Modelling the Effect of Temperature on the Initial Decline during the Lag Phase of Geotrichum candidum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism and Preparation of Inoculum
2.2. Estimation of G. candidum Cell Viable Counts
2.3. Modelling of Lag Death Rate of G. candidum
2.4. Statistical Evaluation
3. Results and Discussion
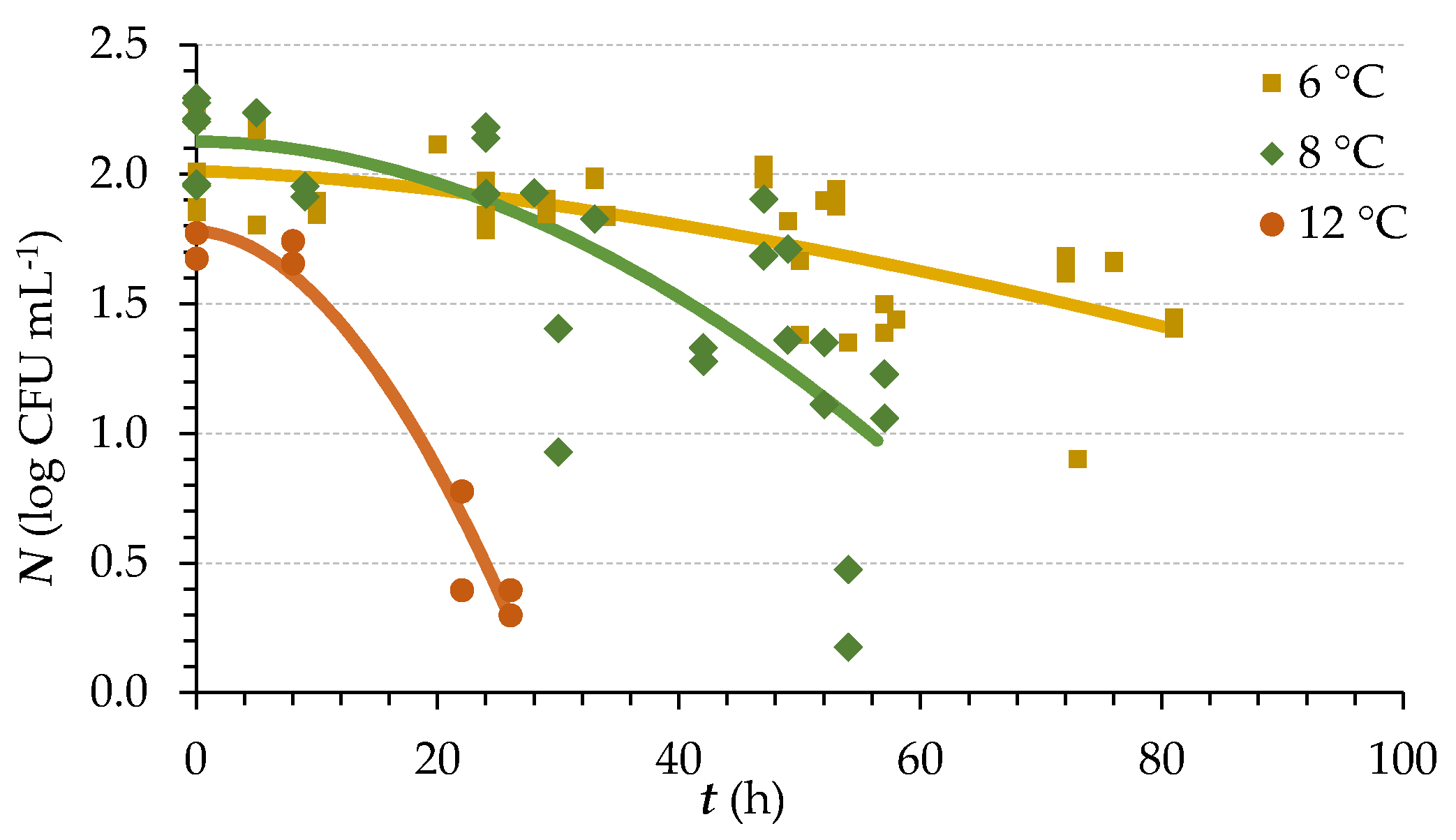
3.1. Lag Death Rate Primary Modelling
3.2. Lag Death Rate Secondary Modelling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Hoog, G.S.; Guarro, J.; Gené, J.; Ahmed, S.A.; Al-Hatmi, A.M.S.; Figueras, M.J.; Vitale, R.G. Atlas of Clinical Fungi, The Ultimate Benchtool for Diagnostics, 4th ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2020; pp. 457–459. [Google Scholar]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Thomidis, T.; Prodromou, I.; Farmakis, A.; Zambounis, A. Effect of temperature on the growth of Geotrichum candidum and chemical control of sour rot on tomatoes. Trop. Plant. Pathol. 2021. [Google Scholar] [CrossRef]
- Fröhlich-Wyder, M.T.; Arias-Roth, E.; Jakob, E. Cheese yeasts. Yeast 2018, 36, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Jaster, H.; Judacewski, P.; Ribeiro, J.C.B.; Zielinski, A.A.F.; Demiate, I.M.; Los, P.R.; Alberti, A.; Nogueira, A. Quality assessment of the manufacture of new ripened soft cheese by Geotrichum candidum: Physicochemical and technological properties. Food Sci. Technol. 2019, 39, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Šipošová, P.; Koňuchová, M.; Valík, Ľ.; Medveďová, A. Growth dynamics of lactic acid bacteria and dairy microscopic fungus Geotrichum candidum during their co-cultivation in milk. Food Sci. Technol. Int. 2020, in press. [Google Scholar] [CrossRef]
- Boutrou, R.; Guéguen, M. Interests in Geotrichum candidum for cheese technology. Int. J. Food Microbiol. 2005, 102, 1–20. [Google Scholar] [CrossRef]
- Garnier, L.; Valence, F.; Mounier, J. Diversity and control of spoilage fungi in dairy products: An update. Microorganisms 2017, 5, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eliskases-Lechner, F.; Guéguen, M.; Panoff, J.M. Geotrichum candidum. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Fox, P.F., McSweeney, P.L.H., Eds.; Academic Press: London, UK, 2011; pp. 765–771. [Google Scholar]
- Koňuchová, M.; Valík, Ľ. Modelling the radial growth of Geotrichum candidum: Effects of temperature and water activity. Microorganisms 2021, 9, 532. [Google Scholar] [CrossRef] [PubMed]
- Laurenčík, M.; Sulo, P.; Sláviková, E.; Piecková, E.; Seman, M.; Ebringer, L. The diversity of eukaryotic microbiota in the traditional Slovak sheep cheese—Bryndza. Int. J. Food Microbiol. 2008, 127, 176–179. [Google Scholar] [CrossRef]
