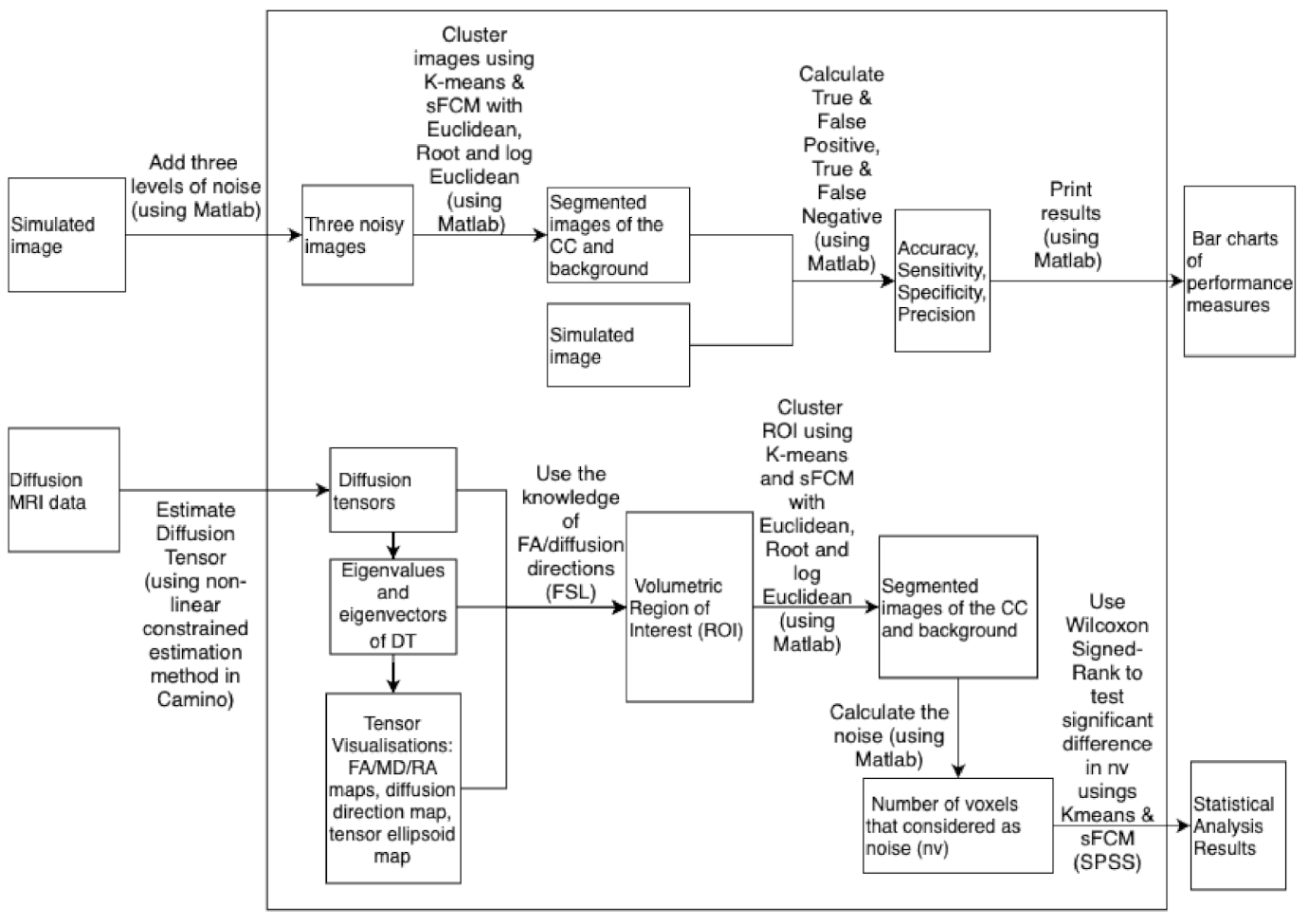
In this section, the results obtained from experiments in the form of two simulation studies and one study using real brain data are described. The two simulation studies were conducted to evaluate the performance of sFCM and K-means for segmenting the CC, in the presence of increasing levels of noise. The CC in the simulation studies is defined, three levels of noise are added and then the segmentation methods are used to segment the CC with performance measures being used to evaluate the quality of the segmentations. Considering images that are slices of a whole brain, there can be multiple regions that makes segmentation more difficult than the consideration of a single region (such as the CC).
