Electrochemical Sandwich Assays for Biomarkers Incorporating Aptamers, Antibodies and Nanomaterials for Detection of Specific Protein Biomarkers
Abstract
:1. Introduction
2. Electrochemical Sandwich Assays for Thrombin Using Aptamers
3. Other Electrochemical Sandwich Assays Using Two Aptamers
4. Electrochemical Sandwich Assays Combining Aptamers and Antibodies
5. Electrochemical Sandwich Assay Combining Aptamers with Other Materials
6. Discussion
Funding
Conflicts of Interest
References
- Ziółkowski, R.; Jarczewska, M.; Górski, L.; Malinowska, E. From Small Molecules toward Whole Cells Detection: Application of Electrochemical Aptasensors in Modern Medical Diagnostics. Sensors 2021, 21, 724. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.B.; Gu, M.B. Aptamer-based sandwich-type biosensors. J. Biol. Eng. 2017, 11, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yáñez-Sedeño, P.; Campuzano, S.; Pingarrón, J. Pushing the limits of electrochemistry toward challenging applications in clinical diagnosis, prognosis, and therapeutic action. Chem. Commun. 2019, 55, 2563–2592. [Google Scholar] [CrossRef]
- Rusling, J.F. Multiplexed electrochemical protein detection and translation to personalized cancer diagnostics. Anal. Chem. 2013, 85, 5304–5310. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-Y.; Wang, J.; Liu, G.; Wu, H.; Wai, C.M.; Lin, Y. A nanoparticle label/immunochromatographic electrochemical biosensor for rapid and sensitive detection of prostate-specific antigen. Biosens. Bioelectron. 2008, 23, 1659–1665. [Google Scholar] [CrossRef] [PubMed]
- Khoshroo, A.; Mazloum-Ardakani, M.; Forat-Yazdi, M. Enhanced performance of label-free electrochemical immunosensor for carbohydrate antigen 15-3 based on catalytic activity of cobalt sulfide/graphene nanocomposite. Sens. Actuat. B Chem. 2018, 255, 580–587. [Google Scholar] [CrossRef]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, J.; Liu, J.; Wang, X.; Chen, B. Disease-related detection with electrochemical biosensors: A review. Sensors 2017, 17, 2375. [Google Scholar] [CrossRef] [PubMed]
- Hianik, T.; Wang, J. Electrochemical Aptasensors—Recent Achievements and Perspectives. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2009, 21, 1223–1235. [Google Scholar] [CrossRef]
- Cox, J.C.; Ellington, A.D. Automated selection of anti-protein aptamers. Bioorg. Med. Chem. 2001, 9, 2525–2531. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Nikolaus, N.; Strehlitz, B. Capture-SELEX: Selection of DNA aptamers for aminoglycoside antibiotics. J. Anal. Methods Chem. 2012, 415697, 1–14. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zu, Y. A highlight of recent advances in aptamer technology and its application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Tan, W. Aptamers generated from cell-SELEX for molecular medicine: A chemical biology approach. Acc. Chem. Res. 2010, 43, 48–57. [Google Scholar] [CrossRef] [Green Version]
- De Girolamo, A.; McKeague, M.; Pascale, M.; Cortese, M.; DeRosa, M.C. Immobilization of Aptamers on Substrates. In Aptamers for Analytical Applications: Affinity Acquisition and Method Design; Dong, Y., Ed.; Wiley: New York, NY, USA, 2018; pp. 85–126. [Google Scholar]
- Meini, N.; Farre, C.; Chaix, C.; Kherrat, R.; Dzyadevych, S.; Jaffrezic-Renault, N. A sensitive and selective thrombin impedimetric aptasensor based on tailored aptamers obtained by solid-phase synthesis. Sens. Actuat. B Chem. 2012, 166, 715–720. [Google Scholar] [CrossRef]
- Jolly, P.; Batistuti, M.R.; Ustuner, S.; Mulato, M.; Arya, S.K.; Estrela, P. Nucleic Acid-Based Aptasensors for Cancer Diagnostics: An Insight into Immobilisation Strategies. In Next Generation Point-of-Care Biomedical Sensors Technologies for Cancer Diagnosis; Chandra, P., Tan, Y.N., Singh, S., Eds.; Springer: New York, NY, USA, 2017; pp. 205–231. [Google Scholar]
- Li, X.; Shen, L.; Zhang, D.; Qi, H.; Gao, Q.; Ma, F.; Zhang, C. Electrochemical impedance spectroscopy for study of aptamer–thrombin interfacial interactions. Biosens. Bioelectron. 2008, 23, 1624–1630. [Google Scholar] [CrossRef]
- Tabrizi, M.A.; Shamsipur, M.; Saber, R.; Sarkar, S.; Sherkatkhameneh, N. Flow injection amperometric sandwich-type electrochemical aptasensor for the determination of adenocarcinoma gastric cancer cell using aptamer-Au@ Ag nanoparticles as labeled aptamer. Electrochim. Acta 2017, 246, 1147–1154. [Google Scholar] [CrossRef]