- Marcellino, N.; Benson, D.R. The good, the bad, and the ugly: Tales of mold-ripened cheese. Microbiol. Spectr. 2013, 1, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Uraz, T.; Özer, B.H. Molds employed in food processing. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortello, M.-R., Eds.; Academic Press: Cambridge, UK, 2014; pp. 522–528. [Google Scholar]
- Šipošová, P.; Koňuchová, M.; Valík, Ľ.; Trebichavská, M.; Medveďová, A. Quantitative characterization of Geotrichum candidum growth in milk. Appl. Sci. 2021, 11, 4619. [Google Scholar] [CrossRef]
- Huang, L. Simulation and evaluation of different statistical functions for describing lag time distributions of bacterial growth curve. Microb. Risk Anal. 2016, 1, 47–55. [Google Scholar] [CrossRef]
- Robinson, T.P.; Ocio, M.J.; Kaloti, A.; Mackey, B.M. The effect of the growth environment on the lag phase of Listeria monocytogenes. Int. J. Food Microbiol. 1998, 44, 83–92. [Google Scholar] [CrossRef]
- Yates, G.T.; Smotzer, T. On the lag phase and initial decline of microbial growth curves. J. Theor. Biol. 2007, 244, 511–517. [Google Scholar] [CrossRef]
- Bertrand, R.L. Lag phase is a dynamic, organized, adaptive, and evolvable period that prepares bacteria for cell division. J. Bacteriol. 2019, 201, e00697-18. [Google Scholar] [CrossRef] [PubMed]
- Rolfe, M.D.; Rice, C.J.; Lucchini, S.; Pin, C.; Thompson, A.; Cameron, A.D.; Alston, M.; Stringer, M.F.; Betts, R.P.; Baranyi, J.; et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J. Bacteriol. 2012, 194, 686–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valík, Ľ.; Ačai, P. Predictive Microbiology and Microbiological Risk Assessment, 1st ed.; SUT: Bratislava, Slovakia, 2016; pp. 18–141. [Google Scholar]
- Hamill, P.; Stevenson, A.; McMullan, P.; Williams, J.; Lewis, A.; Sudharsan, S.; Stevenson, K.E.; Farnsworth, K.D.; Khroustalyova, G.; Takemoto, J.Y.; et al. Microbial lag phase can be indicative of, or independent from, cellular stress. Sci. Rep. 2020, 10, 1–20. [Google Scholar] [CrossRef]
- Baty, F.; Delignette-Muller, M.-L. Estimating the bacterial lag time: Which model, which precision? Int. J. Food Microbiol. 2004, 91, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, I.A.; Bernaerts, K.; Dens, E.J.; Geeraerd, A.H.; Van Impe, J.F. Predictive modelling of the microbial lag phase: A review. Int. J. Food Microbiol. 2004, 94, 137–159. [Google Scholar] [CrossRef]