In simulation study 1, images are considered with multiple regions and a low to moderate level of noise (methods will likely degrade too much in the multiple region case at high noise levels). In practice, clinicians may provide a manual identification of a single region (such as the CC) and use an image zoomed-in on the region. In simulation study 2, a real data slice with a known region of interest (the CC having been manually identified) is used so that a single region versus background clustering instead of multiple region clustering can be adopted, thereby, enabling the consideration of effects of moderate to high noise levels.
The simulations have signal-to-noise ratios that are consistent with real data. Finally, unannotated real data images are considered (i.e., the CC has not already been annotated/indicated by a clinician), drawn from healthy and SCA2 patients, and the fact that the CC is well-known to be a well-connected single white tract is used in order to determine the efficacy of the segmentation method in practice; since there should not be extra small regions or outliers appearing in the same cluster, disconnected voxels in the segmentation results can be considered as noise.
3.1. Simulation Study
This paper adopts the basic tenet of using synthetic tensors with added noise to test the robustness of segmentation methods as in [
20,
21], for example. Adding noise directly to diffusion tensors can cause the resulting matrix to not be positive semidefinite, to ensure the matrix is positive semidefinite noise is added to the Cholesky decomposition (see
Appendix A) of each tensor instead, following [
6]; that is, the noise is added to the lower triangular matrix first using Cholesky decomposition.
Therefore, in terms of experimental design, we will select three values of noise to be added to the Cholesky decomposition of a tensor, and compare the effects on the segmentation of the CC. To compare the quality of segmentation methods (K-means or sFCM), together with a choice of distance metric (the Euclidean, log Euclidean, and root Euclidean), the standard performance measures (with contextual interpretations to follow) of the accuracy, sensitivity, specificity, precision, F-measure, and Gmean are computed. These measures are computed at each noise level to enable an evaluation of robustness.
Let
, be the Cholesky decomposition of tensor
, with
, and let
be a random matrix with an independent and identically distributed (i.i.d.) normal distribution with expected value
and standard deviation
, for each
and
. Thus, we have [
6]:
To create three levels of noisy tensors, three values of
are selected for each simulation study. The number of simulated tensors is
(from using a
image with size
). The region of interest is then clustered into five clusters with the CC being one of the clusters ([
17] found that the best cluster size for segmentation of the CC was 5). In order to segment the CC, cluster label 1 is assigned to the CC, whilst 0 is assigned to the other four clusters (i.e., we take the logical image with 1 as the CC and 0 as the background).
To evaluate the performance of the Euclidean, Log Euclidean, and Root Euclidean metrics and the segmentation methods (K-means or sFCM) for the segmentation of the CC, we use the performance measures of accuracy, sensitivity, specificity, precision, F-measure, and Gmean. These are standard performances measures used in prediction [
22], that are recalled here, but to use them, the concepts of True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN) need to be interpreted suitably in our context. Take TP and TN to be the numbers of voxels in the CC and in the background (i.e., segmented as any other cluster except the CC), respectively, that are segmented correctly. Then, FP and FN are the numbers of tensors in the background and in the CC that are incorrectly segmented, respectively. The basic standard measures [
22] are:
Using the interpretation given, it can be seen that: (i) accuracy is the ratio of the correctly predicted number of tensors to the total number of tensors (i.e., the ability to select all of the tensors in the CC and reject all the tensors that are not in the CC); (ii) sensitivity (sometimes called recall or true positive rate) is the ratio of the number of correctly predicted tensors in the CC to the number of all tensors in the actual CC (i.e., the ability to select all of the tensors in the CC); (iii) specificity (sometimes called the true negative rate) is the ratio of the number of correctly predicted tensors as being not in the CC to the number of all tensors that are not in the actual CC (i.e., the ability to reject all of the tensors that are not in the CC); and (iv) precision is the ratio of the number of correctly predicted tensors in CC to the number of total predicted tensors in the CC.