- Chen, D.; Feng, H.; Li, J. Graphene oxide: Preparation, functionalization, and electrochemical applications. Chem. Rev. 2012, 112, 6027–6053. [Google Scholar] [CrossRef] [PubMed]
- Hayat, A.; Sassolas, A.; Marty, J.-L.; Radi, A.-E. Highly sensitive ochratoxin A impedimetric aptasensor based on the immobilization of azido-aptamer onto electrografted binary film via click chemistry. Talanta 2013, 103, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, S.; Obubuafo, A.; Soper, S.A.; Spivak, D.A. Surface immobilization methods for aptamer diagnostic applications. Anal. Bioanal. Chem. 2008, 390, 1009–1021. [Google Scholar] [CrossRef]
- Di Cera, E. Thrombin. Mol. Asp. Med. 2008, 29, 203–254. [Google Scholar] [CrossRef] [PubMed]
- Ireson, C.R.; Kelland, L.R. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006, 5, 2957–2962. [Google Scholar] [CrossRef] [Green Version]
- Padmanabhan, K.; Padmanabhan, K.; Ferrara, J.; Sadler, J.E.; Tulinsky, A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993, 268, 17651–17654. [Google Scholar] [CrossRef]
- Li, Y.; Guo, L.; Zhang, F.; Zhang, Z.; Tang, J.; Xie, J. High-sensitive determination of human α-thrombin by its 29-mer aptamer in affinity probe capillary electrophoresis. Electrophoresis 2008, 29, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-H.; Chen, R.-H.; Lee, C.-H.; Chang, Y.; Chen, C.-S.; Chen, W.-Y. Studies of the binding mechanism between aptamers and thrombin by circular dichroism, surface plasmon resonance and isothermal titration calorimetry. Coll. Surf. B 2011, 88, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Trapaidze, A.; Hérault, J.-P.; Herbert, J.-M.; Bancaud, A.; Gué, A.-M. Investigation of the selectivity of thrombin-binding aptamers for thrombin titration in murine plasma. Biosens. Bioelectron. 2016, 78, 58–66. [Google Scholar] [CrossRef]
- Deng, B.; Lin, Y.; Wang, C.; Li, F.; Wang, Z.; Zhang, H.; Li, X.-F.; Le, X.C. Aptamer binding assays for proteins: The thrombin example—A review. Anal. Chim. Acta 2014, 837, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, X.; Shi, S.; Zhang, Y.; Yao, T. Three label-free thrombin aptasensors based on aptamers and [Ru (bpy)2 (o-mopip)]2+. J. Mater. Chem. B 2016, 4, 1361–1367. [Google Scholar] [CrossRef]
- Crawley, J.; Zanardelli, S.; Chion, C.; Lane, D. The central role of thrombin in hemostasis. J. Thromb. Haemost. 2007, 5, 95–101. [Google Scholar] [CrossRef]
- Huntington, J.A. How Na+ activates thrombin–a review of the functional and structural data. Biol. Chem. 2008, 389, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Walt, D.R. A fiber-optic microarray biosensor using aptamers as receptors. Anal. Biochem. 2000, 282, 142–146. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Lubin, A.A.; Heeger, A.J.; Plaxco, K.W. Label-Free Electronic Detection of Thrombin in Blood Serum by Using an Aptamer-Based Sensor. Angew. Chem. 2005, 44, 5456–5459. [Google Scholar] [CrossRef]
- Lin, S.-F.; Ding, T.-J.; Liu, J.-T.; Lee, C.-C.; Yang, T.-H.; Chen, W.-Y.; Chang, J.-Y. A guided mode resonance aptasensor for thrombin detection. Sensors 2011, 11, 8953–8965. [Google Scholar] [CrossRef] [Green Version]
- Baek, S.H.; Wark, A.W.; Lee, H.J. Dual nanoparticle amplified surface plasmon resonance detection of thrombin at subattomolar concentrations. Anal. Chem. 2014, 86, 9824–9829. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Wang, Y.; Wei, H.; Dong, S. Amplified electrochemical aptasensor taking AuNPs based sandwich sensing platform as a model. Biosens. Bioelectron. 2008, 23, 965–970. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Electrochemical Detection of Protein Using a Double Aptamer Sandwich. Anal. Lett. 2004, 37, 2901–2909. [Google Scholar] [CrossRef]
- Numnuam, A.; Chumbimuni-Torres, K.Y.; Xiang, Y.; Bash, R.; Thavarungkul, P.; Kanatharana, P.; Pretsch, E.; Wang, J.; Bakker, E. Aptamer-Based Potentiometric Measurements of Proteins Using Ion-Selective Microelectrodes. Anal. Chem. 2008, 80, 707–712. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, Z.; Zhai, Y.; Zhang, L.; Hong, W.; Zhang, Z.; Dong, S. Label-free aptamer biosensor for thrombin detection based on functionalized graphene nanocomposites. Talanta 2015, 141, 247–252. [Google Scholar] [CrossRef]