- Métris, A.; Le Marc, Y.; Elfwing, A.; Ballagi, A.; Baranyi, J. Modelling the variability of lag times and the first generation times of single cells of E. Coli. Int. J. Food Microbiol. 2005, 100, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Mellefont, L.A.; Ross, T. The effect of abrupt shifts in temperature on the lag phase duration of Escherichia coli and Klebsiella oxytoca. Int. J. Food Microbiol. 2003, 83, 295–305. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. Mathematics of predictive food microbiology. Int. J. Food Microbiol. 1995, 26, 199–218. [Google Scholar] [CrossRef] [Green Version]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Geeraerd, A.H.; Valdramidis, V.P.; Van Impe, J.F. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int. J. Food Microbiol. 2005, 102, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.; Kishony, R. Optimization and control in bacterial Lag phase. BMC Biol. 2013, 11, 120. [Google Scholar] [CrossRef] [Green Version]
- Pangallo, D.; Šaková, N.; Koreňová, J.; Puškárová, A.; Kraková, L.; Valík, Ľ.; Kuchta, T. Microbial diversity and dynamics during the production of May bryndza cheese. Int. J. Food Microbiol. 2014, 170, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, M.; Coutinho, T.; Smith, M.T.; van der Walt, J.P. Species reassignment of Geotrichum bryndzae, Geotrichum phurueaensis, Geotrichum silvicola, and Geotrichum vulgarae based on phylogenetic analyses and mating compatibility. Int. J. Syst. Evol. Microbiol. 2012, 62, 3072–3080. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.S.; Filtenborg, O. Introduction to Food and Airborne Fungi, 6th ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2002; pp. 28–389. [Google Scholar]
- ISO 21527-1:2008. Part 1: Colony count technique in products with water activity greater than 0.95. In Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Yeasts and Molds; International Organization for Standardization: Geneva, Switzerland, 2010; pp. 1–12. [Google Scholar]
- Mafart, P.; Couvert, O.; Gaillard, S.; Leguerinel, I. On calculating sterility in thermal preservation methods: Application of the Weibull frequency distribution model. Int. J. Food Microbiol. 2002, 72, 107–113. [Google Scholar] [CrossRef] [Green Version]
- André, S.; Leguerinel, I.; Palop, A.; Desriac, N.; Planchon, S.; Mafart, P. Convergence of Bigelow and Arrhenius models over a wide range of heating temperatures. Int. J. Food Microbiol. 2019, 291, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Davey, K.R. A predictive model for combined temperature and water activity on microbial growth during the growth phase. J. Appl. Bacteriol. 1989, 67, 483–488. [Google Scholar] [CrossRef]