These basic measures can be combined in pairs to give performance measures [
22] called the F-measure (F1 score), which takes into account both the FP and FN values (it is the harmonic mean of precision and sensitivity), and the Gmean (Geometric mean), which combines both the true positive rate and true negative rate. They are defined as follows [
22]:
Those two measures are often used to evaluate performance when the dataset used is imbalanced (i.e., the number of objects assigned to each cluster is different).
The step-by-step calculations for the following two simulation studies are summarised in a block diagram (
Figure A1 in
Appendix B).
3.1.1. Simulation Study 1
Tensors in the CC have small size, horizontal diffusion direction (i.e., the water diffuses between right and left hemisphere of the brain) and high FA. Regions nearby to the CC consist of other white matter (WM) tissues, grey matter (GM) and cerebrospinal fluid (CSF). Tensors in some WM regions have similar sizes and FA to that of the CC, whilst tensors in GM and CSF have larger sizes and smaller FA than that of the CC (since the diffusion is anisotropic in WM and isotropic in GM and CSF). Therefore, tensors are initially simulated from multiple regions, mimicking a real brain image, with differing FA and sizes of tensors around the CC (see
Figure 1a).
Then, three levels of noise
(noise1a),
(noise2a) and
(noise3a) are added to the simulated (original) region (see
Figure 1b–d). The signal-to-noise ratios (SNR) of the three level of noises are 21, 18, and 15. The results of segmentation of the three noisy regions are shown in
Figure 2. The figures visibly demonstrate that sFCM improved the segmentation by reducing the background noise as compared to K-means.
To provide more detailed comparisons, all of the performance measures considered for the six cases are shown in
Figure 3. It can be seen that sCFM with each metric (Euclidean, log Euclidean, and root Euclidean) almost always outperformed K-means with the same metric for all performances measures; the only exceptions are the equality of sensitivity for log Euclidean at noise level 3a, and both root and log Euclidean at noise level 1a.
Furthermore, sCFM with root Euclidean or log Euclidean generally outperform sCFM with Euclidean. In detail: (i) at noise level 1a, root and log Euclidean produce the same results, and thus yield equality for all performance measures, and their measures all outperform Euclidean except for the (equality of) sensitivity; (ii) at noise levels 2a and 3a, root Euclidean has the highest accuracy, sensitivity, F-measure, and Gmean, whilst log Euclidean has the highest specificity and precision. Euclidean has the lowest values for accuracy, F-measure, specificity, and precision, but the same sensitivity as root Euclidean, and a higher Gmean than log Euclidean.
3.1.2. Simulation Study 2
In Simulation Study 1, multiple regions with increasing noise levels were used, which covers a low to moderate range of noise (since the detection of the regions becomes problematic when considering high levels of noise). In Simulation Study 2, consideration of robustness in the face of moderate to high levels of noise is enabled by simulating a homogenous region of the CC and a background only (as per the logical image mentioned earlier); this is because the CC is still visible in this case.
Initial tensors are simulated such that: the tensors have the same determinants (sizes), FA values, and eigenvalues, and they only differ in their orientation (i.e., the eigenvectors); the diffusion directions of the tensors in the simulated CC shape are parallel to y-axis, while the diffusion directions of other tensors are parallel to x-axis. Then, the three levels of noise chosen are: (noise1b), (noise2b), and (noise3b).
These are added to the simulated region (see
Figure 4). The signal-to-noise ratio (SNR) of the three levels of noise are 13, 8, and 5. The results of the segmentation of the three noisy images are shown in
Figure 5. When using Log Euclidean with noise3b (in
Figure 5c), the CC is not visible. The performance measures were calculated and are shown in
Figure 6. Similar to Simulation Study 1, it can be seen that root Euclidean generally provided the highest values of performance measures as compared to the other methods.