- Qiu, H.; Sun, Y.; Huang, X.; Qu, Y. A sensitive nanoporous gold-based electrochemical aptasensor for thrombin detection. Coll. Surf. B 2010, 79, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, T.; Kim, J.; Shannon, C.; Easley, C.J. Quantitation of Femtomolar Protein Levels via Direct Readout with the Electrochemical Proximity Assay. J. Am. Chem. Soc. 2012, 134, 7066–7072. [Google Scholar] [CrossRef] [Green Version]
- Centi, S.; Tombelli, S.; Minunni, M.; Mascini, M. Aptamer-Based Detection of Plasma Proteins by an Electrochemical Assay Coupled to Magnetic Beads. Anal. Chem. 2007, 79, 1466–1473. [Google Scholar] [CrossRef]
- Centi, S.; Messina, G.; Tombelli, S.; Palchetti, I.; Mascini, M. Different approaches for the detection of thrombin by an electrochemical aptamer-based assay coupled to magnetic beads. Biosens. Bioelectron. 2008, 23, 1602–1609. [Google Scholar] [CrossRef]
- Ding, C.; Ge, Y.; Lin, J.-M. Aptamer based electrochemical assay for the determination of thrombin by using the amplification of the nanoparticles. Biosens. Bioelectron. 2010, 25, 1290–1294. [Google Scholar] [CrossRef]
- Gao, F.; Du, L.; Zhang, Y.; Zhou, F.; Tang, D. A sensitive sandwich-type electrochemical aptasensor for thrombin detection based on platinum nanoparticles decorated carbon nanocages as signal labels. Biosens. Bioelectron. 2016, 86, 185–193. [Google Scholar] [CrossRef]
- He, B. Sandwich electrochemical thrombin assay using a glassy carbon electrode modified with nitrogen-and sulfur-doped graphene oxide and gold nanoparticles. Microchim. Acta 2018, 185, 344. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens. Bioelectron. 2005, 20, 2168–2172. [Google Scholar] [CrossRef]
- Kang, Y.; Feng, K.-J.; Chen, J.-W.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Electrochemical detection of thrombin by sandwich approach using antibody and aptamer. Bioelectrochemistry 2008, 73, 76–81. [Google Scholar] [CrossRef]
- Yeh, F.-Y.; Liu, T.-Y.; Tseng, I.-H.; Yang, C.-W.; Lu, L.-C.; Lin, C.-S. Gold nanoparticles conjugates-amplified aptamer immunosensing screen-printed carbon electrode strips for thrombin detection. Biosens. Bioelectron. 2014, 61, 336–343. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Y.; Li, H.; Wen, Y.; Fan, X.; Lin, F.; Tan, L.; Yao, S. Ultrasensitive electrochemical aptasensor for thrombin based on the amplification of aptamer-AuNPs-HRP conjugates. Biosens. Bioelectron. 2011, 26, 2297–2303. [Google Scholar] [CrossRef]
- Chung, S.; Moon, J.-M.; Choi, J.; Hwang, H.; Shim, Y.-B. Magnetic force assisted electrochemical sensor for the detection of thrombin with aptamer-antibody sandwich formation. Biosens. Bioelectron. 2018, 117, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Fan, G.; Chen, W.; Liu, Q.; Zhang, X.; Zhang, X.; Liu, Q. Electrochemical sandwich-type thrombin aptasensor based on dual signal amplification strategy of silver nanowires and hollow Au–CeO2. Biosens. Bioelectron. 2020, 150, 111846. [Google Scholar] [CrossRef]
- Fang, L.-X.; Huang, K.-J.; Liu, Y. Novel electrochemical dual-aptamer-based sandwich biosensor using molybdenum disulfide/carbon aerogel composites and Au nanoparticles for signal amplification. Biosens. Bioelectron. 2015, 71, 171–178. [Google Scholar] [CrossRef]
- Lorenzo-Gómez, R.; Fernández-Alonso, N.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Unravelling the lipocalin 2 interaction with aptamers: May rolling circle amplification improve their functional affinity? Talanta 2019, 197, 406–412. [Google Scholar] [CrossRef]
- Aydoğdu Tığ, G.; Pekyardımcı, Ş. An electrochemical sandwich-type aptasensor for determination of lipocalin-2 based on graphene oxide/polymer composite and gold nanoparticles. Talanta 2020, 210, 120666. [Google Scholar] [CrossRef] [PubMed]
- Salimi, A.; Khezrian, S.; Hallaj, R.; Vaziry, A. Highly sensitive electrochemical aptasensor for immunoglobulin E detection based on sandwich assay using enzyme-linked aptamer. Anal. Biochem. 2014, 466, 89–97. [Google Scholar] [CrossRef]