- Ratkowsky, D.A.; Olley, J.; McMeekin, T.A.; Ball, A. Relationship between temperature and growth rate of bacterial cultures. J. Bacteriol. 1982, 149, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Wells-Bennik, M.H.; Jansen, P.W.; Klaus, V.; Yang, C.; Zwietering, M.H.; den Besten, H.M. Heat resistance of spores of 18 strains of Geobacillus stearothermophilus and impact of culturing conditions. Int. J. Food Microbiol. 2019, 291, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H.J.; Christopoulos, A. Fitting Models to Biological Data Using Linear and Nonlinear Regression. A practical Guide to Curve Fitting, 1st ed.; GraphPad Software: San Diego, CA, USA, 2003; pp. 13–346. [Google Scholar]
- Baranyi, J. Stochastic modelling of bacterial lag phase. Int. J. Food Microbiol. 2002, 73, 203–206. [Google Scholar] [CrossRef]

| T (°C) | δ (h) | p | log N0 (log CFU mL−1) | n | RMSE (log CFU mL−1) | R2 |
|---|---|---|---|---|---|---|
| 6 | 111.94 | 1.53 | 2.01 | 46 | 0.200 | 0.491 |
| 8 | 52.35 | 1.90 | 2.13 | 30 | 0.349 | 0.608 |
| 12 | 20.96 | 1.85 | 1.78 | 8 | 0.164 | 0.959 |
| 15 | 17.96 | 3.57 | 1.93 | 6 | 0.086 | 0.989 |
| 18 | 9.91 | 5.91 | 1.97 | 8 | 0.052 | 0.972 |
| 21 | 7.71 | 2.03 | 1.93 | 8 | 0.087 | 0.984 |
| 25 | 6.14 | 1.27 | 1.97 | 7 | 0.377 | 0.788 |
| Model Parameters | ln k* | C0 | Ea (kJ mol−1) | TK* (K) | b (°C−1 h−0.5) | Tmin (°C) | RMSE (log CFU mL−1) | R2 | AIC |
|---|---|---|---|---|---|---|---|---|---|
| Modified ARH (Equation (3)) | −4.453 | − | 100.88 | 273.02 | − | − | 0.346 | 0.927 | 9.20 |
| Modified ARH (Equation (4)) | − | 39.988 | 100.88 | − | − | − | 0.309 | 0.927 | −4.80 |
| RTK | − | − | − | − | 0.016 | −0.720 | 0.028 | 0.978 | −38.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valík, Ľ.; Šipošová, P.; Koňuchová, M.; Medveďová, A. Modelling the Effect of Temperature on the Initial Decline during the Lag Phase of Geotrichum candidum. Appl. Sci. 2021, 11, 7344. https://doi.org/10.3390/app11167344
Valík Ľ, Šipošová P, Koňuchová M, Medveďová A. Modelling the Effect of Temperature on the Initial Decline during the Lag Phase of Geotrichum candidum. Applied Sciences. 2021; 11(16):7344. https://doi.org/10.3390/app11167344
Chicago/Turabian StyleValík, Ľubomír, Petra Šipošová, Martina Koňuchová, and Alžbeta Medveďová. 2021. "Modelling the Effect of Temperature on the Initial Decline during the Lag Phase of Geotrichum candidum" Applied Sciences 11, no. 16: 7344. https://doi.org/10.3390/app11167344
APA StyleValík, Ľ., Šipošová, P., Koňuchová, M., & Medveďová, A. (2021). Modelling the Effect of Temperature on the Initial Decline during the Lag Phase of Geotrichum candidum. Applied Sciences, 11(16), 7344. https://doi.org/10.3390/app11167344







