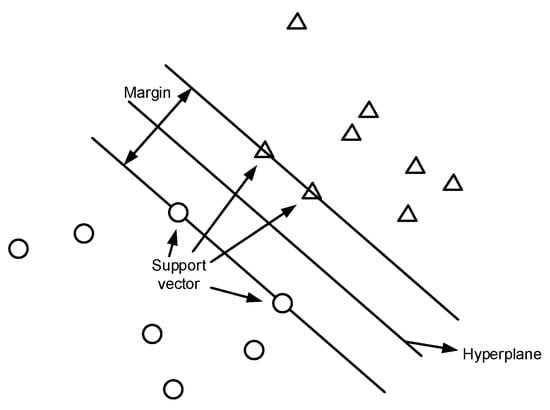
Figure 2.
SVM binary classification.
3. Proposed Model of Deep Transfer Learning Based on SVM Classifier
3.1. Histopathology Image Dataset
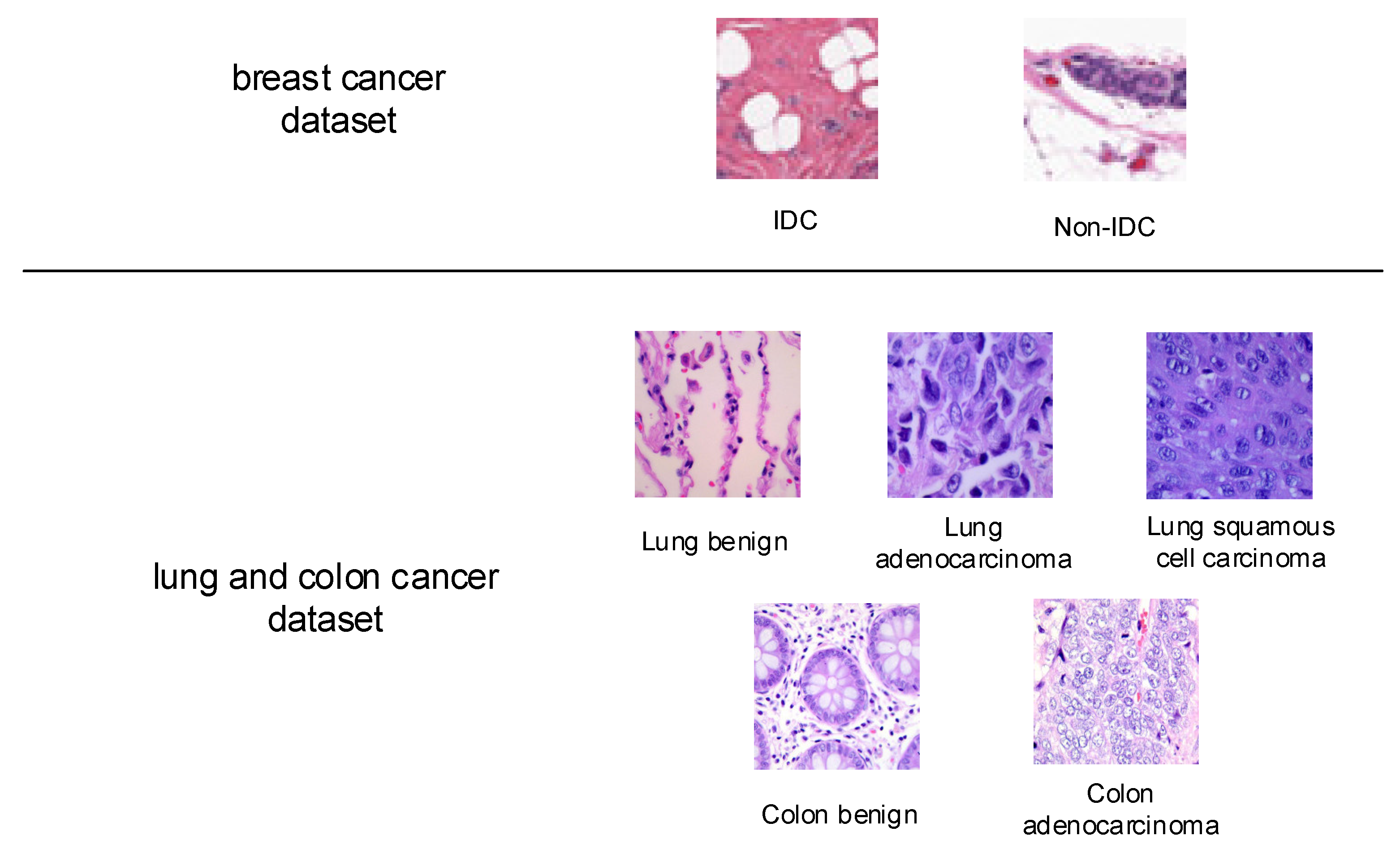
Accurately identifying cancerous tissue and benign tissue is an essential clinical task. The first histopathology image dataset for study in this paper is a breast cancer dataset from [22,23], it contains 198,738 Invasive Ductal Carcinoma (IDC) images and 78,786 non-IDC images. The images are small patches of 50 × 50 pixels extracted from 162 whole mount slide images of breast cancer specimens. The IDC is the most common subtype of all breast cancers, and the regions containing it are usually the focus of pathologists to assign an aggressiveness grade to the whole mount sample. Some sample images from this dataset are given in Figure 3.
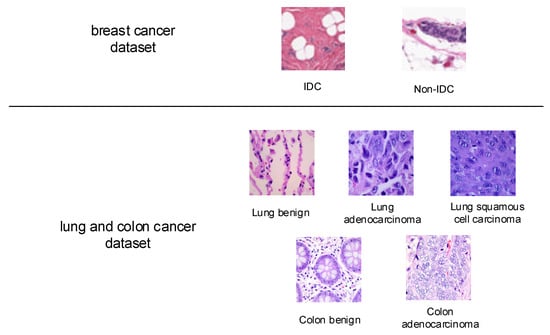
Figure 3.
Sample images from breast cancer dataset, and lung and colon cancer dataset.
The second studied histopathology image dataset from [24] contains 25,000 histopathology images, with a size of 768 × 768 pixels, and five classes, including benign lung tissue, lung adenocarcinomas, and lung squamous cell carcinomas, benign colon tissue, and colon adenocarcinomas, each with 5000 images. The sample images are also shown in Figure 3.
3.2. Deep Learning Architectures for Histopathology Image Classification
In order to classify cancer histopathology images from the above datasets, two deep CNN-based transfer learning networks are first constructed. The off-the-shelf AlexNet pre-trained on the ImageNet dataset is used as the backbone of the transferred network.
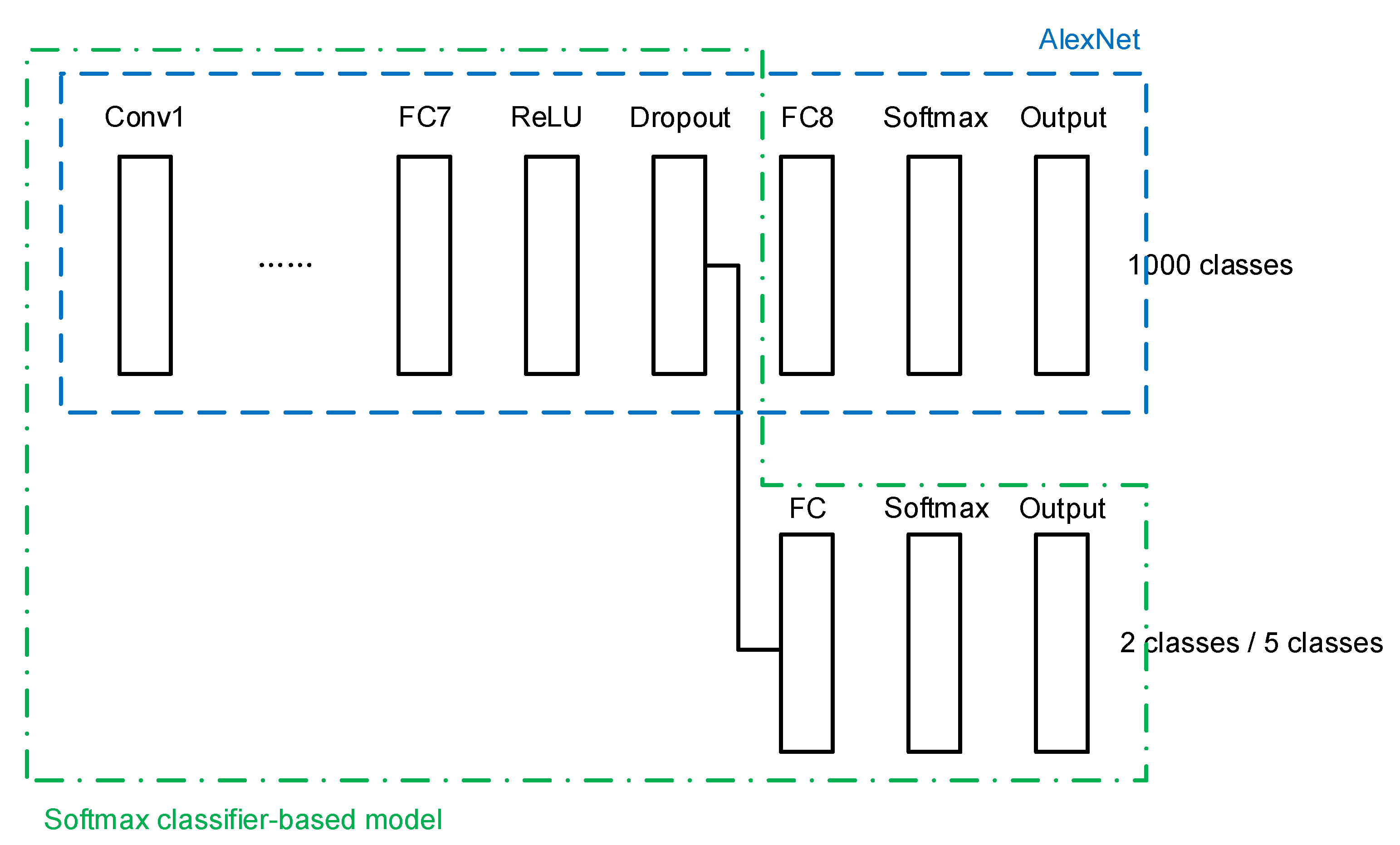
A transfer learning network architecture is built using the conventional softmax classifier, as shown in Figure 4. The last three layers of the original AlexNet, namely a fully connected (FC) layer, a softmax layer, and the output layer, are cut out. A new FC layer, a new softmax layer, and a new output layer are added, of which the newly added FC layer is connected to a dropout layer in the transferred AlexNet.