The findings indicated that: (i) at noise level 1b, all of the six methods yield the same results; (ii) at noise level 2b, sFCM yields the same results using log, root, and Euclidean (and hence the same values of all performance measures), whilst sFCM with each metric almost always outperforms K-means with the same metric (with exceptions that sFCM and K-means with the Euclidean metric have the same specificity and precision, and sFCM and K-means with root Euclidean have the same sensitivity); (iii) at noise level 3b, the log Euclidean metric fails to even detect the CC, whilst sFCM with root Euclidean outperforms sFCM with Euclidean, and sFCM with either metric (root or Euclidean) outperforms K-means with the same metric.
From both studies, it can be seen that the sFCM method improved the segmentation of the CC, almost always providing better performance measures than the corresponding K-means method, especially in noisy images. One can observe that root Euclidean with sFCM almost always outperformed Euclidean and log Euclidean in the segmentation of the CC. With the largest level of noise (i.e., noise3b in Study 2), log Euclidean failed to even detect the shape of CC even when the cluster size was increased. This is likely to be because it is highly affected by outliers, as shown by the following example.
Take three examples of tensors
,
, and
from the CC region of a healthy brain, as follows:
To calculate the log and root Euclidean distances between these tensors, the eigenvalues
and
of each tensor are needed (see
Table 1 and
Appendix A). The eigenvalues, together with the FA values are shown in
Table 3, and then the distances between the tensors are shown in
Table 4.
The distances between
and
, and
and
using log Euclidean are very large in comparison with the distance between
and
. This is due to the use of the log function and the smaller eigenvalue
of
in comparison with the other eigenvalues. All three tensors have high FA (see
Table 3; recall FA ranges from 0 to 1), and hence all of them are expected to be part of the CC. When clustering, all the three tensors are part of the CC using the root Euclidean and Euclidean metrics, but
is excluded from the CC using the log Euclidean metric. This explains the holes in the CC that appear using the log Euclidean metric. This example demonstrates that the distance between tensors with similar FA values can be very large when using the log Euclidean metric.
3.2. Real Brain Image Data Studies
In this section, the application in practice, using real brain image data, is considered, demonstrating that sCFM performs significantly better than K-means for the segmentation of the CC. Subsequently, using the same data set, a brief demonstration that the methods can be used effectively for classification as well as segmentation is provided. Furthermore, it is demonstrated (using sFCM, which ensures robustness to noise) that existing DTI indices (fractional anisotropy, mean diffusivity, and radial diffusivity), as well as the determinant, are all suitable DTI indexes that can be used to distinguish between healthy and SCA2 subjects and are sensitive to ageing effects.
The data consists of nine SCA2 subjects (six males and three females) and sixteen age-matched healthy subjects (nine males and seven females). This data is taken from [
23]. On the same MRI scanner, the subjects have been imaged twice:
years apart (SCA2 patients) and
years apart (control subjects). For more details about the data and MRI acquisition procedures see [
23]. In this work, diffusion weighted data is corrected for eddy current-induced distortions (using FSL). Then, the diffusion tensor imaging is fitted using a non-linear constrained estimation method [
24,
25] in Camino. Matlab and SPSS are used for segmentation and data analysis respectively. The block diagram of the calculations is presented in
Figure A1 in
Appendix B.
3.2.1. 3D Segmentation of the CC
A volumetric region of interest (ROI) from the middle of the brain is chosen as input to the K-means and sFCM (with parameters used
and
) algorithms. The ROI is clustered into five clusters, with CC being one of those clusters, using the root Euclidean metric. To visualise the CC, the cluster labels are binarised (i.e., cluster labels for the CC cluster are all 1 and 0 is used for labels in the other four clusters). Examples of the results of the segmentation of the CC using K-means and sFCM are shown in
Figure 7. To evaluate how much better sFCM is in reducing the noise around the CC as compared to K-means, the number of voxels (nv) that are considered as noise is calculated (i.e., the voxels around the CC that have the same cluster labels as the CC but are not actually part of the CC).