- Eissa, S.; Zourob, M. Aptamer-based label-free electrochemical biosensor array for the detection of total and glycated hemoglobin in human whole blood. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Fernández, A.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Rodríguez, E.F.; Lobo-Castañón, M.J. Focusing aptamer selection on the glycan structure of prostate-specific antigen: Toward more specific detection of prostate cancer. Biosens. Bioelectron. 2019, 128, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Fernândez, A.; Miranda-Castro, R.; Diaz, N.; Suârez, D.; de-los-Santos-Álvarez, N.; Lobo-Castañon, M. Aptamers targeting protein-specific glycosylation in tumor markers: General selection, characterization and structural modeling. Chem. Sci. 2020, 11, 9402–9413. [Google Scholar] [CrossRef]
- Kashefi-Kheyrabadi, L.; Mehrgardi, M.A. Aptamer-based electrochemical biosensor for detection of adenosine triphosphate using a nanoporous gold platform. Bioelectrochemistry 2013, 94, 47–52. [Google Scholar] [CrossRef]
- Sun, D.; Lin, X.; Lu, J.; Wei, P.; Luo, Z.; Lu, X.; Chen, Z.; Zhang, L. DNA nanotetrahedron-assisted electrochemical aptasensor for cardiac troponin I detection based on the co-catalysis of hybrid nanozyme, natural enzyme and artificial DNAzyme. Biosens. Bioelectron. 2019, 142, 111578. [Google Scholar] [CrossRef]
- Luo, Z.; Sun, D.; Tong, Y.; Zhong, Y.; Chen, Z. DNA nanotetrahedron linked dual-aptamer based voltammetric aptasensor for cardiac troponin I using a magnetic metal-organic framework as a label. Microchim. Acta 2019, 186, 374. [Google Scholar] [CrossRef]
- Villalongo, A.; Estabiel, I.; Perez-Calabuig, A.M.; Mayol, B.; Parrado, C.; Villalongo, R. Amperometric aptasensor with sandwich-type architecture for troponin I based on carboxyethylsilanetriol-modified graphene oxide coated electrodes. Biosens. Bioelectron. 2021, 183, 113203. [Google Scholar] [CrossRef]
- Jo, H.; Her, J.; Lee, H.; Shim, Y.-B.; Ban, C. Highly sensitive amperometric detection of cardiac troponin I using sandwich aptamers and screen-printed carbon electrodes. Talanta 2017, 165, 442–448. [Google Scholar] [CrossRef]
- Ou, D.; Sun, D.; Lin, X.; Liang, Z.; Zhong, Y.; Chen, Z. A dual-aptamer-based biosensor for specific detection of breast cancer biomarker HER2 via flower-like nanozymes and DNA nanostructures. J. Mat. Chem. B 2019, 7, 3661–3669. [Google Scholar] [CrossRef]
- Jiang, B.; Wang, M.; Chen, Y.; Xie, J.; Xiang, Y. Highly sensitive electrochemical detection of cocaine on graphene/AuNP modified electrode via catalytic redox-recycling amplification. Biosens. Bioelectron. 2012, 32, 305–308. [Google Scholar] [CrossRef]
- Zhu, X.; Yang, J.; Liu, M.; Wu, Y.; Shen, Z.; Li, G. Sensitive detection of human breast cancer cells based on aptamer–cell–aptamer sandwich architecture. Anal. Chim. Acta 2013, 764, 59–63. [Google Scholar] [CrossRef]
- Yazdanparast, S.; Benvidi, A.; Banaei, M.; Nikukar, H.; Tezerjani, M.D.; Azimzadeh, M. Dual-aptamer based electrochemical sandwich biosensor for MCF-7 human breast cancer cells using silver nanoparticle labels and a poly(glutamic acid)/MWNT nanocomposite. Microchim. Acta 2018, 185, 405. [Google Scholar] [CrossRef]
- Abbaspour, A.; Norouz-Sarvestani, F.; Noori, A.; Soltani, N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of staphylococcus aureus. Biosens. Bioelectron. 2015, 68, 149–155. [Google Scholar] [CrossRef]
- Kim, S.H.; Nam, O.; Jin, E.; Gu, M.B. A new coccolith modified electrode-based biosensor using a cognate pair of aptamers with sandwich-type binding. Biosens. Bioelectron. 2019, 123, 160–166. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H. An antifouling interface integrated with HRP-based amplification to achieve a highly sensitive electrochemical aptasensor for lysozyme detection. Analyst 2019, 144, 5794–5801. [Google Scholar] [CrossRef]
- Diba, F.S.; Kim, S.; Lee, H.J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015, 72, 355–361. [Google Scholar] [CrossRef]
- Ocaña, C.; Hayat, A.; Mishra, R.; Vasilescu, A.; del Valle, M.; Marty, J.-L. A novel electrochemical aptamer–antibody sandwich assay for lysozyme detection. Analyst 2015, 140, 4148–4153. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Hayati, S.; Kim, S.; Lee, H.J. Determination of protein tyrosine kinase-7 concentration using electrocatalytic reaction and an aptamer-antibody sandwich assay platform. Catal. Today 2021, 359, 76–82. [Google Scholar] [CrossRef]
- Jung, J.-W.; Shin, W.-S.; Song, J.; Lee, S.-T. Cloning and characterization of the full-length mouse Ptk7 cDNA encoding a defective receptor protein tyrosine kinase. Gene 2004, 328, 75–84. [Google Scholar] [CrossRef]