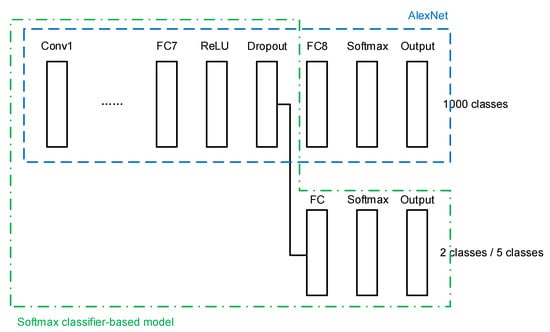
Figure 4.
Softmax classifier-based model.
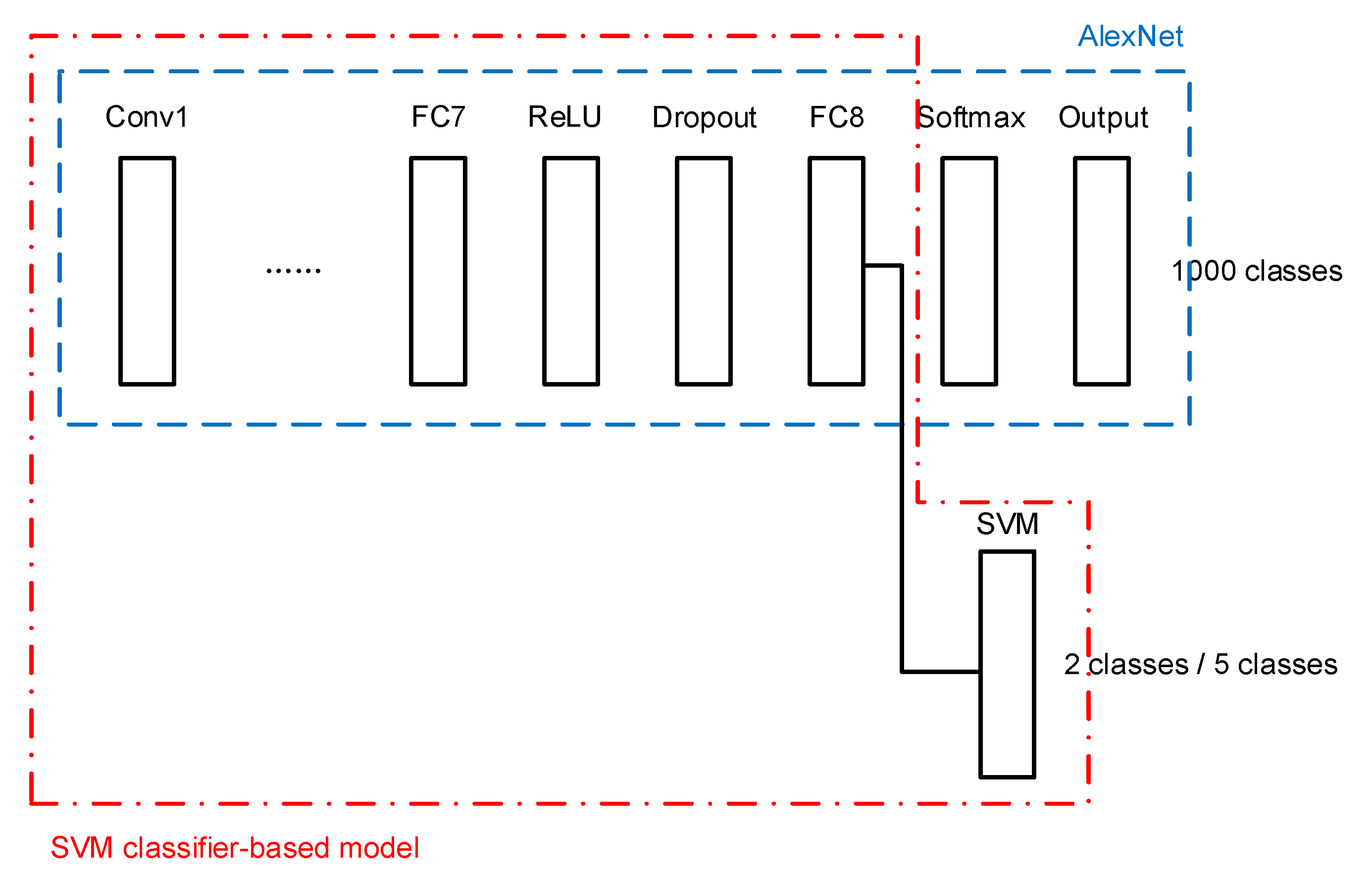
The second architecture consists of the transferred AlexNet and an SVM classifier as the final stage. The SVM classifier is connected to the last FC layer of AlexNet, as shown in Figure 5.
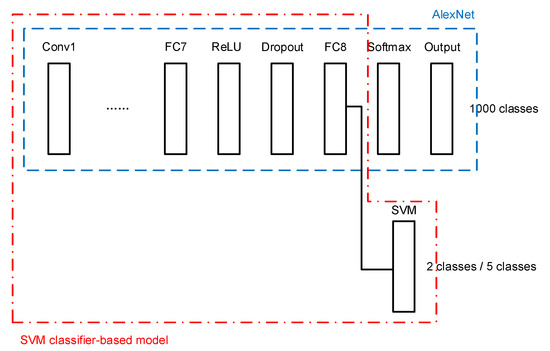
Figure 5.
SVM classifier-based model.
The above two architectures are used for transfer learning in other researches. Some claim that the SVM-based model can lower classification error than the softmax-based model, while others claim that the former model does not show superiority compared with the latter one.
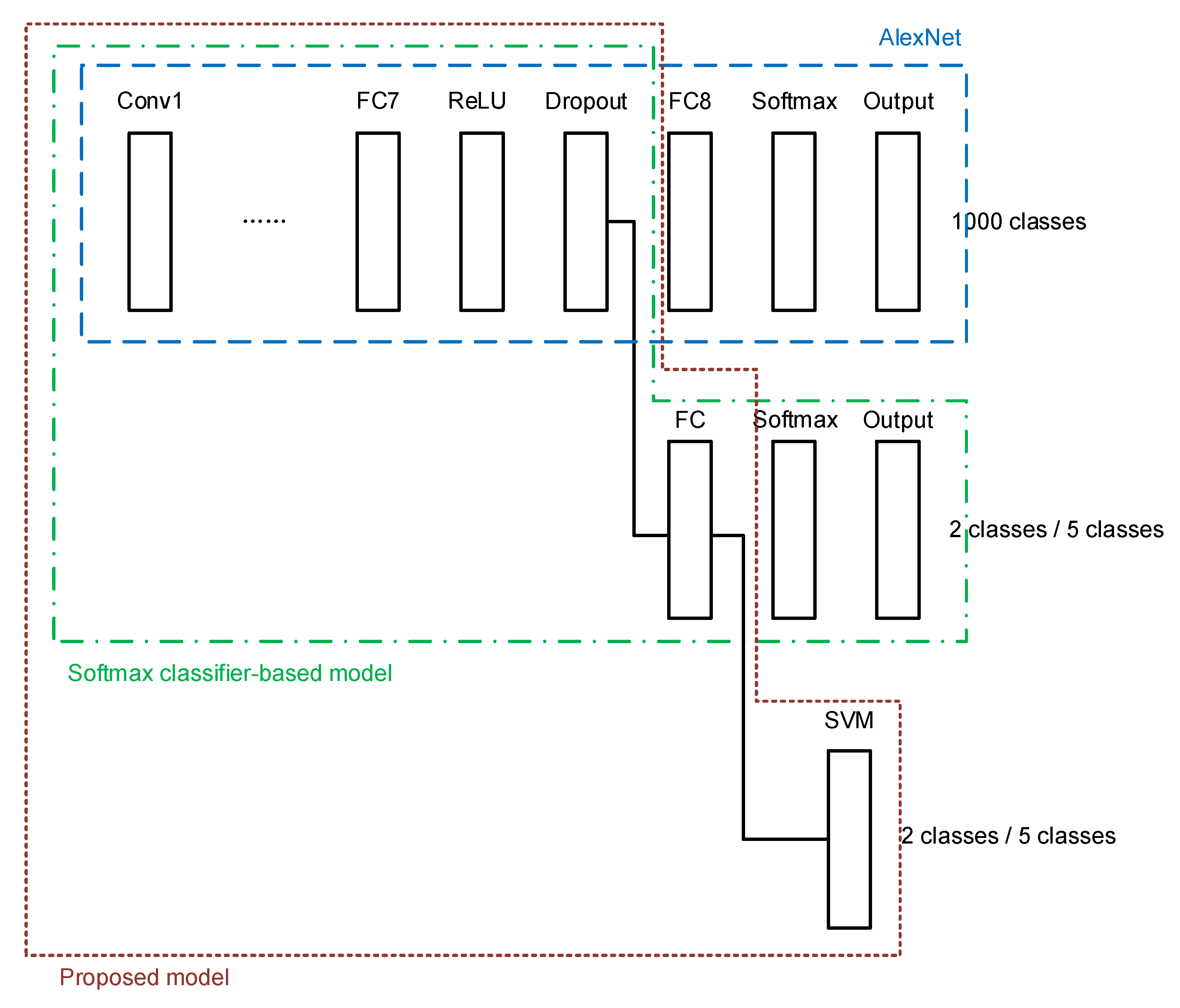
A combination of the above two models is proposed in this paper. After constructing the softmax-based model, the network is trained using the target breast cancer dataset described in the previous section. The weights in the transferred layers borrowed from AlexNet are frozen, and only the newly added layers in the softmax classifier are trained. Then the features extracted by the newly added FC layer are used to feed a new SVM classifier. The proposed architecture is shown in Figure 6. With the proposed architecture, the target dataset is used again to train the added SVM classifier. The key of the proposed architecture is that the SVM classifier is connected to the FC layer that has been trained by the softmax classifier-based transfer learning network using the target dataset.