The Wilcoxon Signed-Rank test is used to test the significant difference in nv values produced by using K-means and sFCM. The results show that the nv values are significantly smaller using sFCM as compared to K-means (see
Table 5) for both the baseline and post baseline data. This confirms that the use of sFCM instead of K-means significantly reduced the amount of noise in these images.
3.2.2. Generalisation to Classification of Brain Images
These new methods can be used for the more general problem of classifying a brain image into white matter, grey matter, and cerebrospinal fluid, using a whole axial slice of the brain image. Since the image contains both the brain and its background, the image is clustered into four clusters for white matter, grey matter, cerebrospinal fluid, and the background of brain (see
Figure 8, where the background is shown in deep blue) using the root Euclidean metric. The CC is not segmented here, but is included as part of the white matter. To segment the CC, the cluster size needs to be 5 (as in
Section 3.2.1); however, this demonstrates that sFCM can be used for classification purposes.
3.2.3. Clinical Applications of Segmentations with sFCM
In clinical studies, DTI indices, such as Fractional anisotropy (FA), mean diffusivity (MD), and Radial diffusivity (RD), are used in the comparison of healthy and non-healthy brain images (often manually segmented). The efficacy of the new methods presented in the paper is demonstrated by using one of our automatically created segmentations (via sFCM with root Euclidean) and demonstrating that these indices can distinguish between healthy and SCA2 subjects, and they are sensitive to ageing effects.
This reaffirms the results in the literature [
12,
13,
15], but with the extra knowledge that the use of sFCM will have reduced the impact of noise. In addition to this, it can be seen that the determinant (DET) of the tensors, which is easy to compute, can also be used to distinguish between healthy and SCA2 subjects and is sensitive to ageing effects. That is, DET is shown to be a viable DTI index.
First, recall the definitions of the DTI indices, which are functions of the eigenvalues of the diffusion tensors. FA measures the deviations from isotropic diffusion of water inside a voxel in the brain, and it is a fraction with FA equal to 1 for diffusion that is highly anisotropic (i.e., water diffuses in one direction) and FA equal to 0 for isotropic diffusion (i.e., water diffuses in all directions). Let
and
be the eigenvalues of diffusion tensor
D and assume that
is the largest eigenvalue. Then, FA [
5] can be calculated as follows:
MD measures the average water diffusivity in a voxel in the brain. It is calculated as follows [
5]:
Radial diffusivity measures the perpendicular diffusion to the main diffusion of water. It is calculated as follows [
5]:
These DTI indices, together with the DET are computed. The Mann–Whitney test is used to test the significant difference in FA, MD, RD, and DET between healthy and SCA2 subjects at the significance level 0.05. The results of FA, MD, RD, and DET are all significant at both baseline and post baseline. In detail:
FA values in SCA2 subjects are significantly lower than in healthy subjects (p-value at baseline = , p-value at post baseline = ).
MD values in SCA2 subjects are significantly increased as compared to healthy subjects (p-value at baseline = , p-value at post baseline = ).
RD values in SCA2 subjects are significantly increased as compared to healthy subjects (p-value at baseline = , p-value at post baseline = ).
DET values in SCA2 subjects are significantly larger than in healthy subjects (p-value at baseline = , p-value at post baseline = ).
These results show that FA, MD, RD, and DET distinguish well between healthy and SCA2 subjects. The rate of change can be calculated as follows:
The rates of change of FA, MD, RD, and DET in SCA2 subjects are not significantly different from the rates of change in healthy subjects.
The Wilcoxon Signed-Rank test is used to test the significant difference in FA, MD, RD, and DET at baseline and post baseline. The results of FA, RD, and DET were all significant. However, MD values were not significantly different at baseline and post baseline. The details are as follows:
FA values at post baseline were significantly lower than at baseline (p-value = ).
RD values at post baseline were significantly increased as compared to RD values at baseline (p-value = ).
DET values at post baseline were significantly larger than at baseline (p-value = ).
These results show that FA reduced while RD and size of tensors (DET) increased with age.