- Qureshi, A.; Gurbuz, Y.; Niazi, J.H. Capacitive aptamer–antibody based sandwich assay for the detection of VEGF cancer biomarker in serum. Sens. Actuators B Chem. 2015, 209, 645–651. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, H.; Liu, L.; Li, C.; Chang, Z.; Zhu, X.; Ye, B.; Xu, M. Fabrication of an antibody-aptamer sandwich assay for electrochemical evaluation of levels of β-amyloid oligomers. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Shui, B.; Tao, D.; Cheng, J.; Mei, Y.; Jaffrezic-Renault, N.; Guo, Z. A novel electrochemical aptamer-antibody sandwich assay for the detection of tau-381 in human serum. Analyst 2018, 143, 3549–3554. [Google Scholar] [CrossRef]
- Wang, J.; Munir, A.; Li, Z.; Zhou, H.S. Aptamer-Au NPs conjugates-accumulated methylene blue for the sensitive electrochemical immunoassay of protein. Talanta 2010, 81, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, J.; Zhang, J.; Zhang, W.; Zhang, Y. RNA aptamer-based electrochemical aptasensor for C-reactive protein detection using functionalized silica microspheres as immunoprobes. Biosens. Bioelectron. 2017, 95, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Centi, S.; Bonel Sanmartin, L.; Tombelli, S.; Palchetti, I.; Mascini, M. Detection of C Reactive Protein (CRP) in Serum by an Electrochemical Aptamer-Based Sandwich Assay. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2009, 21, 1309–1315. [Google Scholar] [CrossRef]
- Zhu, G.; Lee, H.J. Electrochemical sandwich-type biosensors for α−1 antitrypsin with carbon nanotubes and alkaline phosphatase labeled antibody-silver nanoparticles. Biosens. Bioelectron. 2017, 89, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Nie, X.-M.; Gan, S.-L.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Electrochemical immunosensor of platelet-derived growth factor with aptamer-primed polymerase amplification. Anal. Biochem. 2008, 382, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Taleat, Z.; Cristea, C.; Marrazza, G.; Mazloum-Ardakani, M.; Săndulescu, R. Electrochemical immunoassay based on aptamer–protein interaction and functionalized polymer for cancer biomarker detection. J. Electroanal. Chem. 2014, 717–718, 119–124. [Google Scholar] [CrossRef]
- Lu, L.; Liu, B.; Leng, J.; Ma, X.; Peng, H. Electrochemical mixed aptamer-antibody sandwich assay for mucin protein 16 detection through hybridization chain reaction amplification. Anal. Bioanal. Chem. 2020, 412, 7169–7178. [Google Scholar] [CrossRef] [PubMed]
- Ghalehno, M.H.; Mirzaei, M.; Torkzadeh-Mahani, M. Electrochemical aptasensor for tumor necrosis factor α using aptamer–antibody sandwich structure and cobalt hexacyanoferrate for signal amplification. J. Iran. Chem. Soc. 2019, 16, 1783–1791. [Google Scholar] [CrossRef]
- Gu, Y.; Liu, L.; Guo, J.; Xiao, S.; Fang, F.; Yu, X.; Gopinath, S.C.B.; Wu, J.; Liu, X. Biomolecular assembly on interdigitated electrode nanosensor for selective detection of insulin-like growth factor-1. Art. Cells Nanomed. Biotech. 2021, 49, 30–37. [Google Scholar] [CrossRef]
- You, M.; Yang, S.; An, Y.; Zhang, F.; He, P. A novel electrochemical biosensor with molecularly imprinted polymers and aptamer-based sandwich assay for determining amyloid-β oligomer. J. Electroanal. Chem. 2020, 862, 114017. [Google Scholar] [CrossRef]
- Wang, Q.-L.; Cui, H.-F.; Song, X.; Fan, S.-F.; Chen, L.-L.; Li, M.-M.; Li, Z.-Y. A label-free and lectin-based sandwich aptasensor for detection of carcinoembryonic antigen. Sens. Actuators B Chem. 2018, 260, 48–54. [Google Scholar] [CrossRef]
- Chai, Y.; Li, X.; Yang, M. Aptamer based determination of the cancer biomarker HER2 by using phosphate-functionalized MnO2 nanosheets as the electrochemical probe. Microchim. Acta 2019, 186, 1–6. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
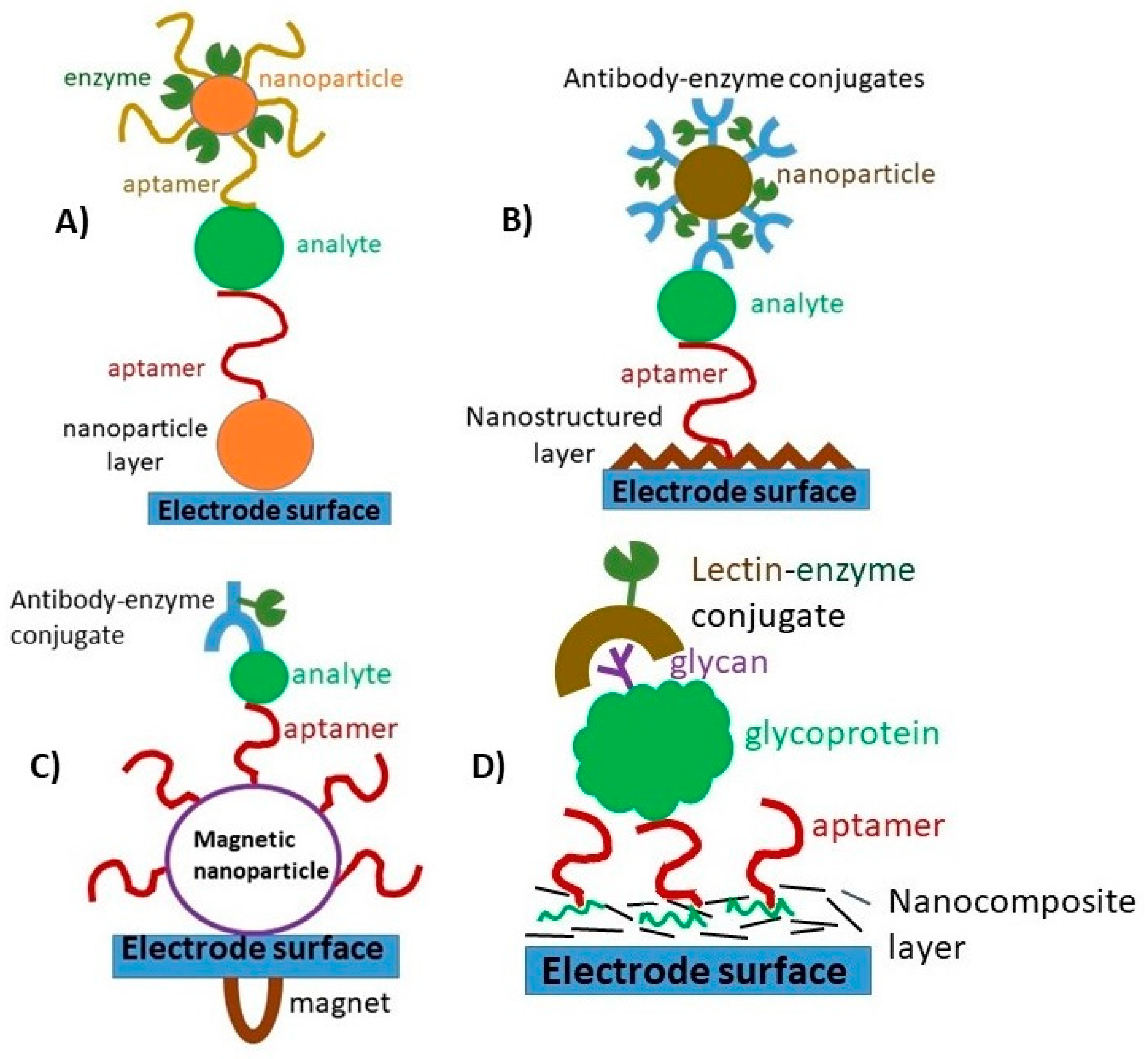

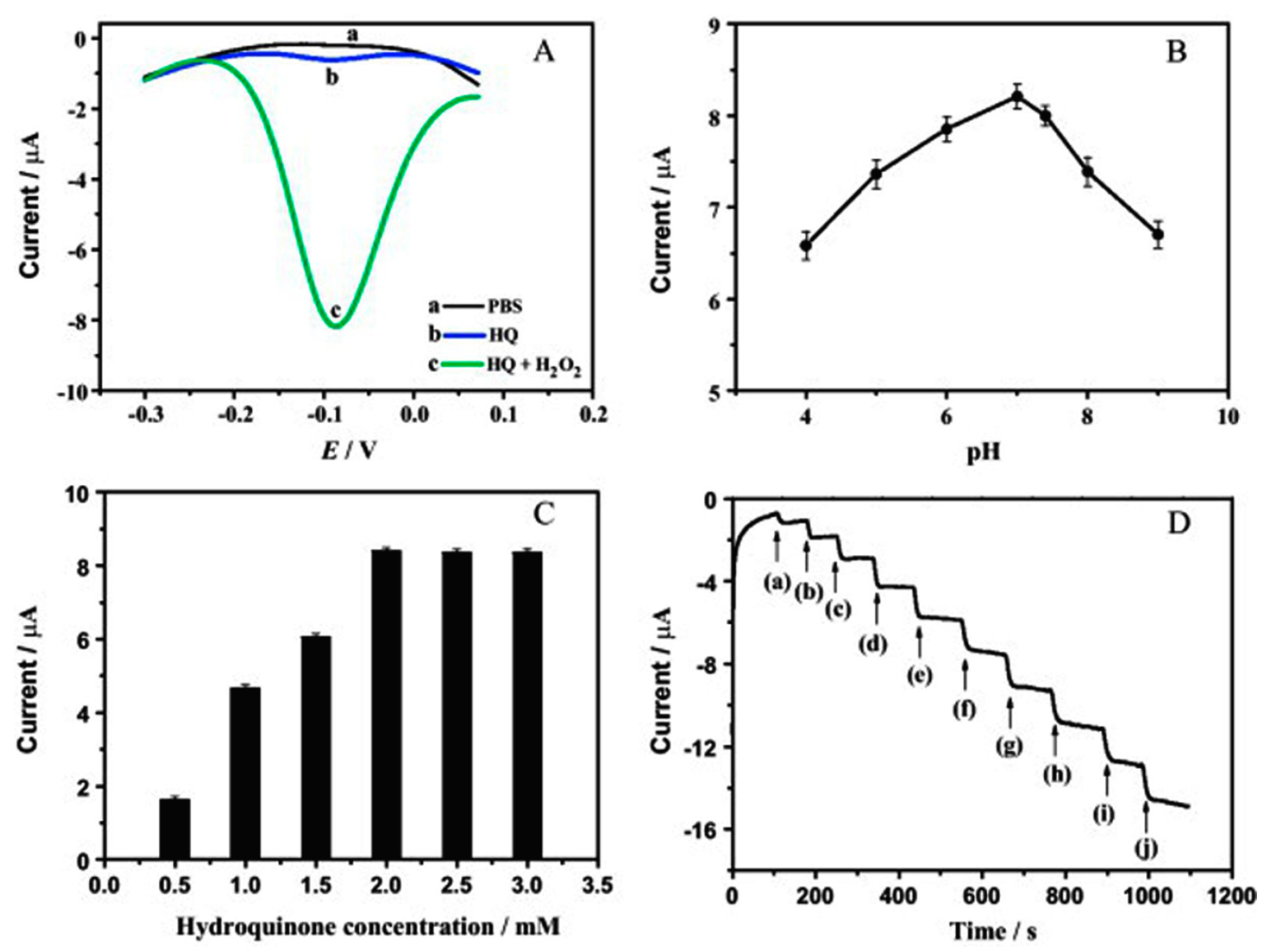
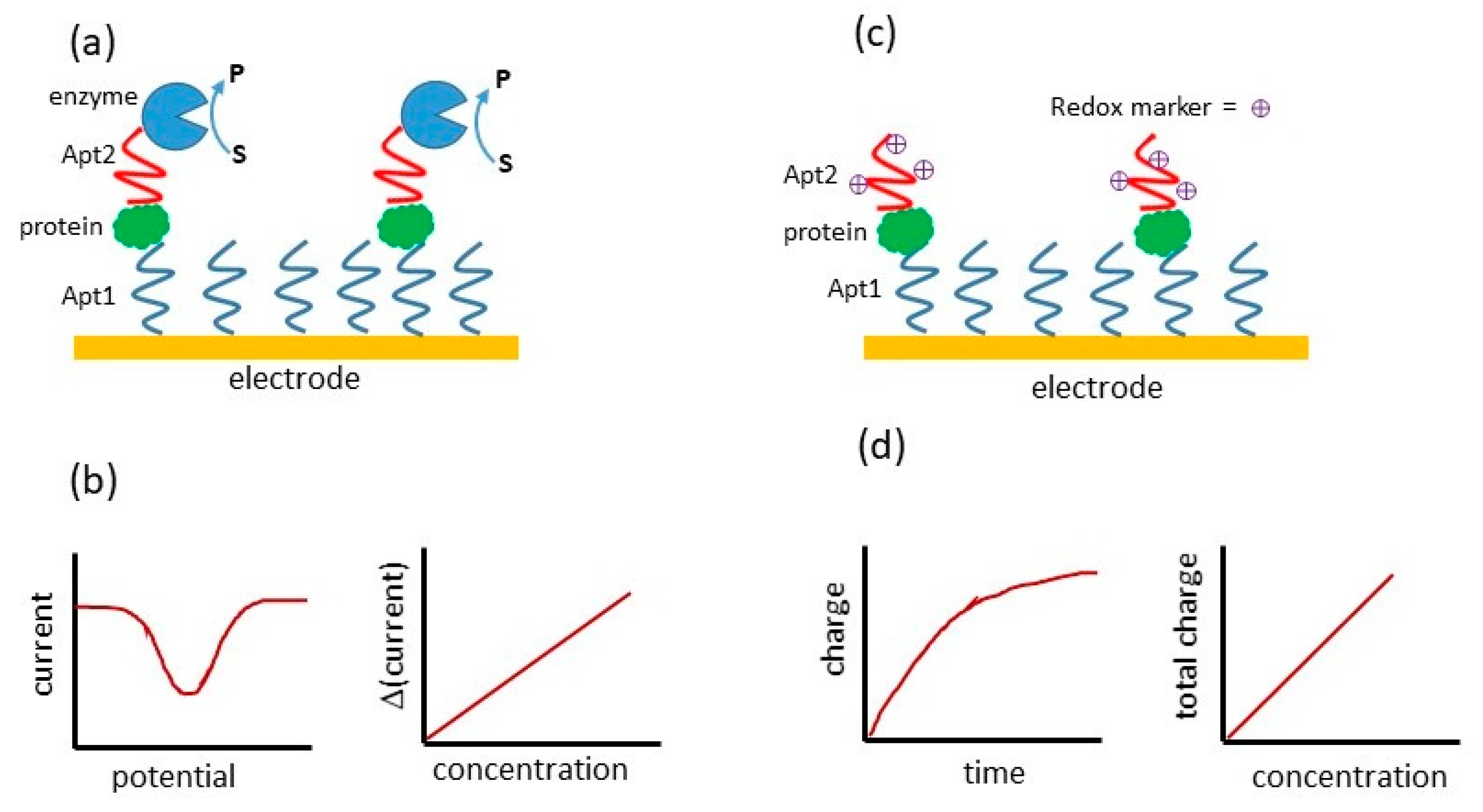

| Components of Sandwich Assay for Thrombin (Th = Thrombin, SAM = Self-Assembled Monolayer) | Method of Detection | Linear Range | Limit of Detection | Ref. |
|---|---|---|---|---|
| Au/SAM/Aptamer covalently attached to methylene blue (not a sandwich assay, for comparison only) | AC voltammetry | 6.4 nM–768 nM | 6.4 nM | [35] |
| Au/thiolated aptamer/Th/aptamer-modified gold nanoparticles with adsorbed rhodamine 6G | Electrochemical Impedance Spectroscopy | 0.05–18 nM | 0.02 nM | [38] |