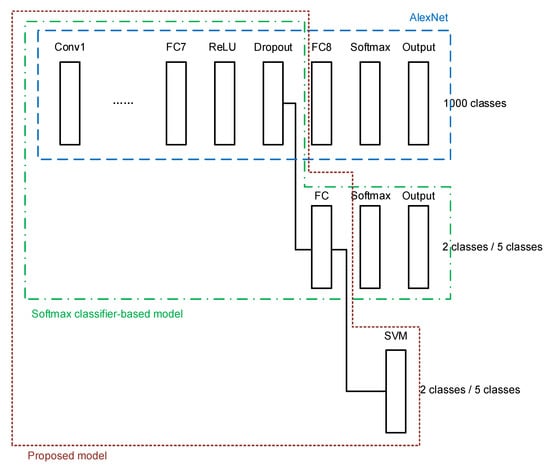
Figure 6.
The proposed transfer learning model for histopathology image classification.
4. Experiments and Discussion
In order to evaluate the classification performance of the softmax classifier-based model, the SVM classifier-based model, and the proposed synthetic model, experiments are first carried out on the breast cancer dataset. In order to have less biased or less optimistic estimate of the models’ performance, four-fold cross-validation setup is used. To simply ensure the balance of each class, 56,000 IDC images and 56,000 non-IDC images are used. The selected images are equally split into four groups with the same number of images for each group. For fold number k, the kth group is used for validation and the other three groups combined are used for training. The process is repeated four times. During the splitting of the dataset, stratified sampling is used in order to eliminate sampling bias, the number of images for each class of each split group are kept the same, i.e., 14,000 IDC images and 14,000 non-IDC images in each group. Same folds are used for all three models. It should be noted that the patient-wise data portioning is not currently considered in this paper, which will be improved in the future work.
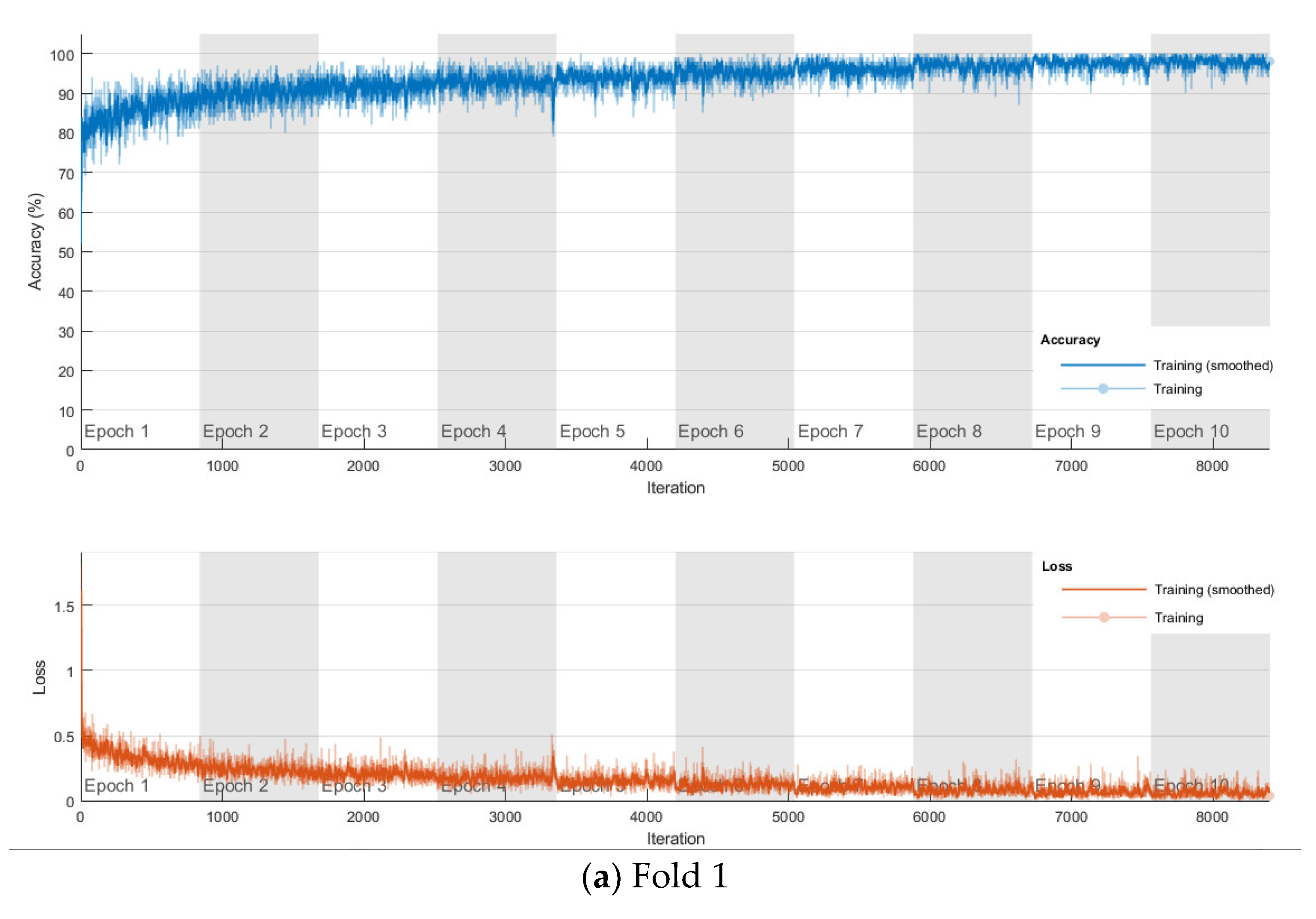
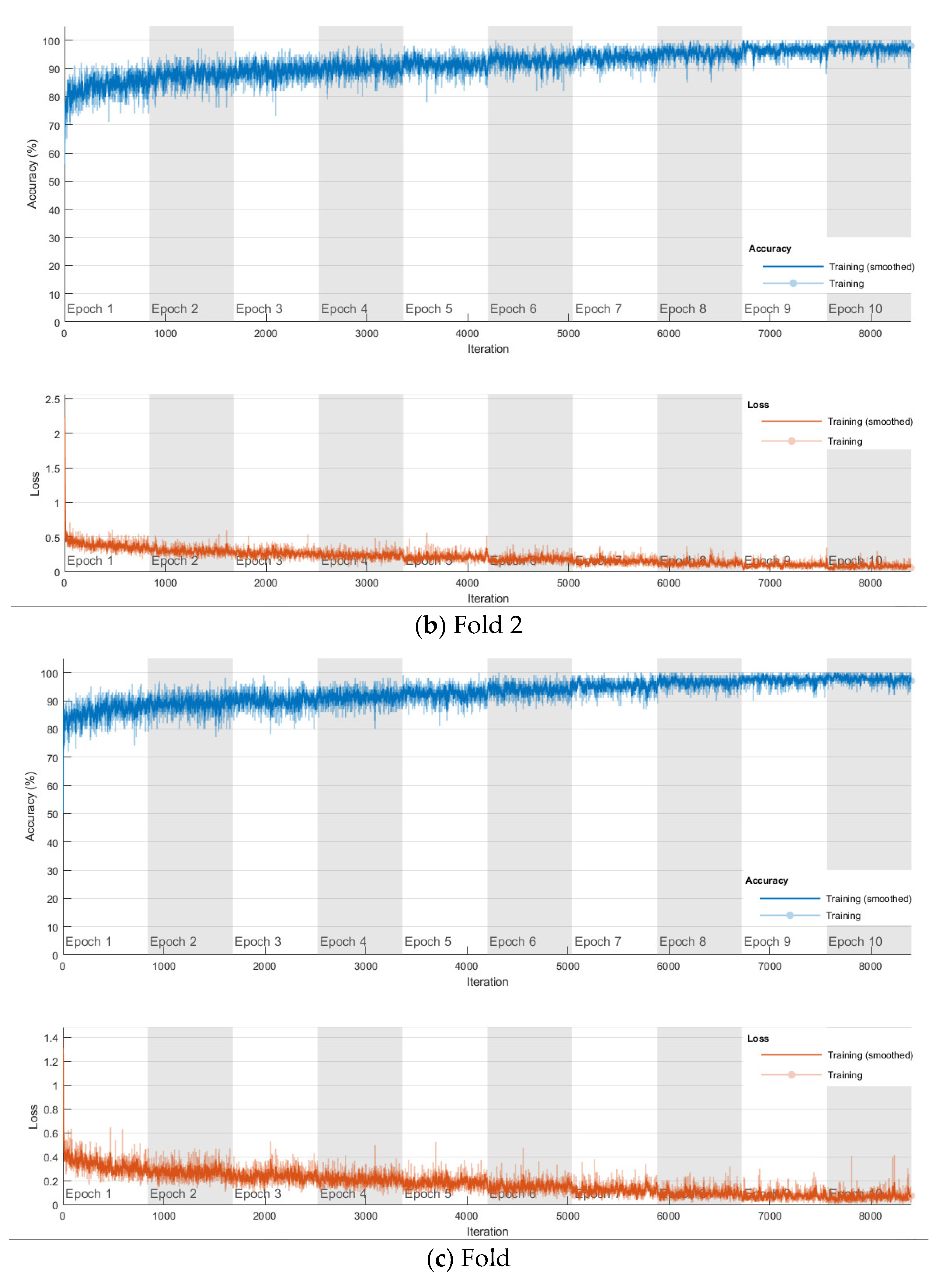
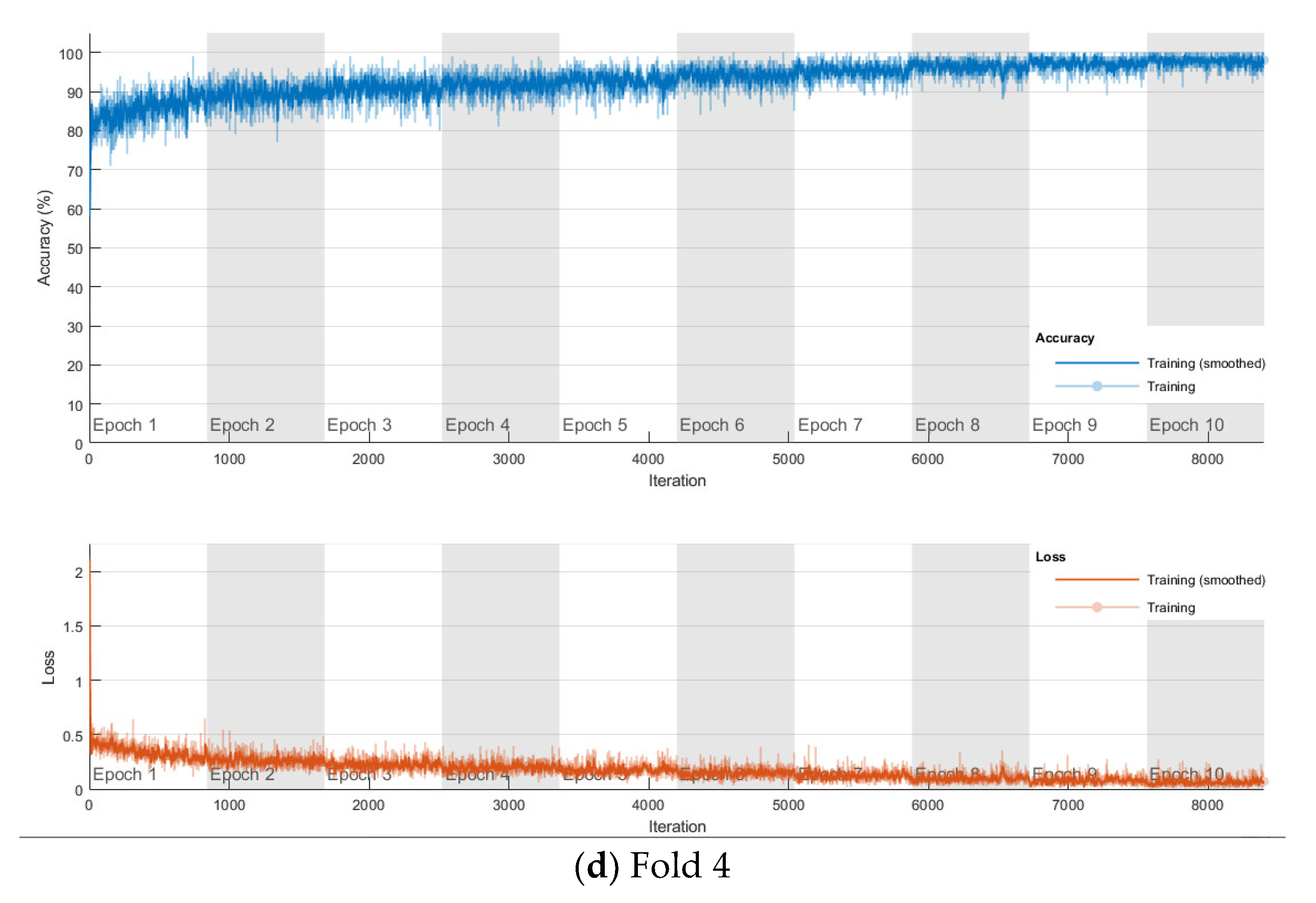
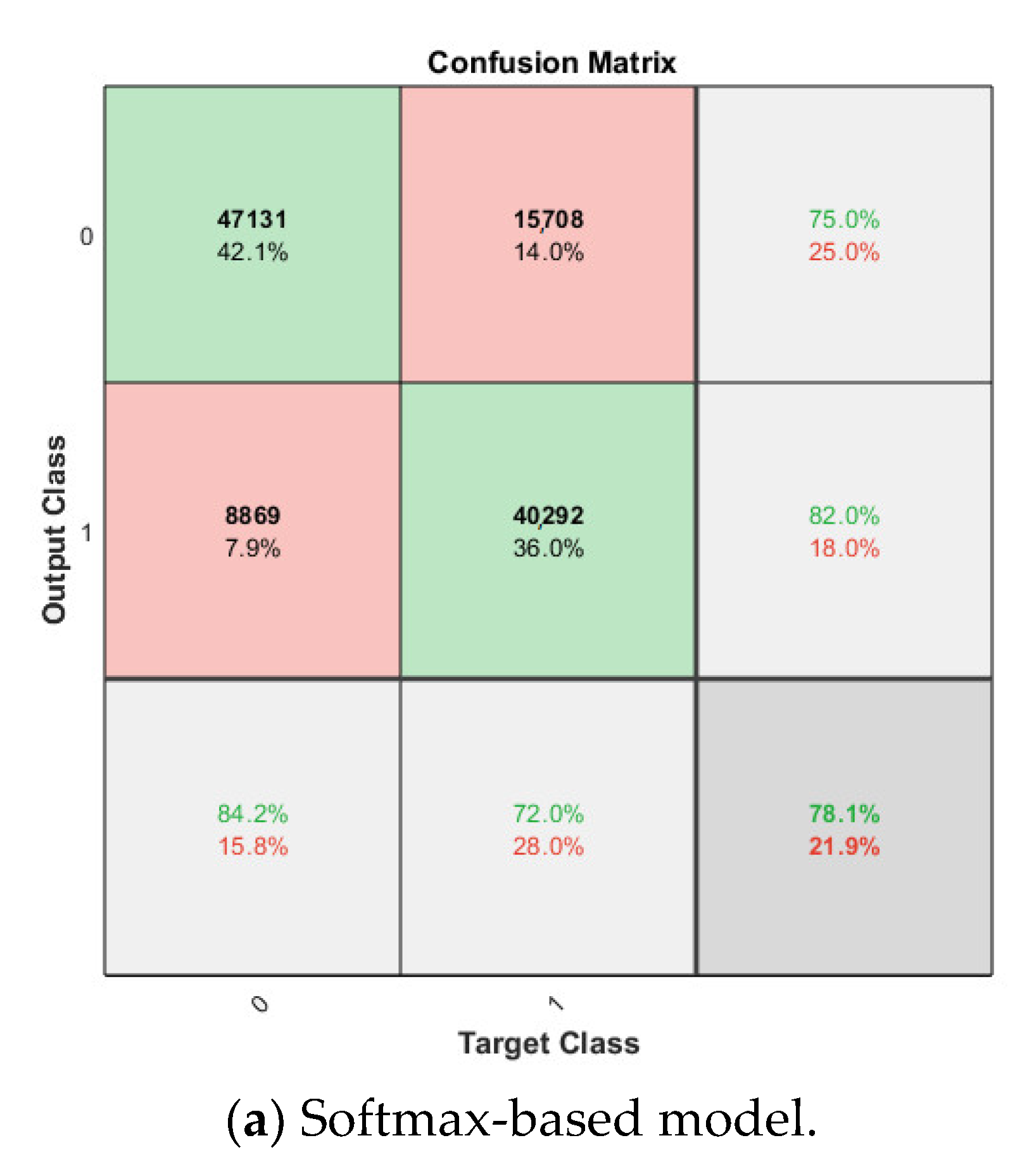
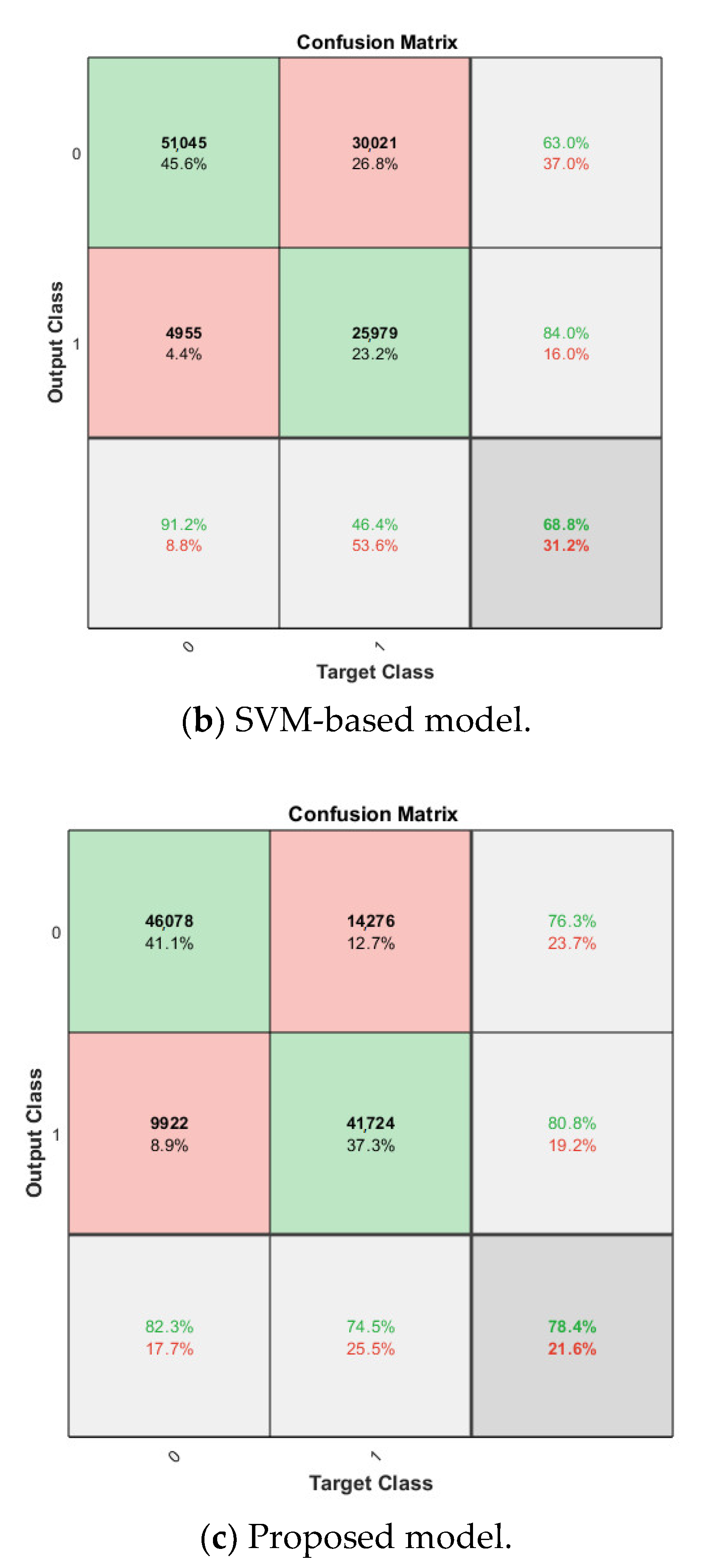
The softmax classifier-based model is first trained using the above cross-validation setup. The images are first resized to 227 × 227 pixels to fit the input size of the transferred AlexNet. The transferred layers are assigned a minimal learning rate to make sure the weights in these layers are frozen, and the features learned from the ImageNet database can be successfully transferred to the target dataset. For each fold of cross-validation, the training is carried out for 10 epochs using the Adam solver. The training set is divided into mini-batches with a size of 100. For each iteration, a mini-batch is used for