| Au/thiolated aptamer/Th/glucose dehydrogenase (GDH)-labeled aptamer (glucose substrate) | Amperometry | nonlinear | 1 μM | [39] |
| Au/mixed SAM with thiolated aptamer/Th/aptamer labeled CdS quantum dots | Potentiometry | 100 pM–1 nM | 0.14 nM | [40] |
| Au electrode/thiolated aptamer/Th/graphene modifed Au nanoparticles with reporter aptamer | Electrochemical Impedance Spectroscopy | 0.3–50 nM | 0.01 nM | [41] |
| Nanoporous Au/thiolated aptamer in mixed SAM/Th/aptamer-modified Au nanoparticles | Chronocoulometry | 0.01–22 nM | 30 fM | [42] |
| Screen-printed carbon electrode/streptavidin coated magnetic beads/5′-biotinylated primary aptamer/Th/5′-biotinylated secondary aptamer/streptavidin–alkaline phosphatase conjugate | Differential pulse voltammetry | 0.1–100 nM (non-linear) | 0.45 nM | [44] |
| Screen-printed carbon electrode, streptavidin-coated magnetic beads, thrombin, biotinylated thrombin, streptavidin–alkaline phosphatase conjugate, β-Ala-Gly-Arg-p-nitroaniline (thrombin substrate) (not sandwich, indirect and direct competitive assay for comparison) | Differential pulse voltammetry | 0–2000 nM (not linear) N/A 100–600 nM studied | 430 nM (direct) not successful (indirect) 175 nM (thrombin product detection) | [45] |
| Au/thiolated aptamer in mixed SAM/Th/Au nanoparticles modified with aptamer and DNA linked CdS nanoparticles | Differential pulse voltammetry | 1.0 fM–10 pM | 0.55 fM | [46] |
| Au/thiolated aptamer/Th/Pt nanoparticle decorated carbon nanocages | Differential pulse voltammetry | 0.05 pM–20 nM | 10 fM | [47] |
| Glassy carbon electrode/graphene oxide sheets doped with nitrogen and sulfur decorated with gold nanoparticles/thiolated aptamer/Th/graphene oxide sheets doped with nitrogen and sulfur decorated with gold nanoparticles modified by thiolated secondary aptamer and thiolated biotinylated DNA/avidin-horseradish peroxidase conjugate | Differential pulse voltammetry | 0.1 pM–10 nM (vs. log[Th]) | 25 fM | [48] |
| Au/thiolated aptamer/Th/pyrroquinoline quinone glucose dehydrogenase labeled secondary aptamer | Chrono-amperometry | 40–100 nM | 10 nM | [49] |
| Glassy carbon electrode/chitosan-nanogold film/cross-linked antibody/Th/secondary aptamer and methylene blue | Differential pulse voltammetry | 1–60 nM | 0.5 nM | [50] |
| Screen-printed carbon electrode/Au nanoparticles and ferrocene dicarboxylic acid mediator/streptavidin/biotinylated aptamer/Th/Au nanoparticles modified with horseradish peroxidase-antibody conjugates | Chrono-amperometry | 10 pM–0.1 μM | 1.5 pM | [51] |
| Au/core–shell Fe3O4/Au magnetic nanoparticles modified with primary aptamer/Th/Au nanoparticles labeled with secondary aptamer and horseradish peroxidase | Differential pulse voltammetry | 0.1–60 pM | 30 fM | [52] |
| Screen-printed carbon electrodes/conducting polymer layer conjugated to primary aptamer/Th/streptavidin–starch coated magnetic nanoparticles and biotinylated secondary aptamer, toluidine blue O | Chrono-amperometry | 1.0–500 nM | 0.49 nM | [53] |
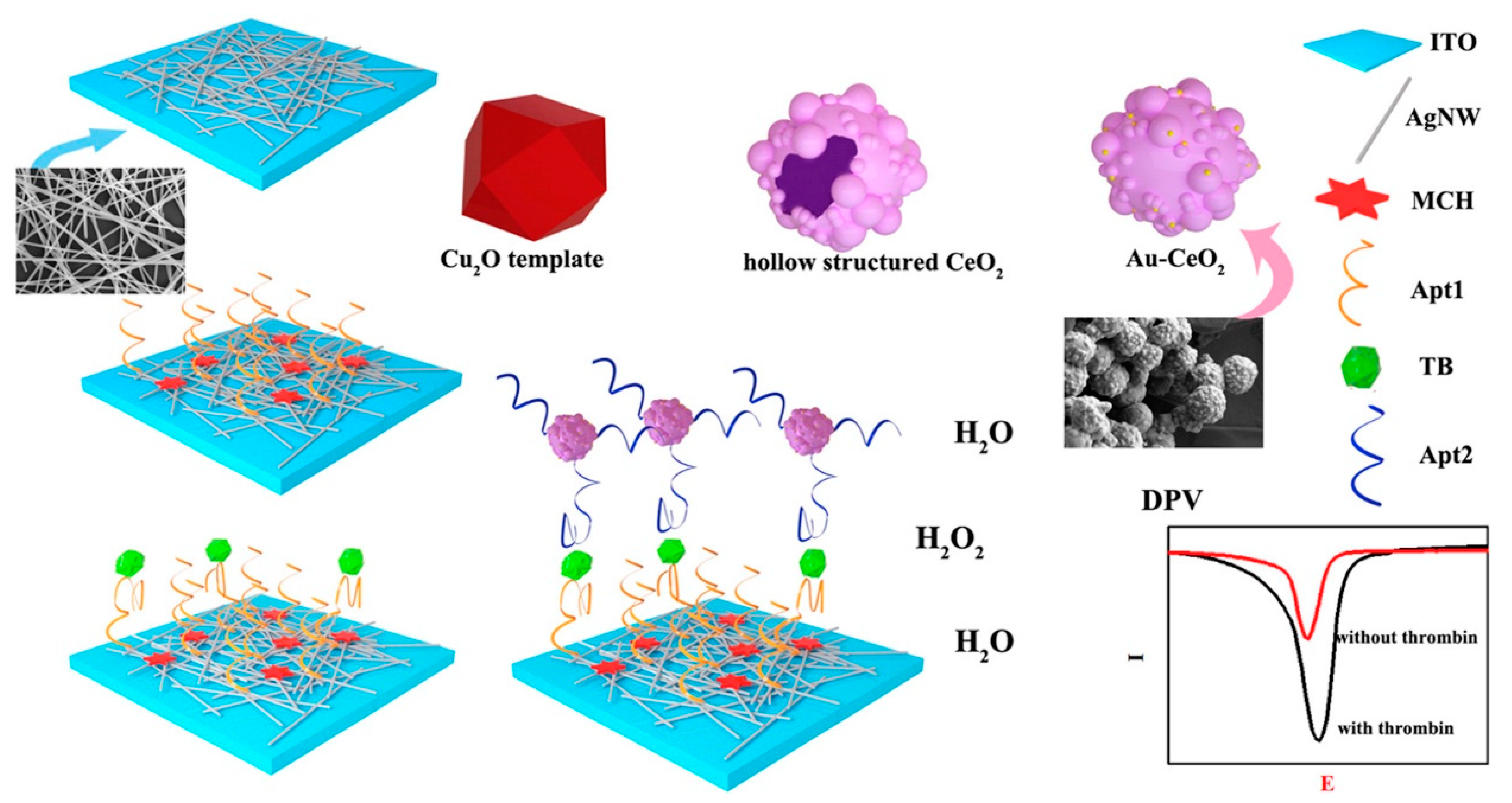
| Indium tin oxide electrodes/Ag nanowires/amino-functionalized primary aptamer/Th/Au nanoparticles on hollow CeO2 structures with secondary aptamer | Differential pulse voltammetry | 0.5 pM–30 nM | 0.25 pM | [54] |
| Analyte | Components of Sandwich Assay | Method of Detection | Linear Range | Limit of Detection | Ref. |
|---|---|---|---|---|---|
| Platelet-Derived Growth Factor BB (PDGF-BB) | Glassy carbon electrode/carbon aerogel-MoS2 composite with Au nanoparticles/primary aptamer/PDGF-BB/Au nanoparticles with secondary aptamer and 6-ferrocenylhexanethiol | Cyclic voltammetry | 0.001–10 nM | 0.3 pM | [55] |
| Lipocalin-2 | Screen-printed Au electrode/magnetic beads coated with capture aptamer/lipocalin-2/reporter aptamer tagged with biotin or 6-carboxyfluorescein /peroxidase label (either by streptavidin conjugate or Fab-antiFITC conjugate) | Chrono-amperometry | N/A | N/A | [56] |
| Lipocalin-2 | Graphite screen-printed electrode/conducting polymer/Au nanoparticles/thiolated aptamer/lipocalin-2/biotinylated aptamer/alkaline phosphatase-streptavidin conjugate | Differential pulse voltammetry | 1–1000 ng/mL | 0.3 ng/mL | [57] |