training, and a different mini-batch is used in the next iteration. The training is conducted throughout the whole training set in 840 iterations or one epoch. The total number of iterations is 8400. The training progress of the four folds of cross-validation is shown in Figure 7, including training accuracy and training loss. The classification accuracy on the validation set for each fold are shown in Table 1, which ranges from 0.6887 to 0.8579, and the average cross-validation accuracy is 0.7806. The confusion matrix of the softmax-based model is shown in Figure 8.
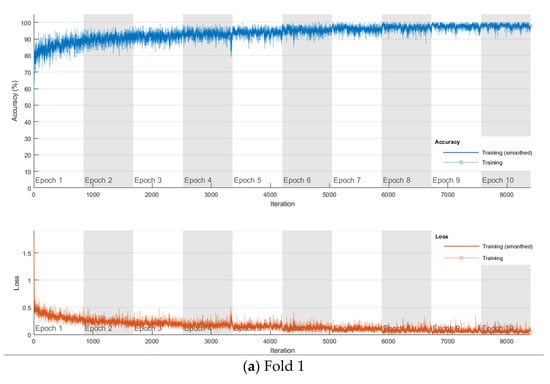
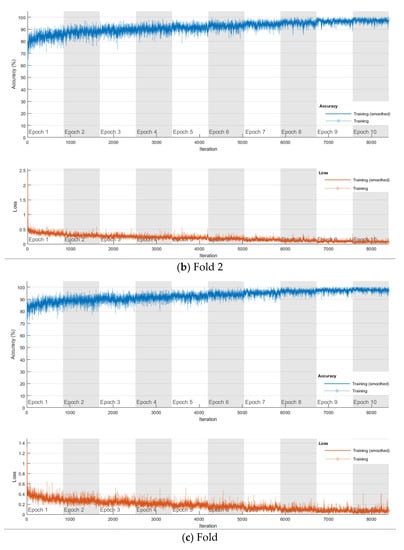
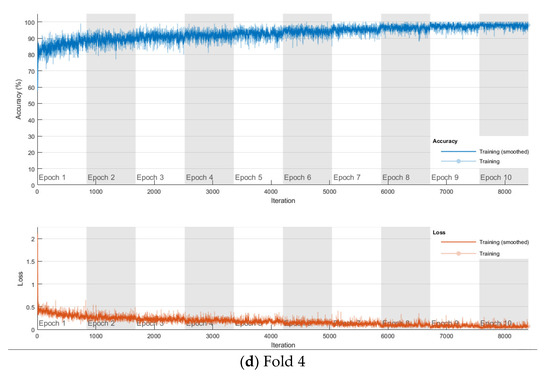
Figure 7.
Training progress of the 4-fold cross-validation of the softmax classifier-based model on breast cancer dataset.

Table 1.
Cross-validation accuracy of the three transfer learning models on breast cancer dataset.
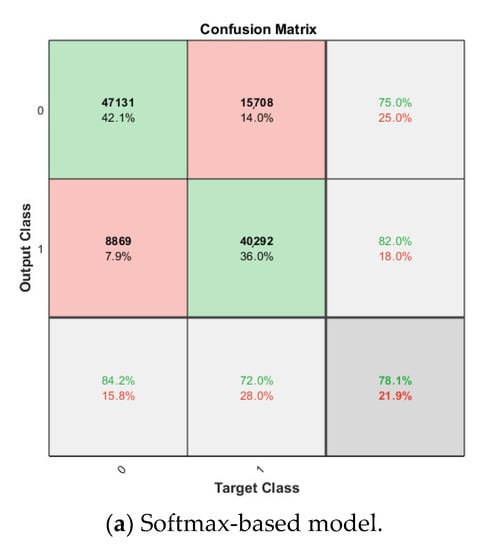
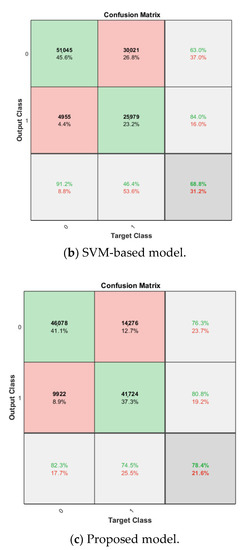
Figure 8.
Confusion matrix on breast cancer dataset.