| Immuno-globulin E (IgE) | Glassy carbon electrode/multi-wall carbon nanotubes + ionic liquid + chitosan composite conjugated to terepthaloyl chloride conjugated to thionine/5′-NH2-IgE aptamer/IgE/biotinylated aptamer/streptavidin–horseradish peroxidase conjugate | Differential Pulse Voltammetry | 0.05–2 nM | 6 pM | [58] |
| Glycated hemoglobin (hbA1c) and total hemoglobin (tHb) | Screen-printed gold electrode/gold nanoparticles modified with thiolated aptamer and mercaptohexanol/hbA1c or tHb | Square wave voltammetry | 100 pg/mL–100 ng/mL | 0.2 ng/mL (tHb); 0.34 ng/mL (hbA1c) | [59] |
| Prostate-specific antigen (PSA) | Screen-printed Au electrode/self-assembled monolayer/streptavidin/biotinylated aptamer/PSA/fluorescein-labeled aptamer/anti-fluorescein–peroxidase conjugate | Chrono-amperometry | Nonlinear, Langmuir fit; 0.66–25 ng/mL in diluted serum; 0.66–62.5 ng/mL in TBS buffer | 0.66 ng/mL | [60] |
| Adenosine tri-phosphate (ATP) | Nanoporous gold on gold recordable CD/capture aptamer/ATP/diamino benzoic acid labeled detection aptamer | Differential pulse voltammetry | Nonlinear, 100 nM to 3 mM | 100 nM | [62] |
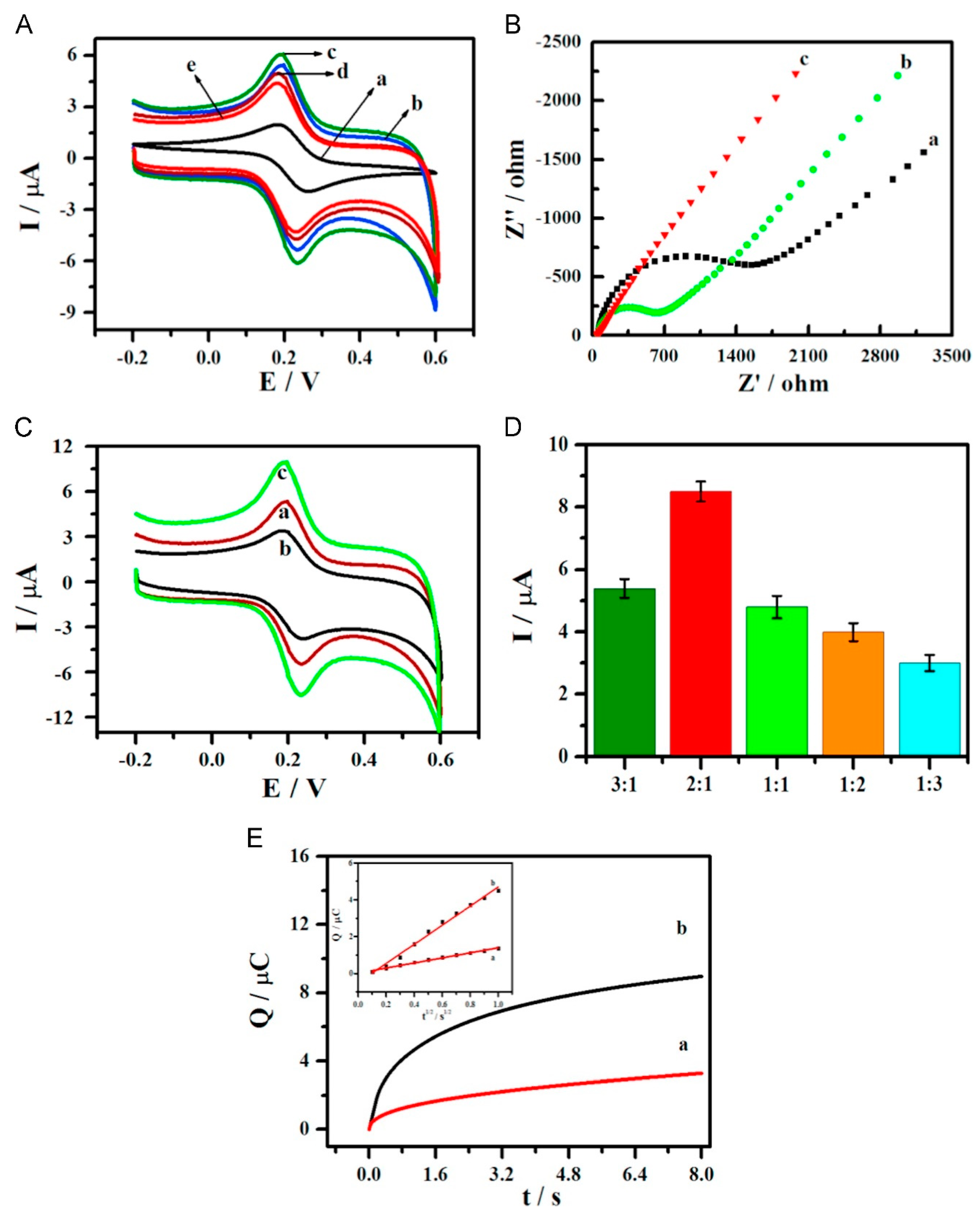
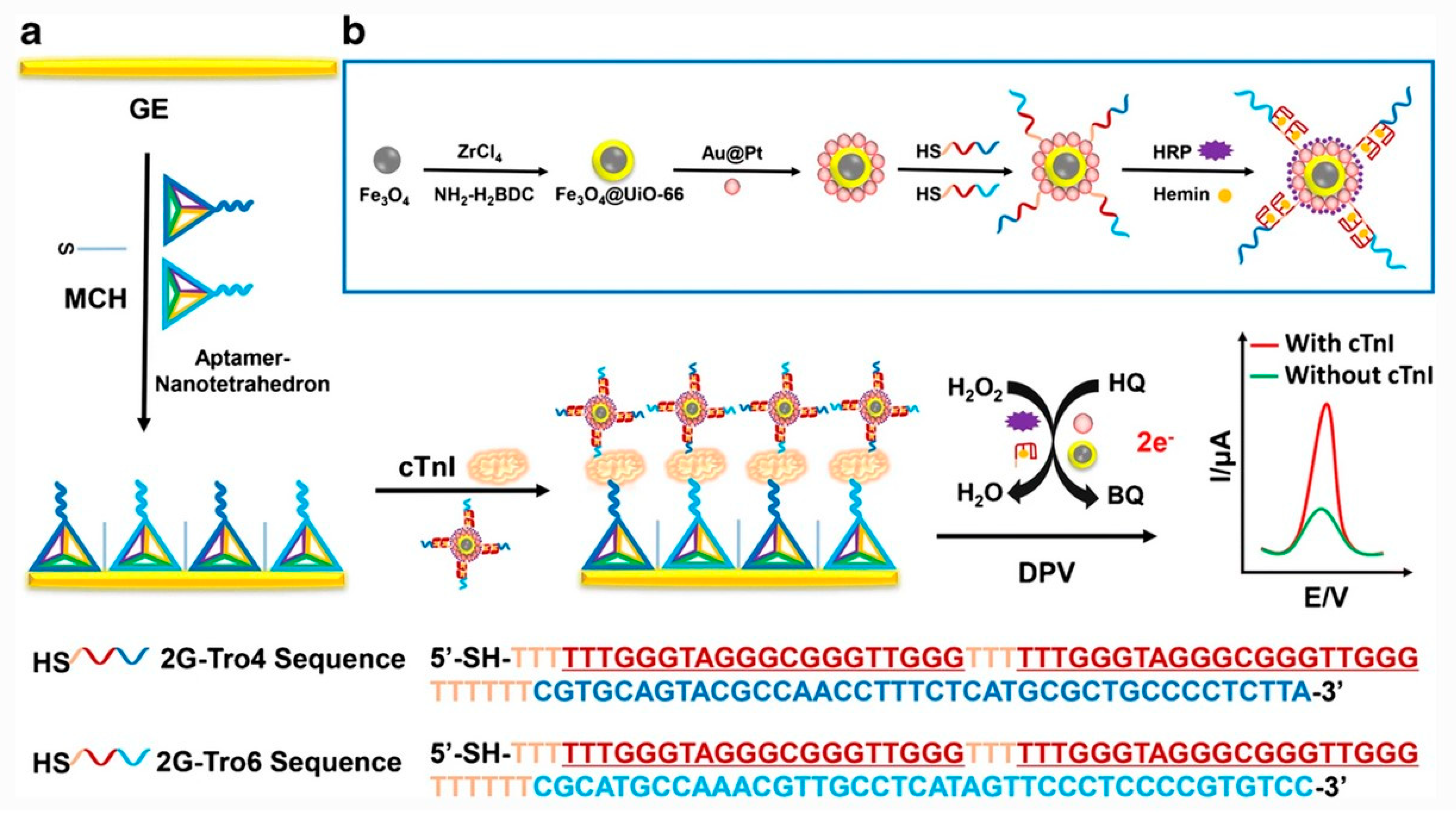
| Cardiac troponin I (cTnI) | Screen-printed gold electrode/DNA nanotetrahedron/aptamer/cTnI/Fe3O4 nanoparticles with aptamer and horseradish peroxidase (HRP), HRP-mimicking Au@Pt nanozymes and G-quadruplex/hemin DNAzyme | Differential pulse voltammetry | 10–105 pg mL−1 | 7.5 pg mL−1 | [63] |
| Cardiac troponin I (cTnI) | Gold electrode/DNA nanotetrahedron/aptamer/cTnI/magnetic metal organic frameworks decorated with Au@Pt nanoparticles, horseradish peroxidase, G-quadruplex/hemin DNAzyme, and aptamers | Differential pulse voltammetry | 10–105 pg mL−1 | 5.7 pg mL−1 | [64] |
| Cardiac troponin I (cTnI) | Screen-printed carbon electrode/carboxyethylsilanetriol-modified graphene oxide/amine-terminated aptamer/cTnI/aptamer–horseradish peroxidase conjugate | Chrono-amperometry | Up to 1.0 μg/mL | 3.4 pg/mL | [65] |
| Cardiac troponin I (cTnI) | Screen-printed carbon electrode/Au nanoparticles/conducting polymer layer/Tro4 aptamer/cTnI/Tro6 aptamer conjugated to hydrazine | Chrono- amperometry | 1–100 pM | 1.0 pM | [66] |
| Human epidermal growth receptor -2 (HER2) | Thiolated DNA nanotetrahedron presenting capture aptamer/HER2/Mn3O4 nanoflowers decorated by Pd@Pt nanoflowers and horseradish peroxidase/complementary signal DNA on Pd@Pt nanoflowers | Differential pulse voltammetry | 0.1–100 ng/mL | 0.08 ng/mL | [67] |
| Cocaine | Screen-printed carbon electrode/graphene and gold nanoparticles/aptamer/cocaine/biotinylated aptamer/streptavidin–alkaline phosphatase conjugate | Differential pulse voltammetry | 1–500 nM | 1 nM | [68] |
| MCF-7 human breast cancer cells | Gold electrode/thiolated apatamer/cells/horseradish peroxidase labeled aptamer | Cyclic voltammetry | 100–107 cells | 100 cells | [69] |
| MCF-7 human breast cancer cells | Glassy carbon electrode/multi-walled carbon nanotube and poly(glutamic acid) composite/MUC-1 aptamer/MCF-7 cell/Ag nanoparticles labeled with MUC-1 aptamer | Differential pulse voltammetry | 2–7 in log(cells mL−1) | 25 cells | [70] |
| S. Aureus bacteria | Streptavidin coated magnetic beads/biotinylated aptamer/bacteria/aptamer-modified silver nanoparticles | Differential pulse voltammetry | 10 to 1 × 106 CFU/mL | 1.0 CFU/mL | [71] |
| Vaspin | Screen-printed Au electrode/gold-sputtered coccoliths/thiolated aptamer/vaspin/aptamer–horseradish peroxidase conjugate | Chrono-amperometry | Up to 18 nM | 298 pM | [72] |
| Lysozyme | Gold electrode/dithiothreitol and aptamer/lysozyme/gold nanoparticle labeled with aptamer and horseradish peroxidase | Differential pulse voltammetry | Linear in log[lys] from 0.01–105 pg/mL | 0.003 pg/mL | [73] |
| Analyte | Components of Sandwich Assay | Method of Detection | Linear Range | Limit of Detection | Ref. |
|---|---|---|---|---|---|
| H5N1 avian influenza virus protein | Screen-printed carbon electrode/Au nanoparticles/aptamer/H5N1/antibody–alkaline phosphatase conjugate | Differential pulse voltammetry | 100 fM–10 pM and 25–100 pM | 100 fM | [74] |
| Lysozyme | Screen-printed carbon electrode/benzoic acid/aptamer/lysozyme/biotinylated antibody/alkaline phosphatase–avidin conjugate | Differential pulse voltammetry | 1 fM–5 nM | 4.3 fM | [75] |
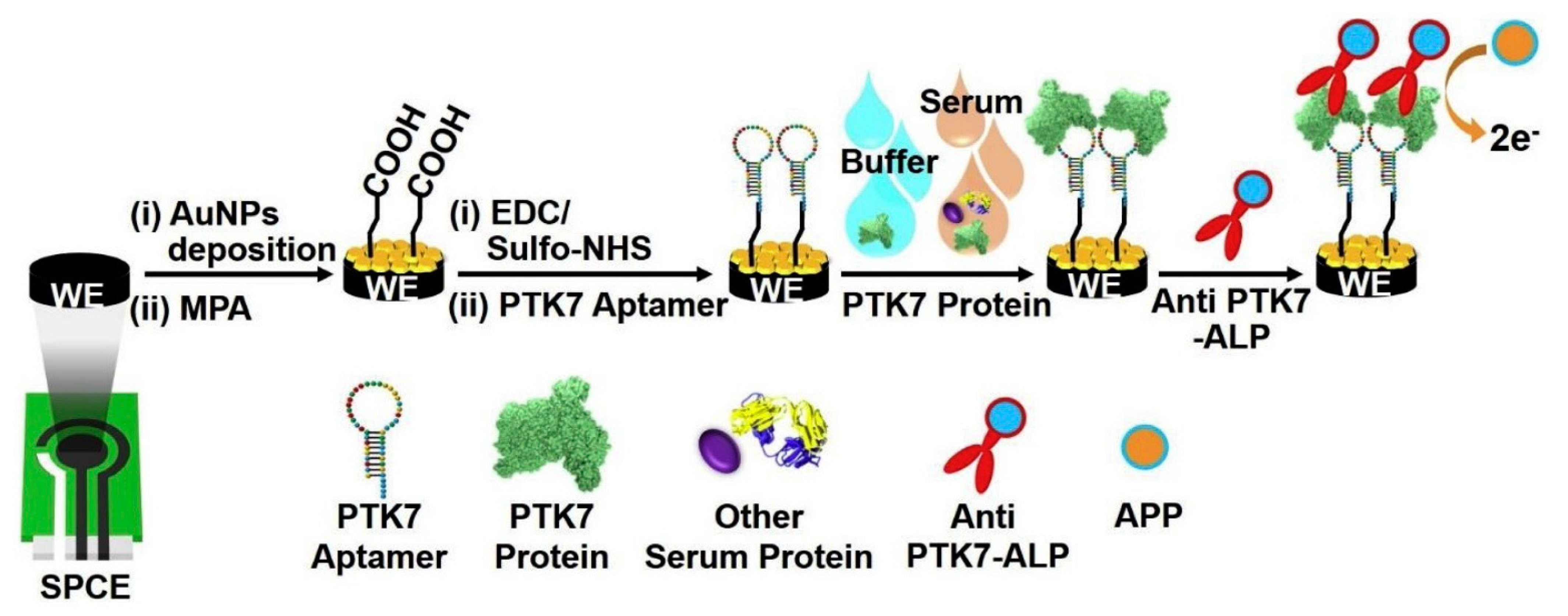