For the cross-validation of the proposed synthetic model, after obtaining the softmax-based model trained on the breast cancer dataset in the nth fold, the newly trained FC layer with the transferred AlexNet in the softmax-based model is used for feature extraction in the nth fold cross-validation of the proposed model. The dataset configuration is exactly the same as in the previous experiments. For the SVM classifier in the proposed approach, linear kernel is used due to its best accuracy on the target dataset compared with other kernels, such as the Gaussian kernel and high-order polynomial kernel. The cross-validation accuracy is also shown in Table 1 for comparison, which ranges from 0.6916 to 0.8558, and the average cross-validation accuracy is 0.7840. The corresponding confusion matrix is shown in Figure 8.
Cross-validation is performed in SVM-based model, linear kernel is used as well. Attached to the frozen transferred AlexNet layers, the SVM classifier is trained on the breast cancer dataset, and the average classification accuracy on the validation set for the four-fold is 0.6877. The confusion matrix is shown in Figure 8.
By comparing the three models for binary breast cancer histopathology image classification, the SVM-based model is less computationally intensive. However, the average cross-validation accuracy of this model is the worst among the three models, which is 11.9% lower than that of the softmax-based model and 12.2% lower than the proposed model. Even though the SVM-based model and the proposed model both feature an identical SVM classifier, the feature extraction using the FC layer trained on the target dataset in the proposed model largely outperforms the feature extraction using only the ImageNet-based AlexNet in the SVM-based model. The proposed model also shows a 0.4% improvement of the average cross-validation accuracy over the softmax-based model after replacing the softmax classifier with the SVM classifier.
To verify the proposed approach, experiments are then conducted on the multiclass lung and colon cancer dataset. Similar to the previous experimental setup, four-fold cross validation is used. The dataset is split into four groups, stratification is used in splitting the dataset, each group contains 1250 images for each class. The ratio of number of images for training and number of images for validation in each fold is 3:1. Same folds are used for all three models.
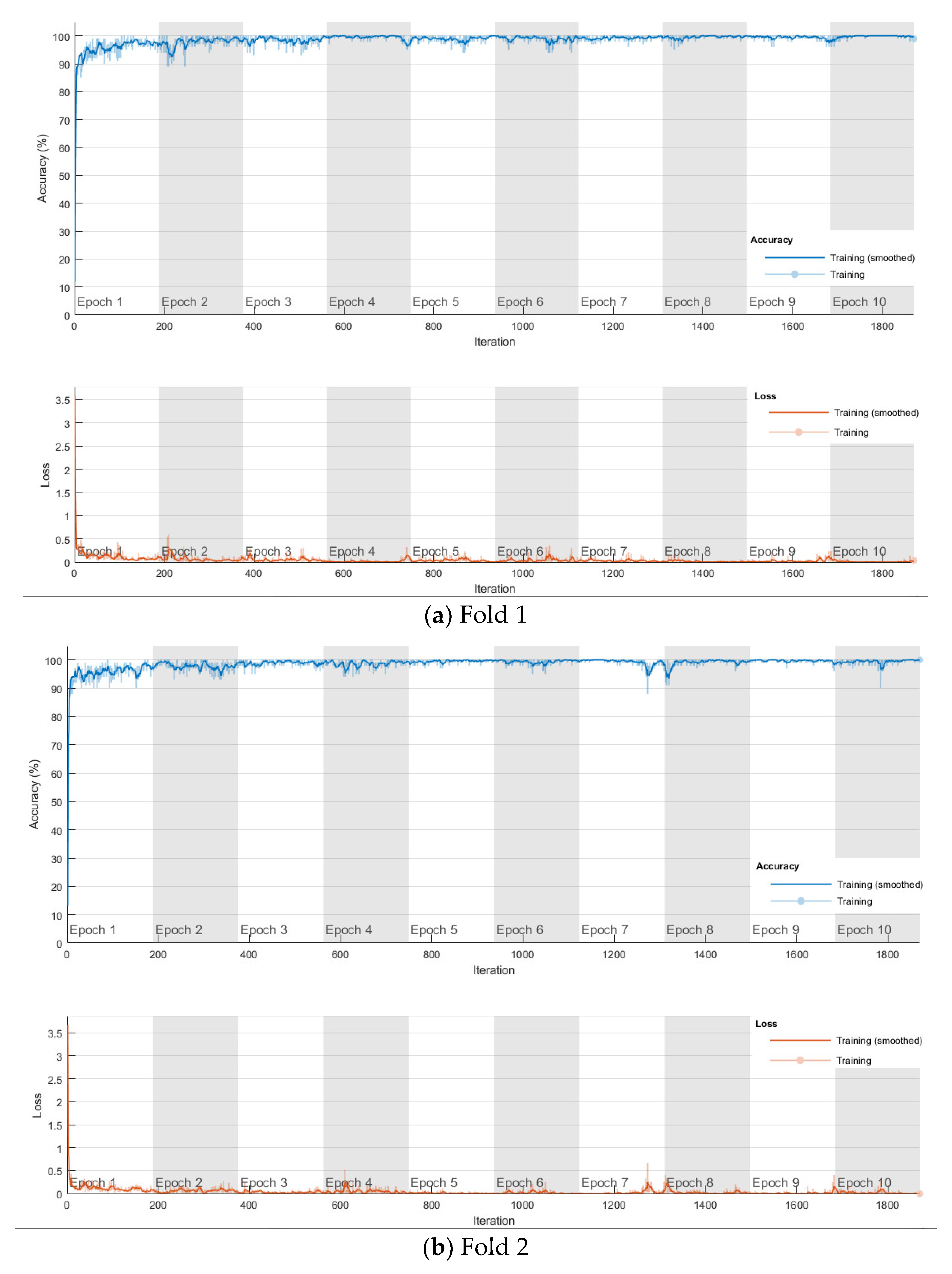
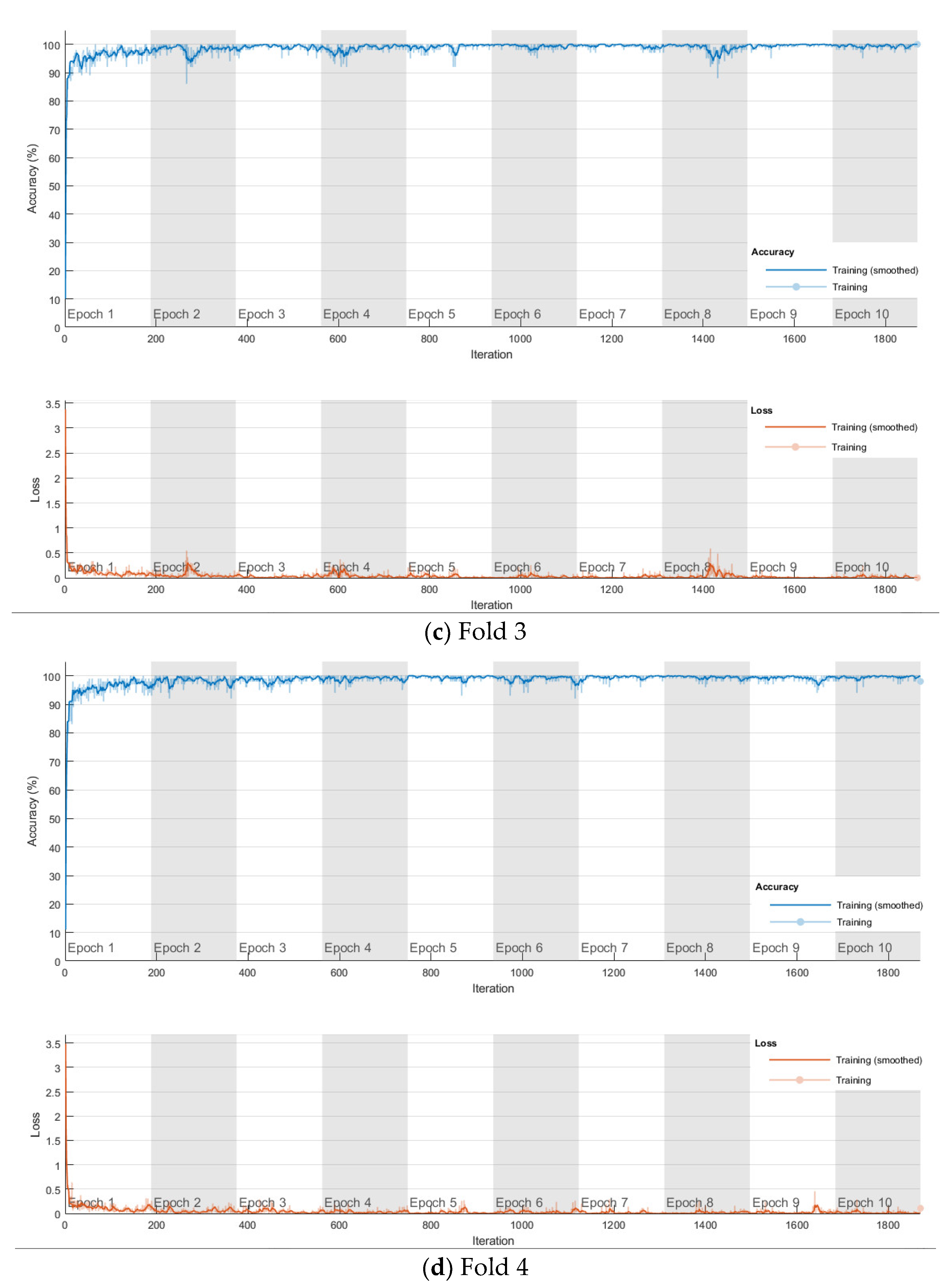
The softmax-based model is first trained using Adam solver for 10 epochs with a mini-batch size of 100 in each fold, and the number of iterations is 1870. The training progress of the four folds are shown in Figure 9. The confusion matrix is shown in Figure 10. The cross-validation accuracy is shown in Table 2, and is above 0.99 for all four folds, and an average accuracy 0.9929 is achieved.
Figure 9.
Training progress of the 4-fold cross-validation of the softmax classifier-based model on lung and colon cancer dataset.
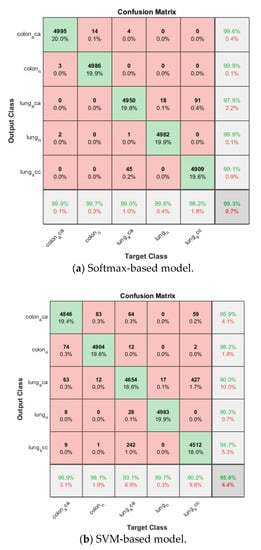
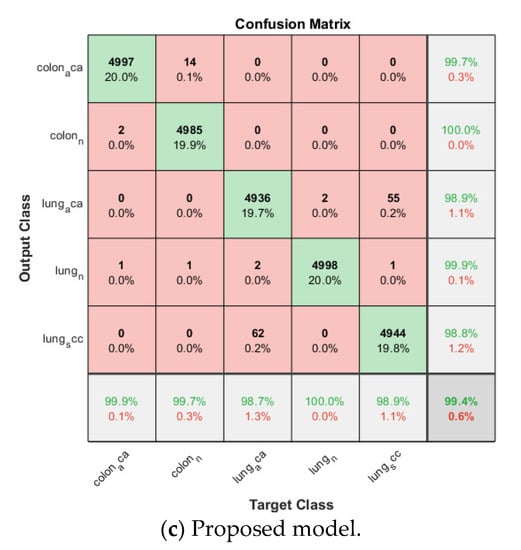
Figure 10.
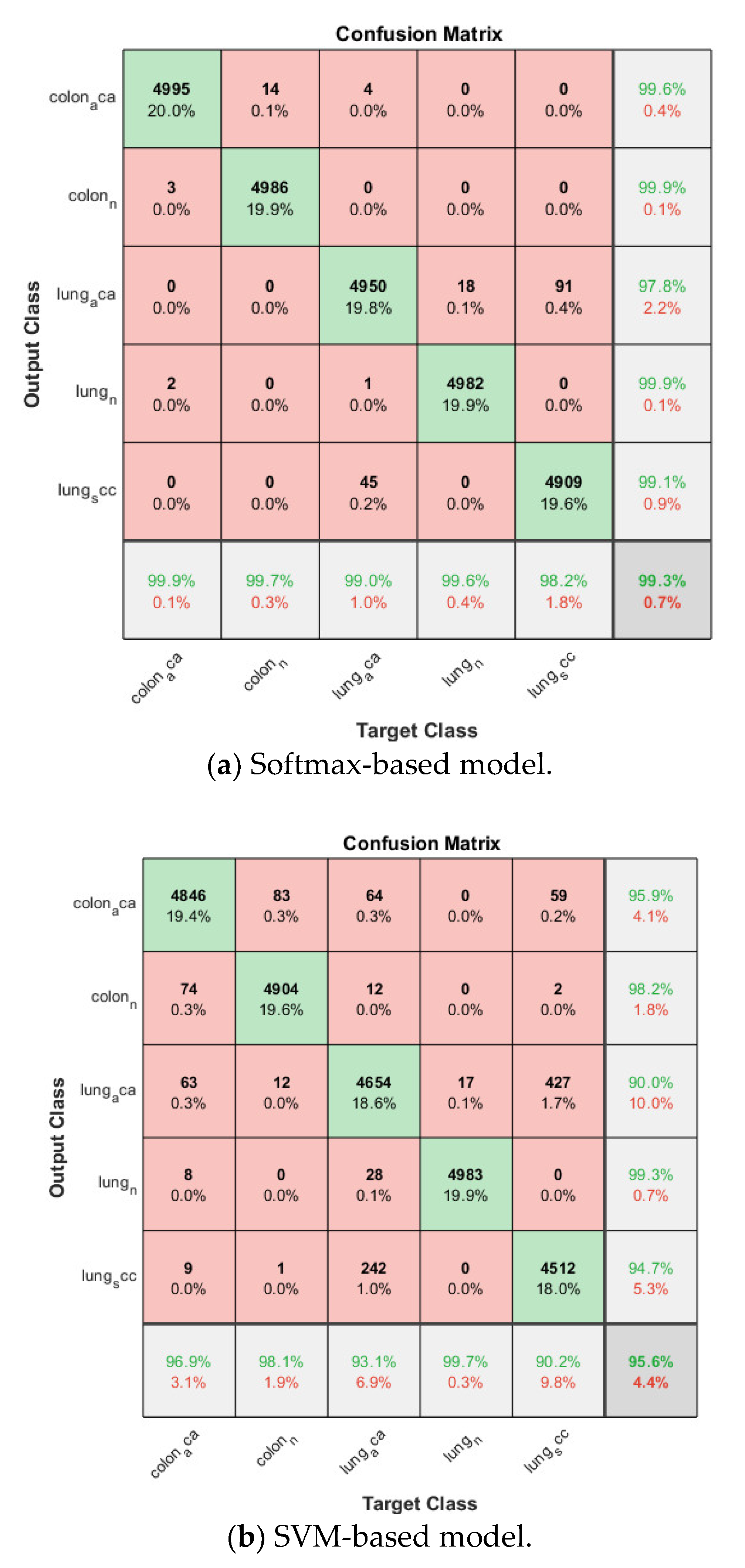
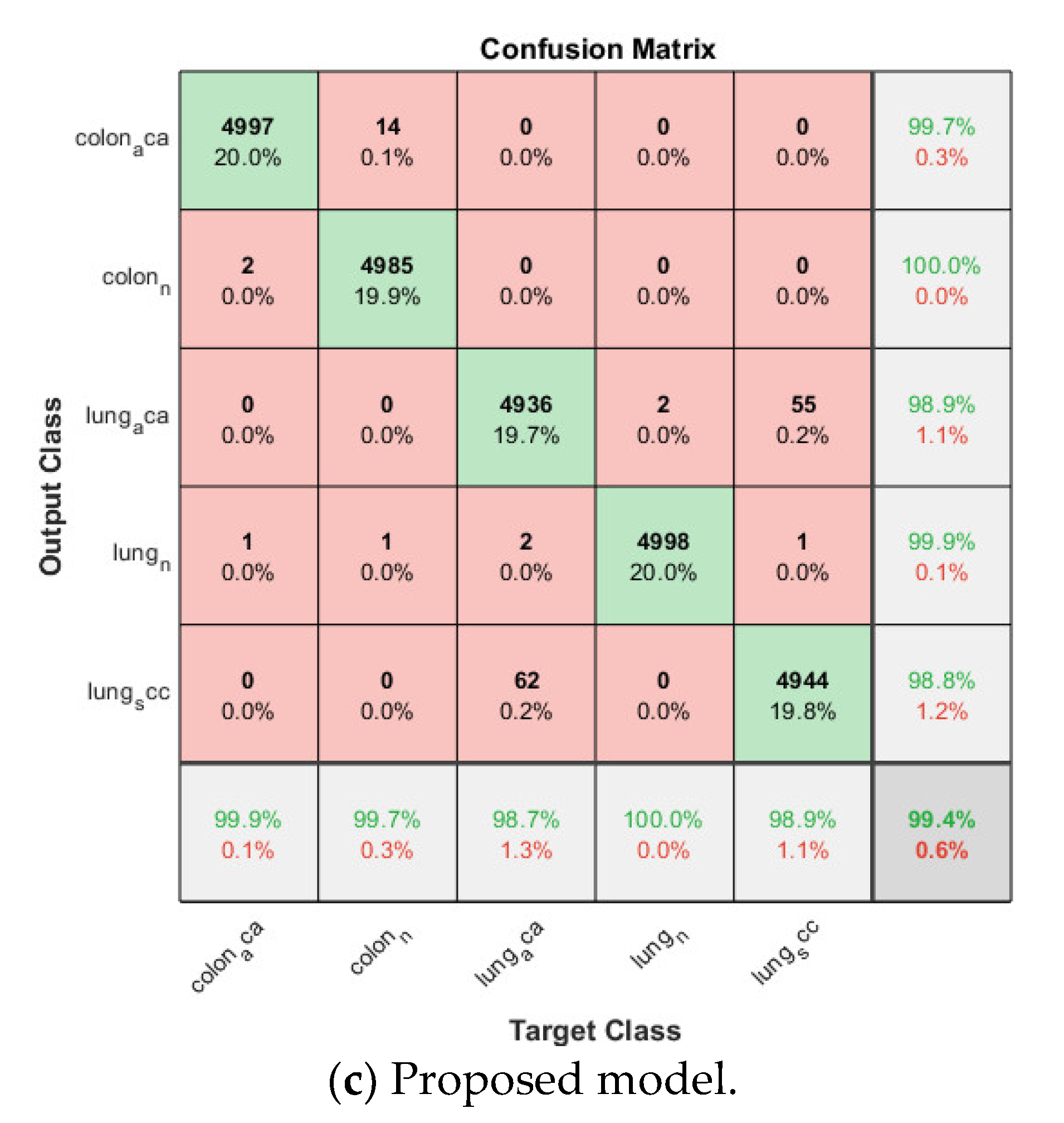
Confusion matrix on lung and colon cancer dataset.