| Protein tyrosine kinase-7 (PTK7) | Screen-printed electrodes/Au nanoparticles/aptamer/PTK7/antibody labeled with alkaline phosphatase | Differential pulse voltammetry | 100 fM–10 pM | 100 fM | [76] |
| Vascular endothelial growth factor-165 (VEGF) | Interdigitated gold electrodes/aptamer and thionine/VEGF/antibody–conjugated magnetic beads | Non-Faradaic impedance spectroscopy | 5 pg/mL–1 ng/mL | 5 pg/mL | [78] |
| Amyloid β-oligomers | Glassy carbon electrode/aptamer/amyloid β/aptamer and thionine-modified Au nanoparticles | Differential pulse voltammetry | 0.5–30 nM | 100 pM | [79] |
| Tau-381 protein | Au electrode/mercaptopropionic acid monolayer/anti-tau antibody/tau-381/Au nanoparticles with aptamer and cysteamine, Fe(CN)63−/4− in solution. | Differential pulse voltametry | 0.5–100 pM | 0.5 pM | [80] |
| Human immunoglobulin E (IgE) | Gold electrode/antibody/aptamer-modified Au nanoparticles with methylene blue | Differential pulse voltammetry | 1–10,000 ng/mL | 0.52 ng/mL | [81] |
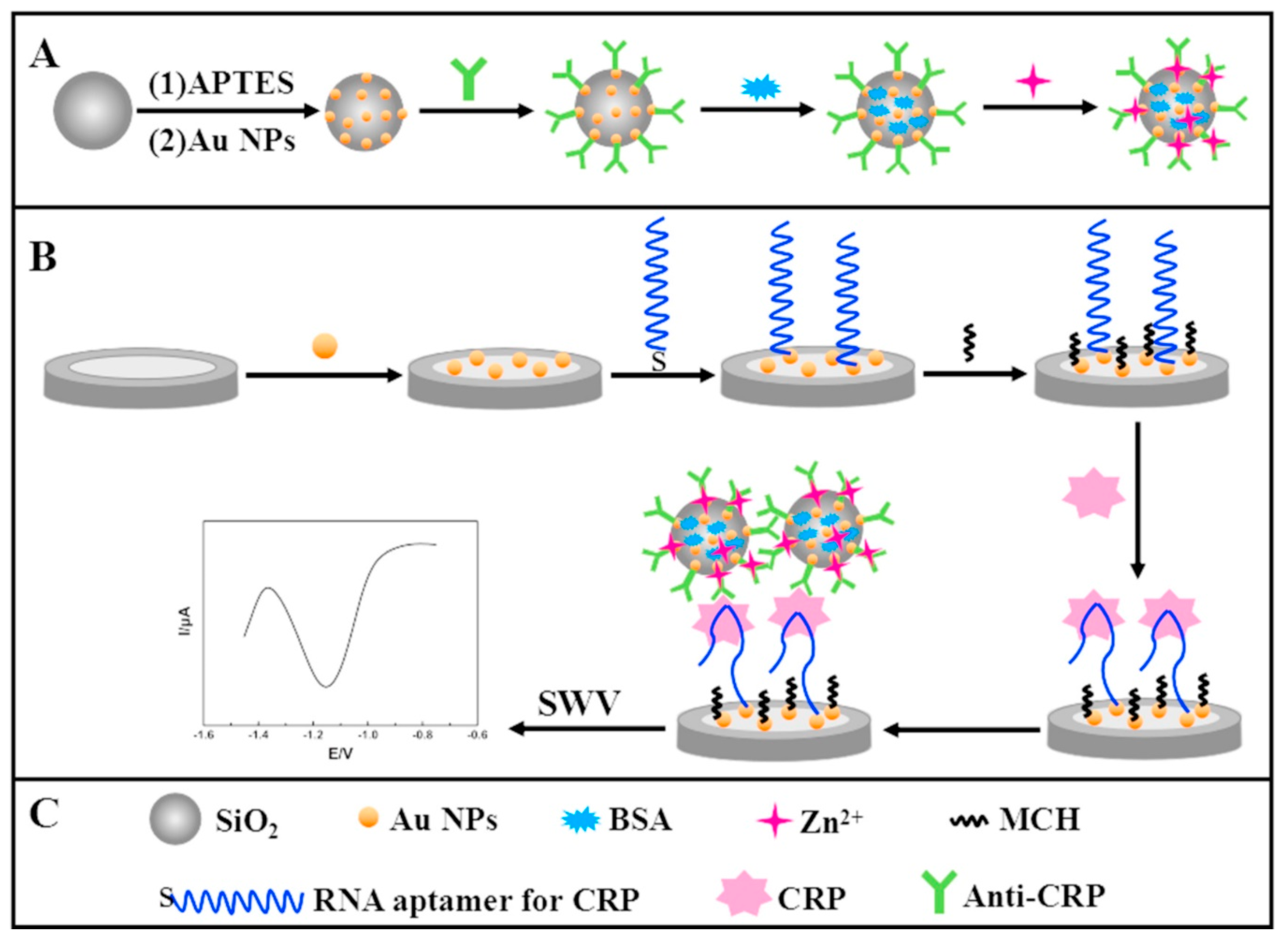
| C-reactive protein | Glassy carbon electrode/Au nanoparticles/aptamer/silica microspheres with Au nanoparticles and Zn2+ | Square-wave voltammetry | 0.005 ng/mL–125 ng/mL | 0.0017 ng/mL | [82] |
| C-reactive protein | Magnet/carbon electrode/streptavidin-coated magnetic beads/biotinylated aptamer/CRP/biotinylated antibody/alkaline phosphatase–streptavidin conjugate | Differential pulse voltammetry | 0.1–50 mg/L (nonlinear response) | 0.054 mg/L | [83] |
| α-1 antitrypsin (AAT) | Screen-printed electrode/carbon nanotubes/aptamer/AAT/Ag nanoparticles modified by antibody–alkaline phosphatase conjugate | Differential pulse voltammetry | 0.05–20 pM | 0.01 pM | [84] |
| Platelet-derived growth factor BB (PDGF-BB) | Gold electrode/antibody/PDGF/aptamer with circular DNA probe/methylene blue | Square-wave voltammetry | 50–500 ng mL−1 | 18 pg mL−1 | [85] |
| Mucin protein-1 (MUC1) | Graphite working electrode/conducting polymer/antibody/MUC1/aptamer/methylene blue | Differential pulse voltammetry | 3–10 ppb | 2.4 ppb | [86] |
| Mucin protein-16 (MUC16) | Indium tin oxide electrode/gold nanoparticles/antibody/MUC16/aptamer and primer tail/DNA/methylene blue | Differential pulse voltammetry | 0.39–200 unit/mL | 0.02 unit/mL | [87] |
| Tumor necrosis factor α (TNFα) | Screen-printed graphite electrode/cobalt hexacyanoferrate/Au nanoparticles/aptamer/TNFa/antibody conjugated to horseradish peroxidase | Differential pulse voltammetry | 1–100 pg/mL | 0.52 pg/mL | [88] |
| Insulin-like growth factor (IGF1) | Interdigitated electrode/streptavidin/biotinylated aptamer/antibody | Amperometry | 10 fM–1 nM | 10 fM | [89] |
| Analyte | Components of Sandwich Assay | Method of Detection | Linear Range | Limit of Detection | Ref. |
|---|---|---|---|---|---|
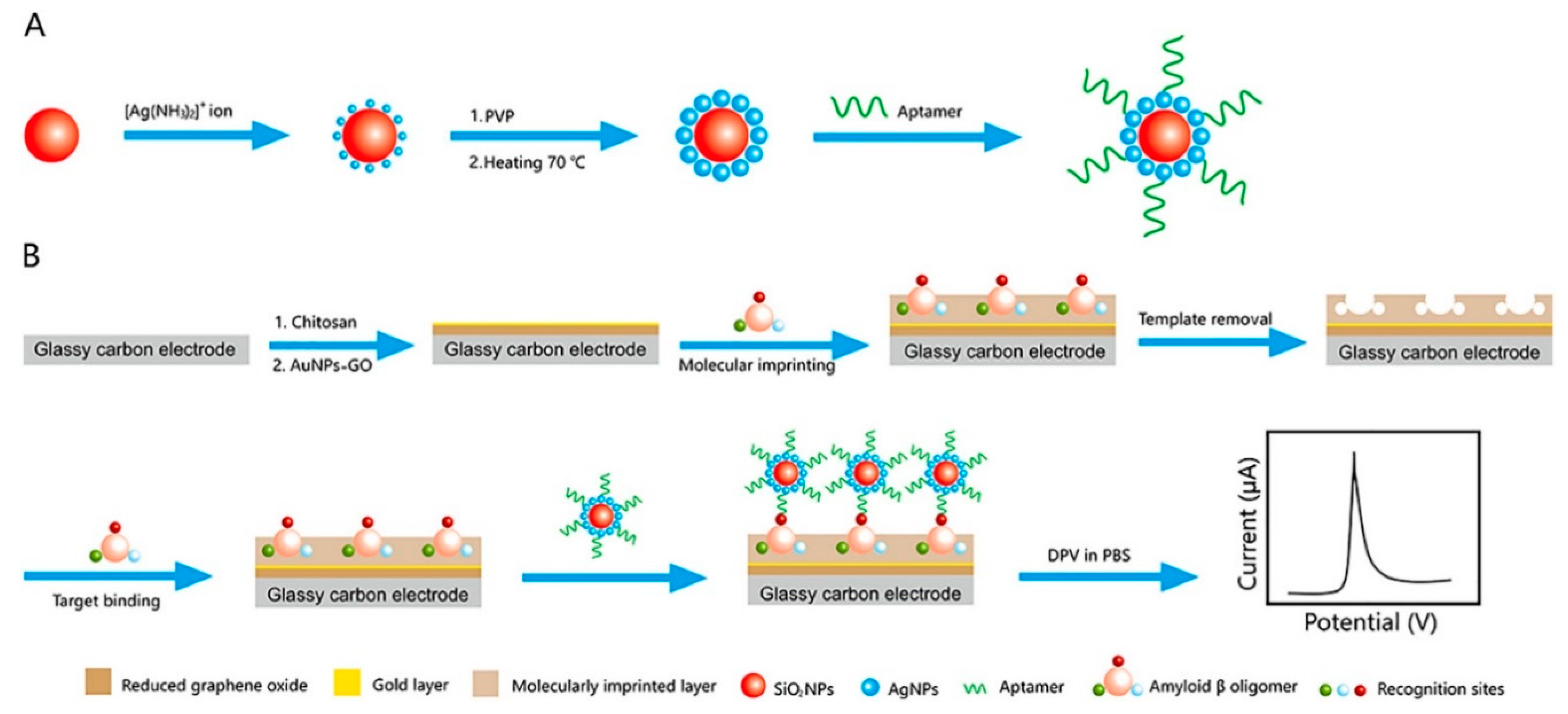
| Amyloid β1–42 oligomers | Glassy carbon electrode/Graphene oxide and Au nanoparticles/Chitosan and molecularly imprinted polymer layer/Aβ oligomer/aptamer-modified Ag nanoparticles bound to SiO2 larger nanoparticle | Differential pulse voltammetry | 5 pg/mL–10 ng mL | 1.22 pg/mL | [90] |
| Carcino-embryonic antigen | Au electrode/thiolated aptamer/CEA/Concanavalin A lectin/horseadish peroxidase | Differential pulse voltammetry | 5–40 ng/mL | 3.4 pg/mL | [91] |
| human epidermal growth factor receptor-2 | Au electrode/HER2 specific peptide/HER2/MnO2 nanosheet with phosphate ions and aptamers, molybdate ions | Square-wave voltammetry | 5 pg/mL–1 ng/mL | 0.05 pg/mL | [92] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neupane, D.; Stine, K.J. Electrochemical Sandwich Assays for Biomarkers Incorporating Aptamers, Antibodies and Nanomaterials for Detection of Specific Protein Biomarkers. Appl. Sci. 2021, 11, 7087. https://doi.org/10.3390/app11157087
Neupane D, Stine KJ. Electrochemical Sandwich Assays for Biomarkers Incorporating Aptamers, Antibodies and Nanomaterials for Detection of Specific Protein Biomarkers. Applied Sciences. 2021; 11(15):7087. https://doi.org/10.3390/app11157087
Chicago/Turabian StyleNeupane, Dharmendra, and Keith J. Stine. 2021. "Electrochemical Sandwich Assays for Biomarkers Incorporating Aptamers, Antibodies and Nanomaterials for Detection of Specific Protein Biomarkers" Applied Sciences 11, no. 15: 7087. https://doi.org/10.3390/app11157087