Table 2.
Cross-validation accuracy of the three transfer learning models on colon and lung cancer dataset.
Then the proposed model is trained based on the already trained softmax-based model in the same fold. The SVM classifier is based on Error-Correcting Output Codes (ECOC) in order to classify multi classes other than binary classification. The validation accuracy for each fold is listed in Table 2, and also above 0.99 for all folds, the average accuracy is 0.9944. The corresponding confusion matrix is shown in Figure 10.
Similarly, ECOC is used in the SVM-based model, and the model is trained four folds, the cross-validation accuracy ranges from 0.9443 to 0.9642 with an average of 0.9560 is obtained. The confusion matrix is also shown in Figure 10.
By comparing the three models for multiclass lung and colon cancer classification, the SVM-based model still has the lowest average cross-validation accuracy even though it is already above 0.95. The proposed model is again the winner among the three models, its average accuracy is 0.2% higher than that of the softmax-based model and 4.0% higher than that of the SVM-based model.
No matter for binary histopathology image classification or for multiclass histopathology image classification, it is proved from the above experimental results that the proposed architecture with training twice on the target dataset enjoys more superior classification accuracy. The feature extraction empowered by the FC layer trained on the target dataset help enhance the classification performance of the proposed model.
5. Conclusions
This paper proposes a transfer learning architecture based on the trained softmax-based model and SVM classifier to perform classification tasks on two histopathology image datasets. The proposed synthetic architecture involves a two-step procedure. The first step is to train a softmax classifier-based network using transfer learning on the target dataset. The second step is to use the already trained FC layer in the first step to connect to an SVM classifier. A pre-trained deep CNN architecture AlexNet is used for feature extraction for transfer learning. Knowing the tissue is cancerous or benign is vital for the doctor to diagnose the cancer of the patients. Thus, improving the classification accuracy of the histopathology image becomes a crucial task for machine learning applications. The softmax-based model, the SVM-based model, and the proposed synthetic model are tested and compared on a binary breast cancer dataset and a multiclass lung and colon cancer dataset. Four-fold cross-validation is used to ensure less bias of the evaluation of the three transfer learning models. Experimental results obtained show that the proposed method achieves the best histopathology cancer image classification accuracy, both in binary and multiclass histopathology image datasets.
Author Contributions
Conceptualization, Y.L. and J.L.; methodology, J.F., J.L. and Y.L; investigation, J.F.; writing—original draft preparation, J.F.; writing—review and editing, J.L.; supervision, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Korea institute of radiological and medical sciences. Grant number 2021-0222.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Restrictions apply to the availability of these data. Data was obtained from https://www.kaggle.com/paultimothymooney/breast-histopathology-images and https://www.kaggle.com/andrewmvd/lung-and-colon-cancer-histopathological-images.
Acknowledgments
This work was supported by Seoul National University of Science and Technology, Seoul, South Korea.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Reihs, R.; Pohn, B.; Zatloukal, K.; Holzinger, A.; Müller, H. NLP for the Generation of Training Data Sets for Ontolo-gy-Guided Weakly-Supervised Machine Learning in Digital Pathology. In Proceedings of the 2019 IEEE Symposium on Computers and Communications (ISCC), Barcelona, Spain, 30 June–3 July 2019; pp. 1072–1076. [Google Scholar]
- Mormont, R.; Geurts, P.; Maree, R. Comparison of Deep Transfer Learning Strategies for Digital Pathology. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Salt Lake City, UT, USA, 18–22 June 2018; pp. 2262–2271. [Google Scholar]
- Foucart, A.; Debeir, O.; Decaestecker, C. SNOW: Semi-Supervised, Noisy And/Or Weak Data for Deep Learning In Digi-tal Pathology. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1869–1872. [Google Scholar]
- Sari, C.T.; Gunduz-Demir, C. Unsupervised Feature Extraction via Deep Learning for Histopathological Classification of Colon Tissue Images. IEEE Trans. Med Imaging 2019, 38, 1139–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, A.I.; Dunn, C.M.; Hale, M.; Hutchins, G.G.A.; Treanor, D.E. The Effect of Quality Control on Accuracy of Digital Pathology Image Analysis. IEEE J. Biomed. Health Inform. 2021, 25, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Zhu, F.; Li, X. Transfer Learning for Visual Categorization: A Survey. IEEE Trans. Neural Networks Learn. Syst. 2015, 26, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Alawad, M.; Gao, S.; Qiu, J.; Schaefferkoetter, N.; Hinkle, J.; Yoon, H.-J.; Christian, J.B.; Wu, X.-C.; Durbin, E.B.; Jeong, J.C.; et al. Deep Transfer Learning Across Cancer Registries for Information Extraction from Pathology Reports. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019; pp. 1–4. [Google Scholar]
- Verlekar, T.T.; Correia, P.; Soares, L. Using transfer learning for classification of gait pathologies. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2376–2381. [Google Scholar]
- Alhussein, M.; Muhammad, G. Voice Pathology Detection Using Deep Learning on Mobile Healthcare Framework. IEEE Access 2018, 6, 41034–41041. [Google Scholar] [CrossRef]
- He, S.; Ruan, J.; Long, Y.; Wang, J.; Wu, C.; Ye, G.; Zhou, J.; Yue, J.; Zhang, Y. Combining Deep Learning with Traditional Features for Classification and Segmentation of Pathological Images of Breast Cancer. In Proceedings of the 2018 11th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 8–9 December 2018; pp. 3–6. [Google Scholar]
- AlTalli, H.; Alhanjouri, M. Chest Pathology Detection in X-Ray Scans Using Social Spider Optimization Algorithm with Generalization Deep Learning. In Proceedings of the 2020 International Conference on Assistive and Rehabilitation Technologies (iCareTech), Gaza, Palestine, 28–29 August 2020; pp. 126–130. [Google Scholar]
- Huang, Y.M.; Du, S.X. Weighted support vector machine for classification with uneven training class sizes. In Proceedings of the 2005 International Conference on Machine Learning and Cybernetics, Guangzhou, China, 18–21 August 2005; Volume 7, pp. 4365–4369. [Google Scholar]
- Wu, C.; Lv, X.; Cao, X.; Mo, Y.; Zhu, J. Classification of Metallogenic Favourability Degree Using Support Vector Ma-chines. In Proceedings of the 2010 International Conference on Intelligent Computation Technology and Automation, Changsha, China, 11–12 May 2010; pp. 932–934. [Google Scholar]
- Mohan, L.; Pant, J.; Suyal, P.; Kumar, A. Support Vector Machine Accuracy Improvement with Classification. In Proceedings of the 2020 12th International Conference on Computational Intelligence and Communication Networks (CICN), Bhimtal, India, 25–26 September 2020; pp. 477–481. [Google Scholar]
- Utkin, L.V.; Chekh, A.I.; Zhuk, Y.A.; Utkin, L.V.; Chekh, A.I.; Zhuk, Y.A. Binary classification SVM-based algorithms with interval-valued training data using triangular and Epanechnikov kernels. Neural Networks 2016, 80, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Al-Shargie, F.; Tang, T.B.; Badruddin, N.; Kiguchi, M.; Al-Shargie, F.; Tang, T.B.; Badruddin, N.; Kiguchi, M. Towards multilevel mental stress assessment using SVM with ECOC: An EEG approach. Med Biol. Eng. Comput. 2018, 56, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, W.; Hu, J.; Guo, J. A Multiclass SVM Method via Probabilistic Error-Correcting Output Codes. In Proceedings of the 2010 International Conference on Internet Technology and Applications, Wuhan, China, 20–22 August 2010; pp. 1–4. [Google Scholar] [CrossRef]
- Gu, X.; Deng, F.; Gao, X.; Zhou, R. An Improved Sensor Fault Diagnosis Scheme Based on TA-LSSVM and ECOC-SVM. J. Syst. Sci. Complex. 2017, 31, 372–384. [Google Scholar] [CrossRef]
- Bizzego, A.; Bussola, N.; Chierici, M.; Maggio, V.; Francescatto, M.; Cima, L.; Cristoforetti, M.; Jurman, G.; Furlanello, C.; Bizzego, A.; et al. Evaluating reproducibility of AI algorithms in digital pathology with DAPPER. PLoS Comput. Biol. 2019, 15, e1006269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Zhang, D.Y. Artificial intelligence and computational pathology. Lab. Investig. 2021, 101, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2021, 124, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Roa, A.; Basavanhally, A.; Gonzalez, F.; Gilmore, H.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In Medical Imaging 2014: Digital Pathology; International Society for Optics and Photonics: San Diego, CA, USA, 2014; Volume 9041. [Google Scholar]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inf. 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Borkowski, A.A.; Bui, M.M.; Thomas, L.B.; Wilson, C.P.; DeLand, L.A.; Mastorides, S.M. Lung and Colon Cancer Histopathological Image Dataset (LC25000). arXiv 2019, arXiv:1912.12142v1. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).